Abstract
When neuroscience’s focus moves from molecular and cellular level to systems level, information technology mixes in and cultivates a new branch neuroinformatics. Especially under the investments of brain initiatives all around the world, brain atlases and connectomics are identified as the substructure to understand the brain. We think it is time to call for a potential interdisciplinary subject, brainsmatics, referring to brain-wide spatial informatics science and emphasizing on precise positioning information affiliated to brain-wide connectome, genome, proteome, transcriptome, metabolome, etc. Brainsmatics methodology includes tracing, surveying, visualizing, and analyzing brain-wide spatial information. Among all imaging techniques, optical imaging is the most appropriate solution to achieve whole-brain connectome in consistent single-neuron resolution. This review aims to introduce contributions of optical imaging to brainsmatics studies, especially the major strategies applied in tracing and surveying processes. After discussions on the state-of-the-art technology, the development objectives of optical imaging in brainsmatics field are suggested. We call for a global contribution to the brainsmatics field from all related communities such as neuroscientists, biologists, engineers, programmers, chemists, mathematicians, physicists, clinicians, pharmacists, etc. As the leading approach, optical imaging will, in turn, benefit from the prosperous development of brainsmatics.
1. Neurosciences Call for Brainsmatics
The brain is the body’s most complex organ. Although many efforts have been devoted to discovering the brain at different levels, many mysteries remain unsolved. As neuroscientists have moved their interests from merely molecular and cellular level to systems level [1], neuronal circuits has become a research hotspot and is emphasized in the The Brain Research through Advancing Innovative Neurotechnologies (BRAIN) initiative [2]. Either network neuroscience or connectomics concerns about the integrated multiscale and multimodality information implied by anatomical and functional connectivity [3,4], but neither of them put enough emphasis on the affiliated spatial information.
Spatial information is important. The word “map” originally refers to surveying the sites and roads diagram of the Earth’s surface. Each site owns unique geographic coordinates, affiliated to a specific street, district, city, province, and country. Each road contains properties not only at the sites at the two ends but also all sites in between. Similarly, the map of the brain represents the wiring diagram inside the brain, reflecting connectivities between synapses, neurons, and brain regions, corresponding to microscopic, mesoscopic, and macroscopic connectomes. The function of a projection is associated with not only the input and output neurons but also the brain regions along the projection.
After the inspiration of the first complete connectome of Caenorhabditis elegans (C. elegans) [5], whole-brain atlases of Drosophila [6], larval zebrafish [7], mouse [8], and adult human [9,10,11] have been obtained worldwide. Other than structural connectivity, functional connectivity like gene expression [12] and transcription [13,14], specific connectivity like the cholinergic system [15], and serotonin system [16], illustrate a broad landscape for future study. Furthermore, unlike the relatively static genome, the connectome of the brain is highly dynamic, adapted from distinct behaviors [17], ages, and diseases. Neural plasticity occurs from milliseconds (molecular events) [18] to years [19]. Such temporal varieties together with structural and functional connectomes require vast information acquisition, processing, storage, and visualization.
Since information science has never been urgently needed in neuroscience as it is today, neuroinformatics has emerged as a branch of neuroscience [20]. After twenty years of rapid and flourishing development, however, neuroinformatics still limits the research scope on data organization, annotation, integration into atlases and models, and simulation [21]. Although the EU’s Human Brain Project has already noticed the needs to integrate information and knowledge of the brain’s physiology and anatomy [22], the efforts to integrate spatial data are still at an early stage [23].
Inspired by geomatics, Luo has proposed another perspective to explore neurosciences [24]. From this point of view, spatial information is the essential property of a molecule, synapse, cell, activity inside the brain. The new term “brainsmatics”, shortened from “brain spatial informatics”, refers to the integrated and systematic approaches of measuring, analyzing, managing, and displaying brain-wide spatial information, which include but are not limited to the molecular, genetic, proteomic, transcriptomic, metabolic, cellular, structural, and functional properties affiliated to unique coordinates inside the brain at a certain moment. Therefore, it may provide a comprehensive understanding of the brain.
The core concept of brainsmatics is brain-wide precise positioning. Why is brain-wide analysis so important? Evidence shows a giant neuron encircles the entire brain [25]. Studies on a certain brain region may probably lead to fragmentary understanding and even an opposite conclusion. Therefore, exploring the whole brain is essential to guarantee the integrity of brain connectivity. The needs for brain-wide analysis in turn put a threshold on imaging technologies. Macroscale Magnetic resonance imaging (MRI) based approaches and mesoscale optical imaging approaches have been applied to obtain a whole-brain connectome of living human brains and mammal animal brains. But electron microscopy (EM)-based microscale and/or nanoscale methods only succeeded in entire brain imaging of C. elegans, Drosophila, and zebrafish larvae [26]. Taking the imaging resolution and the imaging range into consideration, optical imaging approaches are a suitable strategy to achieve a brain-wide connectome at single-cell resolution among the existing techniques. Moreover, the applications of optical imaging in living animals make it a potential tool for dynamic connectome study [27].
Precise positioning requires precise targeting and high resolution locating. Labeling techniques help to target specific cell types or circuits. Based on different omics techniques, connectome, genome, proteome, transcriptome, and metabolome over the whole brain may also be traced and surveyed with affiliated spatial coordinates. The imaging object should be at least in single-neuron resolution and co-localized with landmarks. In this review, we will summarize recent progress of optical imaging related techniques applied in the brainsmatics field, and discuss the current state-of-the-art techniques.
2. Roles of Optical Imaging in Brainsmatics
The scope of brainsmatics includes tracing, surveying, visualizing of brain-wide connectivity, and analyzing temporal and spatial information features extracted from the collected big data. The demands that brainsmatics raises for related techniques lies in following aspects: (1) to be capable of obtaining whole-brain data with high temporal and spatial resolution, (2) the method proceeds in an automated manner and is stable both in the short-term and long-term, (3) the generated data is standardized, and (4) the minimization of the total processing time. Although a majority of current efforts are contributed merely on high voxel resolution on an ex vivo brain, innovative methods developed for monitoring living milliseconds activities in a high voxel resolution throughout the whole brain is the ultimate aim.
Owning a balance of big volume (whole-brain), high voxel resolution (single-neuron resolution), and acceptable time cost, optical imaging is playing a leading role in tracing and surveying processes. Multi-color labeling makes it possible to visualize diverse neurons and projections in the same brain.
