Abstract
Guided mode resonance (GMR)-based sensors have emerged as a promising technology for the early screening of cancer, offering advantages such as sensitivity, specificity, low cost, non-invasiveness, and portability. This review article provides a comprehensive overview of the latest advancements in GMR technology and its applications in biosensing, with a specific focus on cancer. The current state of cancer diagnosis and the critical need for point-of-care (POC) devices to address these challenges are discussed in detail. Furthermore, the review systematically explores various strategies employed in GMR-based cancer detection including design principles and the integration of advanced technologies. Additionally, it aims to provide researchers valuable insights for developing GMR sensors capable of detecting cancer biomarkers outside the laboratory environment.
1. Introduction
According to the World Health Organization (W.H.O.), cancer remains the leading cause of death globally. In 2022, an estimated 20 million new cancer cases were reported, resulting in 9.7 million deaths [1]. The most common cancers are lung, breast, colorectal, and prostate cancers [2]. Cancer research is significant since it contributes directly or indirectly to the well-being of individuals through early detection, better treatments, and enhanced quality of life. However, access to cancer care in low-income areas is challenging due to the need for specialized facilities. Developing POC solutions for the rapid screening and detection of cancer biomarkers can address this issue and facilitate early diagnosis [3]. For example, in the case of lung cancer, traditional imaging methods such as X-rays and CT scans are often inadequate for early detection due to their inherent limitations, such as low sensitivity, false positives or negatives, radiation exposure, and high cost. As an alternative, analyzing breath for volatile organic compounds (VOCs) related to lung cancer can reveal the body’s biochemical status and offer early indications of the disease [4,5]. Therefore, cancer detection is a significant research field that encompasses various approaches including screening methods and diagnostic tools such as biopsies and imaging techniques [6].
Biosensing is a technique used to detect interactions of living entities with specific biorecognition elements such as antibodies, aptamers, DNA, and RNA [7,8], using sensors or detection systems [9,10]. These interactions generate a measurable signal indicating the presence [11,12] of specific biomolecules on the sensor’s surface. Biosensors help in detecting and quantifying these biomolecules in different biological samples like blood, serum, urine, tissue, etc. Based on detection mechanism and applications, these can be classified into different types, as illustrated in Figure 1. The sensors help in diagnosing a disease by detecting and measuring biomarkers at the molecular and cellular levels. Thus, they can facilitate the monitoring of both normal and pathological conditions, as well as the management of life-threatening diseases such as cancer [13,14,15].
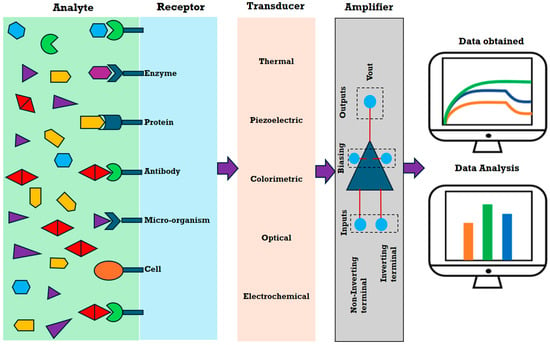
Figure 1.
Schematic illustrating the principle of various types of biosensors.
Among biosensors, optical sensors are particularly well suited for this purpose as they are non-invasive and can detect biomarkers using light. Like all biosensors, an optical biosensor relies on changes in optical properties to detect and quantify the presence of specific biological molecules or reactions. Commonly, optical biosensors operate on principles such as surface plasmon resonance (SPR) [16,17], fluorescence [18], luminescence [19], interferometry [20], and photonic crystals [21,22]. A subclass of photonic crystal sensors, GMR-based sensors, is gaining recognition for their high sensitivity; specificity; rapid detection; portability; cost-effectiveness; multiplexing capabilities; and, most importantly, ease of light coupling. These features make GMR-based sensors particularly suitable for developing POC devices [23].