For whole-brain optical imaging, two issues need to be addressed: one is how to acquire clear images, i.e., how to block/minimize out-of-focus background; the other is how to break the depth limitation of optical imaging. The first issue can be solved by optical sectioning techniques such as confocal microscopy, multi-photon excitation microscopy, and light-sheet microscopy. For the second issue, since the brain is opaque tissue, a physical factor that limits imaging depth is light attenuation caused by absorption and scattering, both of which reduce intensity exponentially with depth. To tackle this issue, one can use chemical clearing to make the brain transparent, or image the shallow tissue and remove the imaged section via physical sectioning.
2.1. Tracing Approaches Prior to Optical Imaging
Without tracing aids, nothing special is captured. The earliest tracing approach applied to the brain tissue was introduced by Golgi in 1873 [28] and was named Golgi staining. Modified and developed by Cajal and others, the Golgi staining methods has become a conventional sparse labeling strategy which aims to reveal individual neuron morphologies at synaptic resolution especially in local neuronal circuits. Unlike Golgi methods, another staining strategy, Nissl staining and its modifications, are capable of labeling nearly all the neuron somas and therefore is widely used to detect brain cytoarchitecture [29]. Though conventional staining method can help to enhance the contrast of fine structures, neither Golgi nor Nissl staining has specificity, i.e., only labeling the structures of interest and with integrity. To target certain molecules, neurons, and circuits, specific labeling approaches have been developed based on chemical or genetic ways.
The tracing process before imaging includes a staining/labeling procedure and sample preparation. To complete brainsmatics’ mission, several bottlenecks should be broken through. For instance, quenching in fluorescence imaging, background noises, and light scattering may affect imaging results. Embedding chemicals may not only determine the samples’ rigidity, resulting in its applicability to the imaging strategy, but also deform the sample and/or affect labeling quality. Less preparation time is also desired.
2.1.1. Specific Labeling
Toxins such as fluorophores and fusion of fluorescent proteins and polypeptide toxins are examples of chemical dyes, which were designed to target receptors like ionotropic acetylcholine (AChR), sodium, and potassium channels [30]. Immunohistochemical staining utilizes antibodies to identify specific proteins and has become a conventional labeling method in biological studies, as well as in brain staining [31]. Co-injecting retrograde and anterograde tracers into separate brain areas makes neuron projections with different information flow patterns available [32].
In the flourishing genome era, a vast number of genetic tools were applied to specific labeling. Transgenic mice expressing red green, yellow, or cyan fluorescent proteins are able to sparsely label entire axons, dendrites, and spines [33]. Genetically modified Cre reporter mice, also expressing fluorescent proteins of different colors, combined with immunohistochemistry and in situ hybridization, may also provide fine dendritic structures and axonal projections of the labeled neurons [34]. The advantages of the Cre-lox recombination system were utilized to stochastically express three or more fluorescent proteins and then create about 90 colors, visualizing distinct adjacent neurons and circuits [35]. Green fluorescent protein (GFP) reconstitution across synaptic partners (GRASP) is based on functional complementation between two nonfluorescent GFP fragments and was successfully applied in the mammalian brain to detect the location of synapses [36].
Fluorescent proteins can be transfected into neurons by viral vectors. This strategy can target specific cell types with monosynaptic and transsynaptic labeling, indicating anterograde and retrograde transport. Among many applicable viruses, adeno-associated virus (AAV) was approved to be a low toxic and stable anterograde transsynaptic tracer for long axonal projections [37,38]. The remarkable progress and advantages of specific genetic tracing methodologies have been previously reviewed in detail [39,40].
Sparse and bright labeling are challenges in this part. It is difficult to label the full morphology of a neuron, especially to label the soma, the thin axons, the complex dendrites, and the long-range terminals with similar signal-to-noise ratio. Bright labeling requests high abundance of fluorescent protein expression in the entire body of neurons. But overexpression of fluorescent proteins will damage the neuron morphology. Sparse labeling is to label a minority of nerve cells, which helps to reduce the disturbance from neighboring neurons and identify the individual morphology and trace the projection in the later reconstruction. A recent attempt employed two AAV vectors and Cre mice and demonstrated a sparse labeling system that can be tuned to achieve the desired density for labeling specific neuron types [41].
2.1.2. Sample Preparation
Following conventional histological strategies, the object tissue sample should be embedded in a supporting medium before treated with a microtome. Plastic embedding such as resins are widely used in large-volume brain tissue with the advantages of its rigidity for fine sectioning aided by a microtome. However, lack of the ability to preserve fluorescence limits their applications. To tackle this problem, Luo’s team developed a modified glycol methacrylate (GMA)-based method that can optimize the fluorescence preservation capability and shorten the penetration time [42]. Another embedding manner is elastic embedding. Take agar as an example, it has minimal detrimental effects on fluorescence, but is relatively soft as an embedded material to be sectioned. Covalent cross-linking of brain-agar after embedding is needed to solve this problem [43].
To avoid the difficulties of 3D reconstruction of mechanical sectioning images, an optical sectioning method was introduced in whole-brain 3D visualization, which inspired a prosperous revolutionary development of tissue clearing techniques [44]. Without mechanical sectioning, the samples are often embedded in hydrogel or just immersed in organic solvents, high refractive index (RI) aqueous solutions, or hyperhydrating solutions [45].
Categorized by the chemical properties of the treated solvents, clearing methods may be divided into two groups: organic solvent-based and aqueous-based. Typical organic solvent-based methods include BABB [44], 3DISCO [46], iDISCO [47], uDISCO [48], vDISCO [49], and FDISCO [50]. Organic solvent-based clearing methods usually achieve a higher level of transparency but easily lose fluorescence, while uDISCO, vDISCO, and FDISCO have improved fluorescence preservation [48,49,50]. Aqueous-based clearing methods may be divided into three sub-groups according to their clearing solutions: ClearT [51], SeeDB [52], and TDE [53] simply immerse the sample in high RI aqueous solutions; Scale [54], CUBIC [55], and ScaleS [56] remove lipids from the sample by hyperhydration reactions in aqueous solutions; CLARITY [57,58] and PACT/PARS [59] embed the sample in hydrogel that preserves fine structure and implements following lipids removal and clearing. Generally, aqueous-based clearing methods preserve fluorescence better but need longer clearing time. Not all the clearing methods are compatible with immunohistochemical staining, so the selection of labeling methods and clearing methods should be considered systematically. The advantages and disadvantages of all the mentioned clearing techniques have been discussed thoroughly in review papers [45,60].