2. Principle, Latest Advancements, and Biological Applications of GMR Technology
A GMR structure, also known as resonant waveguide grating (RWG), typically consists of a grating made from high refractive index (RI) material [24]. The grating can be either fully etched or etched onto a waveguide as shown schematically in Figure 2a [25]. The grating–waveguide structure is typically placed on a substrate, usually a glass plate (RI ≈ 1.45), which has a lower refractive index (n3) compared with the waveguide (nw) and grating material, such as silicon nitride (RI ≈ 2.0) [26].
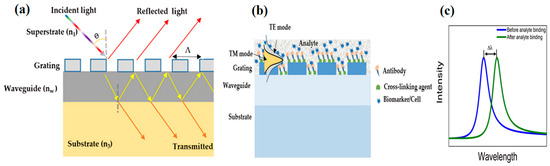
Figure 2.
(a) Schematic diagram of a GMR structure: a periodic grating (ng) is positioned on top of the waveguide (nw), which is placed on a substrate (n3). The incident light is diffracted into various orders with some specific order exciting the waveguide and finally producing a resonance peak response. (b) Schematic of a basic GMR structure operation with the GMR surface prepared to capture the biological entity of interest. The binding of the analyte on the surface leads to change in the effective refractive index, which results in a change in the resonance position. (c) GMR peaks can be obtained for the TE/TM mode. The figure shows the wavelength shift when the analyte binds to the surface in the incident medium.
The resonance in a GMR structure is based on the interaction between light and the grating. When incident light is diffracted by the grating into multiple diffraction orders, one or a range of these orders can become supported guided modes of the underlying waveguide or the grating layer itself [27]. The propagating mode interacts with the grating and leaks out of the waveguide producing a Fano-type filtering response, often referred to as lossy mode resonance [28]. The diffraction of light in the grating is mathematically described by Equation (1) given below. These equations collectively consider the incident and diffracted angles, the refractive indices of the media, and the grating characteristics. In its general form, the diffraction condition is given by Equation (1) [29]:
where θ is the incident angle; θm is the diffracted angle; n1 and nw are the refractive indices of the incident and diffractive media, respectively; m is the diffraction order; λ0 is the free-space wavelength, and Λ is the grating periodicity (i.e., the regular, repeating structure of the grating). In the case of transmission, the phase-matching condition is given by Equation (2):
with k0 (=2π/λ0) as the incident free space wave vector and Kg (=2π/Λ) as the grating vector. In the case of reflection, the analogous condition is given by Equation (3):
n1 sinθ = nw sinθm ± (mλ0/Λ)
k0n1 sinθ + mKg = k0nw sinθm
k0n1 sinθ − mKg = k0n1 sinθm
The propagating mode in the waveguide is classified into two primary types based on the polarization of electromagnetic field: (a) the transverse electric (TE) mode, where the electric field is transverse to the direction of mode propagation, and (b) the transverse magnetic (TM) mode, when the magnetic field is transverse to the direction of mode propagation [30,31]. For a coupled grating-waveguided GMR structure as shown in Figure 2a, the resonance condition is given by the propagation constant () of the waveguide. The azimuthal angle () is the angle between the plane of incidence and the grating vector (the axis along which periodicity is considered). Its value is zero for normal incidence; however, it has a non-zero value under conical diffraction conditions [32].
In a GMR structure, the grating usually has a constant period across the sensor surface. The resonance occurs when the incident wavelength matches the effective grating period and a single resonance wavelength is exhibited across the structure [33]. The detection mechanism relies on the binding of a target biomarker to the biorecognition element attached to the sensor surface [34], as shown in Figure 2b,c. The process can be tracked using a spectrometer or dispersive system following certain surface chemistry protocols [35]. The binding alters the refractive index of the surrounding medium causing minute detectable changes in the optical spectrum, thereby making it an efficient, label-free, in vitro method for detecting cancer biomarkers [36].
GMR devices with uniform grating periodicity are widely used in biosensing for detecting targets like bacteria, proteins, DNA, or cell–surface interactions [37,38,39,40,41]. To perform parallel detections, multiple identical grating sensor arrays are required, which increases the complexity and cost of the system. Nevertheless, it has a solution in the form of chirped GMR, which overcomes these limitations and can operate over a wide range of wavelengths [42]. Chirping is a technique in which the grating period or the fill factor varies gradually along one direction [43]. Another critical advantage of using a chirped GMR is its ability to translate spectral information into spatial information, which can be read out using a low-cost CMOS (complementary metal-oxide-semiconductor) camera instead of a spectrometer. By dividing the chirped grating into finite sensing elements, multi-wavelength sensor arrays can be formed where each array produces a distinct resonance peak. Thus, the technique simplifies the system design, reduces costs, and facilitates simultaneous multi-analyte biosensing without the need for complex scanning or modulation mechanisms.
The performance of GMR sensors depends on a careful balance of several key factors, such as the biorecognition elements used, sensitivity, full-width at half-maximum (FWHM) of the resonance, figure of merit (FOM), and limit of detection (LOD). Each of these factors plays an important role and is discussed below. First, the number of available binding sites per unit area and the strength of interaction between the biorecognition element and the target analyte are crucial. Biorecognition elements such as antibodies, enzymes, nucleic acid, and aptamers are essential for the sensor’s efficiency as they directly influence sensitivity, stability, and specificity. The antibodies offer high specificity through their distinct binding domains. However, they must be immobilized carefully to avoid steric hindrance, which can block binding sites and reduce sensitivity over time. On the other hand, aptamers benefit from precise nucleic acid complementarity, but their negative charge can provoke unintended electrostatic interactions with non-target molecules, causing nonspecific binding. Therefore, the proper engineering and surface chemistry modifications of biorecognition elements are critical for minimizing background noise, ensuring consistent signal transduction, and achieving a robust balance between sensitivity and selectivity [44].
Sensitivity is the ability to detect small refractive index changes that reflect the effectiveness of the sensor in the biomarker detection. However, high sensitivity alone is insufficient if the resonance peak is broad. In such cases, the FWHM becomes equally important as it directly impacts the precision of the sensor. Narrower resonance peaks or lower FWHM values imply higher resolution as they enable the detection of finer shifts in resonant frequency. Another important parameter, the FOM [45], is more useful as it takes into account both sensitivity and FWHM. A high FOM value ensures both strong and sharp resonance peaks. Additionally, the quality factor (Q-factor) describes the sharpness of the resonance. Higher Q-factor values imply a greater ability of the sensor to distinguish between different analyte concentrations. Next, the LOD value of a sensor must be minimized to ensure the detection of analytes at ultra-low concentrations. Although all these parameters are interdependent, the most critical to optimize are sensitivity, FOM, and LOD as they significantly influence the precision, detection capability, and overall performance of the sensor in practical applications. A recent study [46] suggests that LOD optimization requires careful balancing between the Q-factor and resonance amplitude rather than simply maximizing the Q-factor, as was assumed earlier. Experimental validations highlighted the significant impact of optical losses on the resonance amplitude in comparison with the Q-factor. Hence, it can be stated that an optimal trade-off between the Q factor and the resonance amplitude is essential for achieving high performance. In summary, the tunable and high-Q-factor resonance peaks in GMRs enable high-sensitivity detection in biosensing applications [47]. By adjusting the sensor structure parameters, the resonance linewidths can be controlled, which is the optimization of sensor performance for detecting low concentrations of biomarkers.
GMR technology can operate on both intensity-based and phase-based sensing modality, thereby enabling the detection of various substances such as gases, proteins, and disease biomarkers. To mention one of multiple interesting recent applications, Juan-Colas et al. [48] recently demonstrated that GMR can detect proteins very close to the cell itself and even image both the human cells and the proteins upon release. This capability allows observing how cells communicate in both healthy and diseased states. In their work, they applied GMR to map the secretion of the signaling protein thrombopoietin (TPO) from individual human HepG2 cells and also studied variations in TPO expression with changes in the concentration of desialylated platelets [48]. Furthermore, it is important to note that GMR sensors have been successfully utilized for detecting cancer biomarkers [49], cardiac markers [50], and neuropeptides [51], suggesting their significant contribution from a medical diagnostics standpoint.