To summarize this part, the tracing process contains a staining/labeling procedure and sample preparation. Most considerations focus on fluorescence preservation and reducing of the sample preparation time. For instance, chemical reactivation was applied to keep EGFP/EYFP-labeled resin-embedded mouse brain from quenching [61]. An on-line optical clearing was demonstrated to reduce whole-brain imaging time by half [62]. There are plenty of modifications and improvements in the tracing processes developed for different imaging instruments.
2.2. Optical Imaging in Brain Surveying
It is undoubted that higher voxel resolution is always the pursuit of brain connectome studies, as well as brainsmatics. The lateral resolution of optical microscopy is at submicron resolution, which is able to clearly distinguish sparsely labeled cells. However, the axial resolution in the z-direction is restricted by the point spread function of the focused beam, i.e., the full-width half-maximum (FWHM) resolution. Generally, the light penetration depth of microscopy applied in the brain is no more than 500 μm. Therefore, to achieve micron or even submicron resolution in the z-direction, two major strategies are mainly utilized, based on mechanical and optical sectioning.
2.2.1. Mechanical Sectioning Strategy
Inspired by histology methodology, mechanical sectioning, also called tissue sectioning, is combined with light microscopy for whole-brain imaging. In the sectioning part, the microtome is constituted of a fixed cutting tool, e.g., a diamond knife, and the sample is moved toward the knife at a constant speed, and the resin-embedded Golgi-stained mouse brain is sectioned into thin slice with a thickness of 1 μm. Meanwhile, an optical microscope perpendicular to the surface of knife is used to image the thin slice simultaneously, the image data is acquired by a line-scan charge-coupled device (CCD) [63,64]. Therefore the axial resolution is determined by the sectioning thickness.
To perform specific-labeled fluorescence imaging, confocal and two-photon microscopy are employed [65,66]. Those tomography methods can obtain volumetric imaging reconstructed from optical section imaging in a limited thickness of the tissue. However, their inherent laser scanning mode suppresses the scanning speed for a whole imaged plane and therefore prolong the imaging acquisition time. For a 1-micron voxel resolution fluorescent transgenic mouse brain, the imaging acquisition time is about 18 days, thus more considerations should be carried on to keep long-term stability for both sectioning and imaging processed [65].
To speed up the imaging acquisition time, different strategies are applied. Serial two-photon tomography (STPT) applies mosaic field of view (FOV) imaging in two-photon tomography on every optical section and with less sectioning times (50 μm spacing in z-direction) [43]. Brain-wide positioning system (BPS) employs structured illumination microscopy, also holding tomography capability, to perform mosaic scanning on each optical section at 4 μm spacing in the z-direction [67]. Block-face serial microscopy tomography (FAST) utilizes a spinning disk-based confocal microscope, and mosaic FOV images are obtained by a scientific complementary metal oxide semiconductor (sCMOS) camera or an electron-multiplying charge-coupled device (EM-CCD) camera [68].
2.2.2. Optical Sectioning Strategy
The introduction of light sheet fluorescence microscopy (LSFM) to whole-brain imaging aims to overcome the difficulties in mechanical sectioning methods such as 3D reconstruction and mechanical distortions [44]. A thin sheet of excitation light is orthogonally oriented to the detection direction, and the emitted fluorescence signals are acquired from each illuminating plane by a CCD camera. Only when applied to optically transparent objects, LSFM can achieve an integrated imaging atlas. In that case, it can dispense with the time cost by mechanical sectioning, which is another strategy to reduce the imaging acquisition time. Although this method can save the sectioning time, the sample preparation process is complex and time-consuming. Moreover, the axial resolution of LSFM is limited due to the required long working distance for large samples, which compromise the numerical aperture of the objective lens.
Another limitation of clearing methods compatible with LSFM is the morphological changes. Organic solvent-based clearing methods often lead to sample size shrinkage, while aqueous-based methods easily cause sample expansion [60]. However, the expansion side effect of cleared tissue induced by a polymer gel can be utilized and modified to keep the sample isotropically expanded. Then it becomes an advantage of named expansion microscopy (ExM) that results in 70 nm lateral resolution in brain tissue [69].
Also depending on cleared samples, optical projection tomography (OPT), an optical analogue of X-ray computed tomography, was recently applied to whole mouse brain imaging. The acquisition time is extremely fast, only four to eight minutes, but the voxel resolution is only 50 × 50 × 50 μm [70].
As mentioned, the booming development of clearing techniques is intended for matching LSFM. However, when the inspiring demonstrations appeared, those clearing techniques were soon modified and applied in mechanical sectioning imaging strategies, which implement an increased voxel resolution and decreased imaging time in whole-brain connectome acquisition [71].
2.3. Data Processing after Optical Imaging
The huge volume whole-brain mesoscopic data collected by optical imaging systems bring challenges for data processing and analysis [72]. The current practice of whole-brain mesoscopic data processing starts with denoising using classical methods [73] and deep learning methods. Then the data is compressed using an encoder/decoder similar to those for video compression, taking the advantage of the continuity between sectioning slices. This compression greatly reduces the data size while preserving enough information for further analysis. Stitching is also necessary if the image is taken block by block [74]. Then we can extract semantic morphological information through brain region segmentation, neuron projection analysis, and neuron reconstruction [75]. Meanwhile, registration of the collected data to standard brain spatial template can be achieved by nonlinear transformation in order to comparatively study the samples in a fair approach [76]. After these processing procedures, new knowledge may be summarized and contributed to the neuroscience research community.
In summary, the considerations of an integrative strategy of whole-brain fluorescence imaging include but are not limited to voxel resolution, sample preparation time, data acquisition time, data processing time, and specific demonstration. Many relevant techniques have been developed and contributed to the brainsmatics field. To attain a set of single-neuron resolution brain atlases, a proper and complementary combination of techniques of tracing, imaging, and visualization is required. Each technique has specific purpose and features, and is influenced or requested by another employed technique. The influence and restriction relationships between the techniques in a combination is complicated. For better understanding, we summarized a few common relationships in Figure 1 to demonstrate a general consideration for brainsmatics studies.