3. GMR Sensors for Cancer Biomarker Detection
This section describes the state-of-the-art research on GMR-based cancer detection, explaining how biomarker binding events alter the refractive index and generate detectable optical shifts. Theoretical advancements and novel insights that have given new directions to the field and extended the existing framework are covered in the discussion. Table 1 lists various cancer-related biomarkers, along with their sources, the reported values of LOD, and the sensitivity achieved using GMR sensors. Thus, the section evaluates different approaches, highlighting general patterns and variations across various studies, and focuses on overarching trends in applying GMR for biomedical applications.

Table 1.
The various types of biomarkers related to cancer or other kinds of diseases associated with different body parts with their corresponding source, type, LODs, and sensitivity achieved using GMR sensors. (NA—not available.)
Several studies have effectively utilized GMR sensors for the identification of specific cancer biomarkers, which supports the efficacy of these sensors. For instance, Wawro et al. [52] used a GMR-based detection system to identify ovarian cancer biomarkers, specifically calreticulin and ryanodine receptor 3, which exhibit altered levels in blood and serum. By using a surface functionalization technique [63,64], the sensor successfully detected calreticulin concentrations ranging from 8.5 nM to 68 nM through covalent binding to its monoclonal IgG antibody conjugate. In another similar study, Kaja et al. demonstrated that the GMR sensors deliver highly accurate, consistent, and reproducible quantification of protein biomarkers, after validation through quantitative immunoblotting. Their study identified fibronectin, apolipoprotein A1, and tissue inhibitor of metalloproteinases 3 (TIMP3) as key markers for differentiating primary from metastatic ovarian carcinoma. They reported elevated fibronectin levels in primary ovarian carcinoma, which support the notion that this protein is upregulated and secreted early in malignancy [65]. Thus, both studies focused on the early diagnosis of ovarian cancer using GMR: Wawro et al. detected calreticulin and ryanodine receptor 3, while the study by Kaja et al. provided critical insights into the regulation of fibronectin levels in ovarian cancer.
In light of the promising findings presented in these reports, Tu et al. [53] introduced a new method for the detection of a pan-cancer biomarker, β-actin (ACTB). This biomarker may lead to non-Hodgkin lymphoma on mutations and has been linked to a variety of cancers, including liver, melanoma, renal, colorectal, gastric, pancreatic, esophageal, lung, breast, prostate, ovarian cancers, leukemia, and lymphoma. A biomarker linked to multiple cancer types is referred to as a pan-cancer biomarker. In this study, they designed a three-layered grating structure integrated with a microfluidic system for binding β-actin on the sensor surface. As demonstrated in Figure 3, the integration of microfluidic channels with the GMR sensor allowed for the filtration of unwanted substances, thereby improving the sensitivity by preventing false readings. This filtration enabled the sensor to detect concentrations of 2.5 mg/mL with a 2.1 nm peak wavelength shift, whereas the unfiltered sensors failed to function due to non-specific binding on the sensor surface. Compared with other reported results, 2.5 mg/mL is relatively high for sensitive cancer biomarker detection. Furthermore, this study suggests the utility of GMR technology in scenarios when structural protein accumulation signals malignancy. Moreover, its overexpression may indicate cytoskeletal remodeling in lymphoma, influencing cell motility and metastatic potential.
Figure 3.
Schematic of the integration of a microfluidic channel with a three-layer grating structure (right picture). The sample is injected into the lab-on-a-chip (LOC) system via a syringe, filtered to remove cell debris, and then flows to an antibody-immobilized GMR biosensor for analyte detection. The optical setup measures the quantity, and the sample is then directed to the waste container. Reproduced with permission from [53].