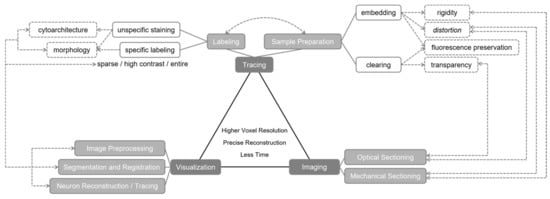
Figure 1.
Relevant optical imaging technique categories and relationships employed in brainsmatics studies. The influence and restriction relationships between technique categories are indicated by dashed lines.
In general, a mechanical sectioning strategy results in higher axial resolution but requires more imaging time, while an optical sectioning strategy reduces imaging time but results in restricted axial resolution. To demonstrate several cutting-edge practices, we compared in Table 1 the features of three typical strategies all utilizing mechanical sectioning methods. To balance the comparison, only reported applications on mouse brain are listed, emphasizing only tracing and imaging procedures. Since brain-wide precise positioning is the core of brainsmatics, strategies of optical sectioning methods are excluded due to the lower voxel resolution and lack of precise location information. As indicated in Table 1, only one practice provided positioning information. We expect more innovative methods to emerge and be applied in whole-brain studies in the near future.

Table 1.
Typical practices of whole-brain fluorescence imaging with single-neuron resolution.
3. Ideal Optical Imaging in Brainsmatics
Brain-wide precise positioning is the core of brainsmatics. To date, optical imaging is apparently the most appropriate solution to obtain a brain-wide mesoscopic atlas. Here we will discuss the prospect and development objectives of optical imaging in brainsmatics.
Higher spatial resolution. Due to the existing diffraction limit, the theoretical best lateral resolution of conventional optical imaging technique is ~300 nm, which is capable of identifying the dendrites and synapses, ensuring the studies on neural projections or neuronal circuits. However, poor axial resolution may lead to problems during 3D reconstruction. Hence, isotropic voxel resolution imaging techniques, such as oblique light-sheet, is getting more and more attention nowadays. In addition, neuroscientists always want to see how synapses contact/communicate, which requires higher resolution than 300 nm. To fulfill this requirement, researchers are spending efforts on two possible ways. One is the super-resolution imaging techniques, such as stochastic optical reconstruction microscopy (STORM), stimulated emission depletion (STED) and photoactivated localization microscopy (PALM) [77,78]. The other is developing deep learning microscopy that can predict the fine structures via neural networks [79,80]. Compared with single-cell resolution, the single-neuron resolution emphasizes the z-direction and the entire morphology display of a neuron. It requires full labeling on somas, axon ends, dendrites, and synapses of a specific neuron. For a specific circuit or network, transsynaptic labeling should be further developed to trace the entire circuit or network of a specific function.
High throughput imaging. The ultimate goal of neuroscientist is to obtain the brain connectome of higher mammals such as marmosets and humans. However, as the sample size increases, the imaging time increases. Take the marmoset as an example, it has a volume 90 times larger than a mouse brain. If we continue with the current techniques, the cost is unendurable. As a result, high throughput imaging techniques are highly needed. One possible way is to develop the array imaging techniques, through multi-point excitation and multi-point detection, one can possibly detect a large FOV at single acquisition [81]. Another possible manner is using large FOV microscopy with relatively low resolution to image the whole brain, then use a deep learning algorithm to regenerate high-resolution images [82].
Multi-dimensional information. To discover distinct cell types in a more precise way, one needs to obtain multi-dimensional information, e.g., morphology, transcriptomic, proteomic, location, and etc. However, to date, there is no such instrument that can perform all the aforementioned measurements. Therefore, it requests a brand-new platform/pipeline that integrates optical imaging with other techniques such as RNA sequencing and mass spectrometry to implement all the measurements. Besides, multi-dimensional information also means massive data. Take the micro-optical sectioning tomography (MOST) technique for example, morphology information for each mouse brain has reached ~10 Terabits alone, not to mention adding other types of information. Hence, it needs to develop a novel neuroinformatics method that can effectively integrate and analyze such big data.
Automation, standardization, and industrialization. Automation of imaging can reduce experimental time and manual uncertainty. Brain spatial information is huge and too much is unknown. However, publications on brain-wide mesoscale imaging and visualization are no more than five per year since 2007. Like the human genome project, the implementation of brainsmatics databank needs the collaborated input from many world-wide groups. Thus, the standardization of applied methods in every step, including the imaging process, is important. After standardization of certain methods, the industrialization of a full chain, including sample preparation, imaging, image processing, and reconstruction, helps to accelerate the data output and facilitate neuroscience research. Neuroscientists have already realized that large-scale, standardized data generation in an industrial manner will change the way neuroscience is done [83].
Brainsmatics is a big universe, which covers all the units in the brain, with both static and dynamic status. Based on the high-resolution brain atlas attained by optical imaging, brainsmatics will play an important role in understanding brain functions, defeating brain diseases, and developing brain-inspired intelligence.
The high-resolution databanks with various brain-wide spatial information, reconstructed from optical imaging data, will provide basic knowledge of cell type and brain connectome for neuroscience research. Comparison of structural and functional differences between normal and abnormal brains will help to discover the mechanisms of brain disease development. Understanding the neuronal circuits reconstructed by optical imaging data helps to mimic and develop new algorithms in artificial intelligence.
Optical imaging has been successfully demonstrated on both ex vivo and in vivo applications. But still, the universe needs more contributions from all the related communities such as neuroscientists, biologists, engineers, programmers, chemists, mathematicians, physicists, clinicians, pharmacists, etc. With the development of brainsmatics, optical imaging will also benefit from global and interdisciplinary collaborations.
Author Contributions
conceptualization, H.S. and Q.L.; writing—original draft preparation, H.S.; writing—review and editing, H.S., Y.G, J.C. and Q.L.; supervision, Q.L.; funding acquisition, Q.L.
Funding
This research was funded by the National Natural Science Foundation of China, grant numbers 81827901, 61721092.
Acknowledgments
The authors would like to thank Hui Gong for valuable comments.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Sudhof, T.C. Molecular Neuroscience in the 21st Century: A Personal Perspective. Neuron 2017, 96, 536–541. [Google Scholar] [CrossRef] [PubMed]
- Jorgenson, L.A.; Newsome, W.T.; Anderson, D.J.; Bargmann, C.I.; Brown, E.N.; Deisseroth, K.; Donoghue, J.P.; Hudson, K.L.; Ling, G.S.; MacLeish, P.R.; et al. The BRAIN Initiative: Developing technology to catalyse neuroscience discovery. Philos. Trans. R. Soc. B Biol. Sci. 2015, 370, 20140164:1–20140164:12. [Google Scholar] [CrossRef] [PubMed]
- Bassett, D.S.; Sporns, O. Network neuroscience. Nat. Neurosci. 2017, 20, 353–364. [Google Scholar] [CrossRef] [PubMed]
- Lichtman, J.W.; Sanes, J.R. Ome sweet ome: What can the genome tell us about the connectome? Curr. Opin. Neurobiol. 2008, 18, 346–353. [Google Scholar] [CrossRef] [PubMed]
- White, J.G.; Southgate, E.; Thomson, J.N.; Brenner, S. The structure of the nervous system of the nematode Caenorhabditis elegans. Philos. Trans. R. Soc. B Biol. Sci. 1986, 314, 1–340. [Google Scholar] [CrossRef] [PubMed]
- Chiang, A.; Lin, C.; Chuang, C.; Chang, H.; Hsieh, C.; Yeh, C.; Shih, C.; Wu, J.; Wang, G.; Chen, Y.; et al. Three-Dimensional Reconstruction of Brain-wide Wiring Networks in Drosophila at Single-Cell Resolution. Curr. Biol. 2011, 21, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Hildebrand, D.G.C.; Cicconet, M.; Torres, R.M.; Choi, W.; Quan, T.M.; Moon, J.; Wetzel, A.W.; Scott Champion, A.; Graham, B.J.; Randlett, O.; et al. Whole-brain serial-section electron microscopy in larval zebrafish. Nature 2017, 545, 345–349. [Google Scholar] [CrossRef] [PubMed]
- Oh, S.W.; Harris, J.A.; Ng, L.; Winslow, B.; Cain, N.; Mihalas, S.; Wang, Q.; Lau, C.; Kuan, L.; Henry, A.M.; et al. A mesoscale connectome of the mouse brain. Nature 2014, 508, 207–214. [Google Scholar] [CrossRef]
- Craddock, R.C.; James, G.A.; Holtzheimer, P.E.; Hu, X.P.; Mayberg, H.S. A whole brain fMRI atlas generated via spatially constrained spectral clustering. Hum. Brain Mapp. 2012, 33, 1914–1928. [Google Scholar] [CrossRef]
- Amunts, K.; Lepage, C.; Borgeat, L.; Mohlberg, H.; Dickscheid, T.; Rousseau, M.E.; Bludau, S.; Bazin, P.L.; Lewis, L.B.; Oros-Peusquens, A.M.; et al. BigBrain: An ultrahigh-resolution 3D human brain model. Science 2013, 340, 1472–1475. [Google Scholar] [CrossRef]
- Fan, L.; Li, H.; Zhuo, J.; Zhang, Y.; Wang, J.; Chen, L.; Yang, Z.; Chu, C.; Xie, S.; Laird, A.R.; et al. The Human Brainnetome Atlas: A New Brain Atlas Based on Connectional Architecture. Cereb. Cortex 2016, 26, 3508–3526. [Google Scholar] [CrossRef] [PubMed]
- Lein, E.S.; Hawrylycz, M.J.; Ao, N.; Ayres, M.; Bensinger, A.; Bernard, A.; Boe, A.F.; Boguski, M.S.; Brockway, K.S.; Byrnes, E.J.; et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature 2007, 445, 168–176. [Google Scholar] [CrossRef] [PubMed]
- Bakken, T.E.; Miller, J.A.; Ding, S.; Sunkin, S.M.; Smith, K.A.; Ng, L.; Szafer, A.; Dalley, R.A.; Royall, J.J.; Lemon, T.; et al. A comprehensive transcriptional map of primate brain development. Nature 2016, 535, 367–375. [Google Scholar] [CrossRef]
- Miller, J.A.; Ding, S.; Sunkin, S.M.; Smith, K.A.; Ng, L.; Szafer, A.; Ebbert, A.; Riley, Z.L.; Royall, J.J.; Aiona, K.; et al. Transcriptional landscape of the prenatal human brain. Nature 2014, 508, 199–206. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Yu, B.; Sun, Q.; Zhang, Y.; Ren, M.; Zhang, X.; Li, A.; Yuan, J.; Madisen, L.; Luo, Q.; et al. Generation of a whole-brain atlas for the cholinergic system and mesoscopic projectome analysis of basal forebrain cholinergic neurons. Proc. Natl. Acad. Sci. USA 2018, 115, 415–420. [Google Scholar] [CrossRef]
- Beliveau, V.; Ganz, M.; Feng, L.; Ozenne, B.; Højgaard, L.; Fisher, P.M.; Svarer, C.; Greve, D.N.; Knudsen, G.M. A High-Resolution in vivo Atlas of the Human Brain’s Serotonin System. J. Neurosci. 2017, 37, 120–128. [Google Scholar] [CrossRef] [PubMed]
- Lin, X.; Lei, V.L.C.; Li, D.; Hu, Z.; Xiang, Y.; Yuan, Z. Mapping the small-world properties of brain networks in Chinese to English simultaneous interpreting by using functional near-infrared spectroscopy. J. Innov. Opt. Heal. Sci. 2018, 11, 1840001:1–1840001:12. [Google Scholar] [CrossRef]
- Lamprecht, R.; LeDoux, J. Structural plasticity and memory. Nat. Rev. Neurosci. 2004, 5, 45–54. [Google Scholar] [CrossRef]
- Burke, S.N.; Barnes, C.A. Neural plasticity in the ageing brain. Nat. Rev. Neurosci. 2006, 7, 30–40. [Google Scholar] [CrossRef] [PubMed]
- Young, M.P.; Scannell, J.W. Brain Structure-Function Relationships: Advances from Neuroinformatics. Philos. Trans. R. Soc. B Biol. Sci. 2000, 355, 3–6. [Google Scholar] [CrossRef][Green Version]