Further, GMR-based sensing was reported for detecting another type of cancer biomarker, TNF-α (Tumor necrosis factor alpha) [66] (Figure 4a(i)), which is associated with oral squamous cell carcinoma and is also found in human chronic wounds. Recently, Bakshi et al. [55] successfully detected and quantified TNF-α along with another key wound health biomarker, IL-6 (Interleukin 6) [67], using a chirped GMR biosensor. IL-6 is also produced in response to infection, injury, or inflammation. The sensor detected both biomarkers at concentrations of 1 ng/mL in hydrogel-based dressings containing clinical wound exudate samples as depicted in Figure 4a(ii). The hydrogel matrix showed no interference with antigen–antibody binding, as confirmed by consistent resonance shifts in TNF-α assays after adjusting for its refractive index effect. The combined analysis of TNF-α and IL-6 levels as reported by Bakshi et al. could reveal inflammatory signatures associated with tumorigenesis. Thus, the work supported the GMR being suitable for detecting biomarkers associated with cancer, as well as chronic wounds.
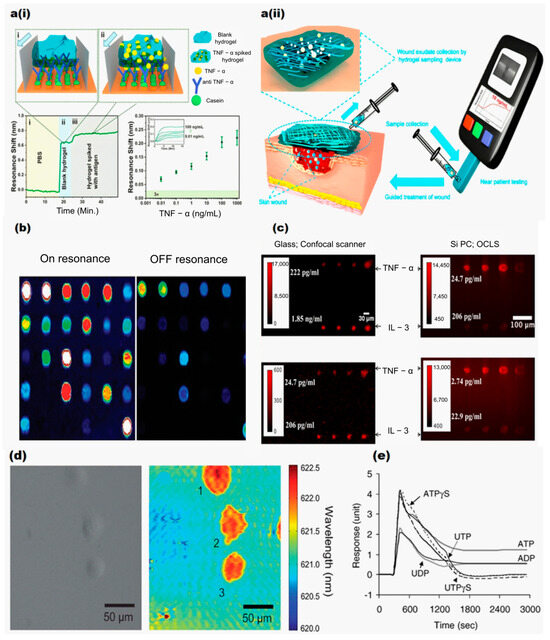
Figure 4.
(a(i),a(ii)) Introduction of blank hydrogel, followed by spiked hydrogel, with arrows indicating flow direction. Biorecognition was performed in blank and spiked hydrogel. Resonance shifts were observed for TNF-α concentrations from 0.01 ng/mL to 1 μg/mL. Reproduced from [55], under CC-BY license. (b) Detection of fluorophore-tagged DNA and protein molecules. (c) Comparison of fluorescence images of microspots on a glass surface and photonic crystal for various concentrations of TNF-α and IL-3. Reproduced with permission from [62]. (d) Bright field and PWV imaging of Panc-1 cells attached to the PC surface. Reproduced with permission from [68]. (e) The DMR responses of quiescent A431 cells measured by stimulation with six P2Y receptor agonists at 20 μM. Reproduced with permission from [41].
In another work, Poulikakos et al. [69] developed a metasurface based on GMR for detecting fibrous tissue and its orientation through colorimetric measurement. The sensor reflects green for isotropic tissue and blue for fibrous tissue, with a blue-to-green transition indicating tissue orientation. They demonstrated TE and TM modes on a GMR metasurface using a 215 nm thick Si3N4 layer (RI ≈ 2.0) and a 75 nm thick SiO2 layer (RI ≈ 1.45) deposited on a SiO2 substrate with a rhombohedral lattice periodicity of 75 nm × 275 nm. The choice of low-loss dielectric materials minimizes absorption losses. Additionally, it was applied to fibrous tissue (modeled as a birefringent medium with a refractive index difference (Δn) of 0.003), and the induced phase delay led to a measurable blue shift in the reflected color. This spectral shift was used to quantitatively map the spatial arrangement and density of collagen fibers. The study provides insights into structural changes linked to cancer progression, which could help in identifying metastatic spread. The metasurface’s colorimetric response (green for isotropic tissue and blue for fibrous tissue) enabled rapid visual distinction between localized tumors (isotropic) and metastatic tumors (fibrous). The work demonstrated the potential of GMR technology combined with calorimetric measurement.
Furthermore, Huang et al. [61] reported another significant advancement by using photonic crystal (PC) surfaces for highly sensitive protein microarrays. The photonic crystal sensor surface was designed to resonate at the excitation wavelength of cyanine-5 (Cy5), enhancing the fluorescent intensity for Cy5-labeled analytes as depicted in Figure 4b. This approach of using a confocal microarray scanner rather than a conventional glass surface improved the signal-to-noise ratio by 3.8–6.6 times and reduced detection limits by 6–89%. Further developments included a silicon-based Photonic Crystal Enhanced Fluorescence (PCEF) platform for multiplexed biomarker assays, as demonstrated by George et al. [62]. In this platform, a cylindrical lens was used to rapidly scan a microarray in under a minute. The compact detection device integrated fluorescence excitation from a semiconductor laser with the photonic crystal’s resonant optical modes. In their study, two cytokines, IL-3 and anti-TNF-α, were used as antibodies on the PC slides. Microspot fluorescent sandwich immunoassays were utilized to demonstrate the platform’s capabilities and successfully detected biomarkers at concentrations as low as 25 pg/mL and 0.21 ng/mL. Additionally, the use of a fluorescent microarray for detecting a breast cancer miRNA biomarker (miR-21) enabled detection at concentrations as low as 0.6 pM. The sensor’s performance was evaluated using an objective-coupled line scanning (OCLS) setup to capture fluorescence images of various concentrations of IL-3 and TNF-α, as illustrated in Figure 4c. Both studies reported by Huang et al. and George et al. harnessed engineered photonic crystal platforms to amplify weak fluorescence signals from cancer biomarkers (e.g., miR-21 and cytokines), thereby enabling the detection of these molecules at extremely low concentrations. The enhanced sensitivity and multiplexing capacity provided by these platforms support detailed molecular profiling of cancer cell biology.
Understanding cell membrane interactions with surfaces is crucial for cancer-related processes such as tumor metastasis differentiation and apoptosis. Traditional methods rely on fluorescent dyes, proteins, or histological stains, which could be cytotoxic and suffer from photobleaching. To overcome these limitations, Chen et al. [68] investigated the dynamics of Pancreatic cancer (Panc-1) cell and mHAT9a cell attachment using a hyperspectral imaging microscope. This technique captured the evolution of cell attachment and morphology during chemotaxis and drug-induced apoptosis. Images of the sensor surface at the GMR peak wavelength value (PWV) were continuously collected. Positive shifts in PWV values indicated cell adherence and extension, with spectra from inside and outside the cellular regions. Figure 4d compares brightfield and PWV images of Panc-1 cells, highlighting the sensors’ capability to detect variations in cell adherence. Fang et al. [40] further expanded this work by demonstrating GMR’s ability to detect cellular responses to stimuli, including changes in cell shape and mass in both vertical and horizontal directions. The study involved human epidermoid carcinoma A431 cells and Chinese hamster ovary (CHO-K1) cells. Their findings showed that as more CHO cells grew together (cell confluency), the resonant peak moved to higher angles. Moreover, exposure to high concentrations of dimethyl sulfoxide caused both a peak shift and a shape change, highlighting the sensor’s high sensitivity to toxic substances. Additionally, they [70] observed dynamic mass redistribution (DMR) in A431 cells when triggered by epidermal growth factor (EGF), showing cellular movement in real time. Figure 4e shows the maximum DMR responses of A431 cells when exposed to six P2Y agonists at 20 μM. These agonists cause different levels of cell activity, showing how cell signaling can vary. They also studied how different G protein-coupled receptors (GPCRs) were activated, detecting unique light patterns for each GPCR group [41]. The sensor’s precision in detecting subtle cellular changes makes it valuable for studying complex environments, with applications in drug discovery, toxicology, and mechanobiology. Altered cell adhesion is a hallmark of cancer progression, and it plays a key role in metastasis. Cancer cells first detach from primary tumors and then invade the extracellular matrix. To investigate these phenomena, Zaytseva et al. combined GMR biosensor technology with total internal reflection fluorescence (TIRF) imaging. Their approach monitored cell adhesion dynamics without the need for labels. They used an engineered HEK293 cell line that expressed fluorescent β2-adrenergic receptors. The study revealed changes in receptor signaling along with cell adhesion, which can aid in cancer detection [71]. The above three studies by Chen et al., Fang et al., and Zaytseva et al. enable real-time monitoring of cancer cell adhesion, morphology, and receptor signaling, which are important for understanding metastasis and cellular responses to therapeutics. The sensitive detection of subtle changes in cellular behavior, including mass redistribution, receptor activation, and adhesion dynamics, facilitate an understanding of molecular mechanisms underlying cancer progression.