- Nayak, L.; Dasgupta, A.; Das, R.; Ghosh, K.; De Rajat, K. Computational neuroscience and neuroinformatics: Recent progress and resources. J. Biosci. 2018, 43, 1037–1054. [Google Scholar] [CrossRef] [PubMed]
- Frackowiak, R.; Markram, H. The future of human cerebral cartography: A novel approach. Philos. Trans. R. Soc. B Biol. Sci. 2015, 370, 20140171:1–20140171:13. [Google Scholar] [CrossRef] [PubMed]
- Bjerke, I.E.; Øvsthus, M.; Papp, E.A.; Yates, S.C.; Silvestri, L.; Fiorilli, J.; Pennartz, C.M.A.; Pavone, F.S.; Puchades, M.A.; Leergaard, T.B.; et al. Data integration through brain atlasing: Human Brain Project tools and strategies. Eur. Psychiat. 2018, 50, 70–76. [Google Scholar] [CrossRef] [PubMed]
- Luo, Q. Brainsmatics—bridging the brain science and brain-inspired artificial intelligence. Sci. Sin. Vitae 2017, 47, 1015–1024. (In Chinese) [Google Scholar] [CrossRef]
- Reardon, S. Giant neuron encircles entire brain of a mouse. Nature 2017, 543, 14–15. [Google Scholar] [CrossRef] [PubMed]
- Zeng, H. Mesoscale connectomics. Curr. Opin. Neurobiol. 2018, 50, 154–162. [Google Scholar] [CrossRef] [PubMed]
- Silvestri, L.; Mascaro, A.A.; Lotti, J.; Sacconi, L.; Pavone, F.S. Advanced optical techniques to explore brain structure and function. J. Innov. Opt. Heal. Sci. 2013, 6, 1230002:1–1230002:15. [Google Scholar] [CrossRef]
- Golgi, C. Sulla struttura della sostanza grigia del cervello. Gazz. Med. Ital. 1873, 33, 244–246. [Google Scholar]
- Ecker, J.R.; Geschwind, D.H.; Kriegstein, A.R.; Ngai, J.; Osten, P.; Polioudakis, D.; Regev, A.; Sestan, N.; Wickersham, I.R.; Zeng, H. The BRAIN Initiative Cell Census Consortium: Lessons Learned toward Generating a Comprehensive Brain Cell Atlas. Neuron 2017, 96, 542–557. [Google Scholar] [CrossRef]
- Kuzmenkov, A.I.; Vassilevski, A.A. Labelled animal toxins as selective molecular markers of ion channels: Applications in neurobiology and beyond. Neurosci. Lett. 2018, 679, 15–23. [Google Scholar] [CrossRef]
- Micheva, K.D.; Smith, S.J. Array tomography: A new tool for imaging the molecular architecture and ultrastructure of neural circuits. Neuron 2007, 55, 25–36. [Google Scholar] [CrossRef] [PubMed]
- Zingg, B.; Hintiryan, H.; Gou, L.; Song, M.Y.; Bay, M.; Bienkowski, M.S.; Foster, N.N.; Yamashita, S.; Bowman, I.; Toga, A.W.; et al. Neural Networks of the Mouse Neocortex. Cell 2014, 156, 1096–1111. [Google Scholar] [CrossRef] [PubMed]
- Feng, G.; Mellor, R.H.; Bernstein, M.; Keller-Peck, C.; Nguyen, Q.T.; Wallace, M.; Nerbonne, J.M.; Lichtman, J.W.; Sanes, J.R. Imaging neuronal subsets in transgenic mice expressing multiple spectral variants of GFP. Neuron 2000, 28, 41–51. [Google Scholar] [CrossRef]
- Madisen, L.; Zwingman, T.A.; Sunkin, S.M.; Oh, S.W.; Zariwala, H.A.; Gu, H.; Ng, L.L.; Palmiter, R.D.; Hawrylycz, M.J.; Jones, A.R.; et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat. Neurosci. 2010, 13, 133–140. [Google Scholar] [CrossRef] [PubMed]
- Livet, J.; Weissman, T.A.; Kang, H.; Draft, R.W.; Lu, J.; Bennis, R.A.; Sanes, J.R.; Lichtman, J.W. Transgenic strategies for combinatorial expression of fluorescent proteins in the nervous system. Nature 2007, 450, 56–62. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Zhao, T.; Petralia, R.S.; Yu, Y.; Peng, H.; Myers, E.; Magee, J.C. mGRASP enables mapping mammalian synaptic connectivity with light microscopy. Nat. Methods 2012, 9, 96–102. [Google Scholar] [CrossRef] [PubMed]
- Chamberlin, N.L.; Du, B.; de Lacalle, S.; Saper, C.B. Recombinant adeno-associated virus vector: Use for transgene expression and anterograde tract tracing in the CNS. Brain Res. 1998, 793, 169–175. [Google Scholar] [CrossRef]
- Zingg, B.; Chou, X.; Zhang, Z.; Mesik, L.; Liang, F.; Tao, H.W.; Zhang, L.I. AAV-Mediated Anterograde Transsynaptic Tagging: Mapping Corticocollicular Input-Defined Neural Pathways for Defense Behaviors. Neuron 2017, 93, 33–47. [Google Scholar] [CrossRef]
- Luo, L.; Callaway, E.M.; Svoboda, K. Genetic Dissection of Neural Circuits. Neuron 2008, 57, 634–660. [Google Scholar] [CrossRef]
- Huang, Z.J.; Zeng, H. Genetic approaches to neural circuits in the mouse. Annu. Rev. Neurosci. 2013, 36, 183–215. [Google Scholar] [CrossRef]
- Lin, R.; Wang, R.; Yuan, J.; Feng, Q.; Zhou, Y.; Zeng, S.; Ren, M.; Jiang, S.; Ni, H.; Zhou, C.; et al. Cell-type-specific and projection-specific brain-wide reconstruction of single neurons. Nat. Methods 2018, 15, 1033–1036. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Hu, B.; Zhang, Y.; Luo, Q.; Gong, H. Development of a Plastic Embedding Method for Large-Volume and Fluorescent-Protein-Expressing Tissues. PLoS ONE 2013, 8, e60877:1–e60877:5. [Google Scholar] [CrossRef] [PubMed]
- Ragan, T.; Kadiri, L.R.; Venkataraju, K.U.; Bahlmann, K.; Sutin, J.; Taranda, J.; Arganda-Carreras, I.; Kim, Y.; Seung, H.S.; Osten, P. Serial two-photon tomography for automated ex vivo mouse brain imaging. Nat. Methods 2012, 9, 255–258. [Google Scholar] [CrossRef] [PubMed]
- Dodt, H.; Leischner, U.; Schierloh, A.; Jährling, N.; Mauch, C.P.; Deininger, K.; Deussing, J.M.; Eder, M.; Zieglgänsberger, W.; Becker, K. Ultramicroscopy: Three-dimensional visualization of neuronal networks in the whole mouse brain. Nat. Methods 2007, 4, 331–336. [Google Scholar] [CrossRef] [PubMed]
- Ariel, P. A beginner’s guide to tissue clearing. Int. J. Biochem. Cell Biol. 2017, 84, 35–39. [Google Scholar] [CrossRef] [PubMed]
- Erturk, A.; Becker, K.; Jahrling, N.; Mauch, C.P.; Hojer, C.D.; Egen, J.G.; Hellal, F.; Bradke, F.; Sheng, M.; Dodt, H.U. Three-dimensional imaging of solvent-cleared organs using 3DISCO. Nat. Protoc. 2012, 7, 1983–1995. [Google Scholar] [CrossRef] [PubMed]
- Renier, N.; Wu, Z.; Simon, D.J.; Yang, J.; Ariel, P.; Tessier-Lavigne, M. iDISCO: A Simple, Rapid Method to Immunolabel Large Tissue Samples for Volume Imaging. Cell 2014, 159, 896–910. [Google Scholar] [CrossRef] [PubMed]
- Pan, C.; Cai, R.; Quacquarelli, F.P.; Ghasemigharagoz, A.; Lourbopoulos, A.; Matryba, P.; Plesnila, N.; Dichgans, M.; Hellal, F.; Ertürk, A. Shrinkage-mediated imaging of entire organs and organisms using uDISCO. Nat. Methods 2016, 13, 859–867. [Google Scholar] [CrossRef]
- Cai, R.; Pan, C.; Ghasemigharagoz, A.; Todorov, M.I.; Förstera, B.; Zhao, S.; Bhatia, H.S.; Parra-Damas, A.; Mrowka, L.; Theodorou, D.; et al. Panoptic imaging of transparent mice reveals whole-body neuronal projections and skull–meninges connections. Nat. Neurosci. 2019, 22, 317–327. [Google Scholar] [CrossRef]
- Qi, Y.; Yu, T.; Xu, J.; Wan, P.; Ma, Y.; Zhu, J.; Li, Y.; Gong, H.; Luo, Q.; Zhu, D. FDISCO: Advanced solvent-based clearing method for imaging whole organs. Sci. Adv. 2019, 5, eaau8355:1–eaau8355:13. [Google Scholar] [CrossRef]
- Kuwajima, T.; Sitko, A.A.; Bhansali, P.; Jurgens, C.; Guido, W.; Mason, C. ClearT: A detergent- and solvent-free clearing method for neuronal and non-neuronal tissue. Development 2013, 140, 1364–1368. [Google Scholar] [CrossRef] [PubMed]
- Ke, M.; Fujimoto, S.; Imai, T. SeeDB: A simple and morphology-preserving optical clearing agent for neuronal circuit reconstruction. Nat. Neurosci. 2013, 16, 1154–1161. [Google Scholar] [CrossRef] [PubMed]
- Costantini, I.; Ghobril, J.; Di Giovanna, A.P.; Mascaro, A.L.A.; Silvestri, L.; Müllenbroich, M.C.; Onofri, L.; Conti, V.; Vanzi, F.; Sacconi, L.; et al. A versatile clearing agent for multi-modal brain imaging. Sci. Rep. 2015, 5, 9808:1–9808:9. [Google Scholar] [CrossRef] [PubMed]
- Hama, H.; Kurokawa, H.; Kawano, H.; Ando, R.; Shimogori, T.; Noda, H.; Fukami, K.; Sakaue-Sawano, A.; Miyawaki, A. Scale: A chemical approach for fluorescence imaging and reconstruction of transparent mouse brain. Nat. Neurosci. 2011, 14, 1481–1488. [Google Scholar] [CrossRef] [PubMed]
- Susaki, E.A.; Tainaka, K.; Perrin, D.; Kishino, F.; Tawara, T.; Watanabe, T.M.; Yokoyama, C.; Onoe, H.; Eguchi, M.; Yamaguchi, S.; et al. Whole-Brain Imaging with Single-Cell Resolution Using Chemical Cocktails and Computational Analysis. Cell 2014, 157, 726–739. [Google Scholar] [CrossRef] [PubMed]
- Hama, H.; Hioki, H.; Namiki, K.; Hoshida, T.; Kurokawa, H.; Ishidate, F.; Kaneko, T.; Akagi, T.; Saito, T.; Saido, T.; et al. ScaleS: An optical clearing palette for biological imaging. Nat. Neurosci. 2015, 18, 1518–1529. [Google Scholar] [CrossRef] [PubMed]
- Chung, K.; Wallace, J.; Kim, S.; Kalyanasundaram, S.; Andalman, A.S.; Davidson, T.J.; Mirzabekov, J.J.; Zalocusky, K.A.; Mattis, J.; Denisin, A.K.; et al. Structural and molecular interrogation of intact biological systems. Nature 2013, 497, 332–337. [Google Scholar] [CrossRef] [PubMed]
- Chung, K.; Deisseroth, K. CLARITY for mapping the nervous system. Nat. Methods 2013, 10, 508–513. [Google Scholar] [CrossRef] [PubMed]
- Yang, B.; Treweek, J.B.; Kulkarni, R.P.; Deverman, B.E.; Chen, C.K.; Lubeck, E.; Shah, S.; Cai, L.; Gradinaru, V. Single-cell phenotyping within transparent intact tissue through whole-body clearing. Cell 2014, 158, 945–958. [Google Scholar] [CrossRef]
- Richardson, D.S.; Lichtman, J.W. Clarifying Tissue Clearing. Cell 2015, 162, 246–257. [Google Scholar] [CrossRef]
- Xiong, H.; Zhou, Z.; Zhu, M.; Lv, X.; Li, A.; Li, S.; Li, L.; Yang, T.; Wang, S.; Yang, Z.; et al. Chemical reactivation of quenched fluorescent protein molecules enables resin-embedded fluorescence microimaging. Nat. Commun. 2014, 5, 3992:1–3992:9. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Yang, X.; Chen, S.; Zhang, L.; Long, B.; Tan, C.; Yuan, J.; Gong, H. On-line optical clearing method for whole-brain imaging in mice. Biomed. Opt. Express 2019, 10, 2612–2622. [Google Scholar] [CrossRef] [PubMed]
- Mayerich, D.; Abbott, L.; McCormick, B. Knife-edge scanning microscopy for imaging and reconstruction of three-dimensional anatomical structures of the mouse brain. J. Microsc. 2008, 231, 134–143. [Google Scholar] [CrossRef] [PubMed]
- Li, A.; Gong, H.; Zhang, B.; Wang, Q.; Yan, C.; Wu, J.; Liu, Q.; Zeng, S.; Luo, Q. Micro-Optical Sectioning Tomography to Obtain a High-Resolution Atlas of the Mouse Brain. Science 2010, 330, 1404–1408. [Google Scholar] [CrossRef] [PubMed]
- Gong, H.; Zeng, S.; Yan, C.; Lv, X.; Yang, Z.; Xu, T.; Feng, Z.; Ding, W.; Qi, X.; Li, A.; et al. Continuously tracing brain-wide long-distance axonal projections in mice at a one-micron voxel resolution. Neuroimage 2013, 74, 87–98. [Google Scholar] [CrossRef] [PubMed]
- Zheng, T.; Yang, Z.; Li, A.; Lv, X.; Zhou, Z.; Wang, X.; Qi, X.; Li, S.; Luo, Q.; Gong, H.; et al. Visualization of brain circuits using two-photon fluorescence micro-optical sectioning tomography. Opt. Express 2013, 21, 9839–9850. [Google Scholar] [CrossRef]
- Gong, H.; Xu, D.; Yuan, J.; Li, X.; Guo, C.; Peng, J.; Li, Y.; Schwarz, L.A.; Li, A.; Hu, B.; et al. High-throughput dual-colour precision imaging for brain-wide connectome with cytoarchitectonic landmarks at the cellular level. Nat. Commun. 2016, 7, 12142:1–12142:12. [Google Scholar] [CrossRef] [PubMed]
- Seiriki, K.; Kasai, A.; Hashimoto, T.; Schulze, W.; Niu, M.; Yamaguchi, S.; Nakazawa, T.; Inoue, K.; Uezono, S.; Takada, M.; et al. High-Speed and Scalable Whole-Brain Imaging in Rodents and Primates. Neuron 2017, 94, 1085–1100. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; Tillberg, P.W.; Boyden, E.S. Expansion microscopy. Science 2015, 347, 543–548. [Google Scholar] [CrossRef]
- Nguyen, D.; Marchand, P.J.; Planchette, A.L.; Nilsson, J.; Sison, M.; Extermann, J.; Lopez, A.; Sylwestrzak, M.; Sordet-Dessimoz, J.; Schmidt-Christensen, A.; et al. Optical projection tomography for rapid whole mouse brain imaging. Biomed. Opt. Express 2017, 8, 5637–5650. [Google Scholar] [CrossRef]
- Economo, M.N.; Clack, N.G.; Lavis, L.D.; Gerfen, C.R.; Svoboda, K.; Myers, E.W.; Chandrashekar, J. A platform for brain-wide imaging and reconstruction of individual neurons. ELife 2016, 5, e10566:1–e10566:22. [Google Scholar] [CrossRef] [PubMed]
- Lichtman, J.W.; Denk, W. The big and the small: Challenges of imaging the brain’s circuits. Science 2011, 334, 618–623. [Google Scholar] [CrossRef] [PubMed]
- Malik, W.Q.; Schummers, J.; Sur, M.; Brown, E.N. Denoising Two-Photon Calcium Imaging Data. PLoS ONE 2011, 6, e20490:1–e20490:11. [Google Scholar] [CrossRef] [PubMed]
- Santi, P.A.; Johnson, S.B.; Hillenbrand, M.; Grandpre, P.Z.; Glass, T.J.; Leger, J.R. Thin-sheet laser imaging microscopy for optical sectioning of thick tissues. Biotechniques 2009, 46, 287–294. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Zeng, T.; Peng, H.; Ji, S. Deep Learning Segmentation of Optical Microscopy Images Improves 3-D Neuron Reconstruction. IEEE Trans. Med. Imaging 2017, 36, 1533–1541. [Google Scholar] [CrossRef] [PubMed]
- Haker, S.; Tannenbaum, A.; Kikinis, R. Mass Preserving Mappings and Image Registration; Niessen, W.J., Viergever, M.A., Eds.; Springer: Berlin/Heidelberg, Germany, 2001; pp. 120–127. [Google Scholar]
- Willig, K.I.; Rizzoli, S.O.; Westphal, V.; Jahn, R.; Hell, S.W. STED microscopy reveals that synaptotagmin remains clustered after synaptic vesicle exocytosis. Nature 2006, 440, 935–939. [Google Scholar] [CrossRef] [PubMed]
- Zhong, H. Applying superresolution localization-based microscopy to neurons. Synapse 2015, 69, 283–294. [Google Scholar] [CrossRef] [PubMed]
- Nehme, E.; Weiss, L.E.; Michaeli, T.; Shechtman, Y. Deep-STORM: Super-resolution single-molecule microscopy by deep learning. Optica 2018, 5, 458–464. [Google Scholar] [CrossRef]
- Wang, H.; Rivenson, Y.; Jin, Y.; Wei, Z.; Gao, R.; Günaydın, H.; Bentolila, L.A.; Kural, C.; Ozcan, A. Deep learning enables cross-modality super-resolution in fluorescence microscopy. Nat. Methods 2019, 16, 103–110. [Google Scholar] [CrossRef]
- Kim, K.H.; Buehler, C.; Bahlmann, K.; Ragan, T.; Lee, W.A.; Nedivi, E.; Heffer, E.L.; Fantini, S.; So, P.T.C. Multifocal multiphoton microscopy based on multianode photomultiplier tubes. Opt. Express 2007, 15, 11658–11678. [Google Scholar] [CrossRef]
- Zhang, X.; Chen, Y.; Ning, K.; Zhou, C.; Han, Y.; Gong, H.; Yuan, J. Deep learning optical-sectioning method. Opt. Express 2018, 26, 30762:1–30762:11. [Google Scholar] [CrossRef] [PubMed]
- Cyranoski, D. China launches brain-imaging factory. Nature 2017, 548, 268–269. [Google Scholar] [CrossRef] [PubMed][Green Version]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).