In another advancement, Chan et al. [72] developed a label-free GMR-based sensing method for detecting breast cancer cells (MCF-7), mapping cell growth and apoptosis in response to the anticancer drugs doxorubicin hydrochloride and curcumin. Cell attachment increased the intensity of the resonantly reflected wavelength, allowing them to map the spatial distribution of cells. After 24 h, MCF-7 cell counts increased by 240% (Figure 5a), but curcumin and doxorubicin reduced cell numbers by 93% and 89%, respectively, through apoptosis. The sensor provided continuous imaging of cell behavior, although it lacked the resolution for detailed single-cell views. The work showed the potential of GMR for drug delivery.
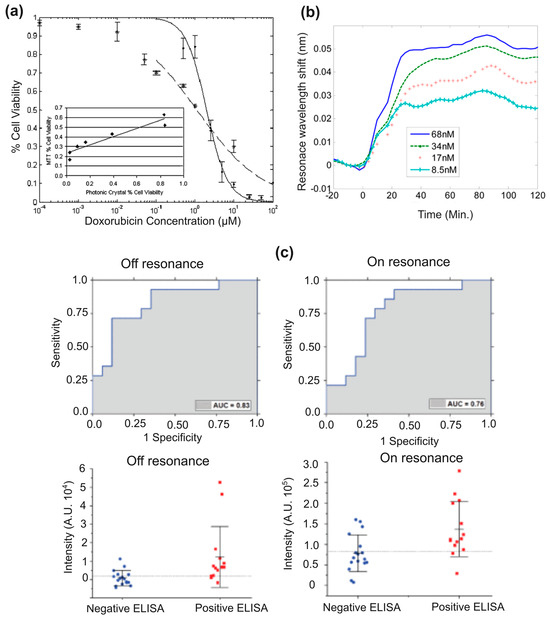
Figure 5.
(a) A comparison of IC50 values for MCF-7 cell viability with doxorubicin, where the photonic crystal biosensor shows an IC50 of 2 µM and the MTT assay measures 1 µM [72]. (b) Resonance wavelength shifts as calreticulin binds to IgG, with binding tracked for 80 min [73]. (c) Results from 14 ELISA-positive and 17 ELISA-negative samples show specificity of 0.71 and sensitivity of 0.79. Reproduced from [59,72,73], under CC-BY license.
To further investigate the detection of TNF-α and calreticulin, Magnusson et al. [73] measured biotin binding to GMR sensors and studied their interactions with their respective antibodies (Figure 5b). By modeling these binding events, they assessed the attributes of the biolayer and its environment. The sensor’s polarization sensitivity was analyzed, revealing that the TM mode exhibited greater sensitivity than the TE mode. This study contributes to the design of sensors that support multiple leaky modes, enabling direct light addressing and simultaneous calibration of variations such as temperature and sample background density. These advancements enhance detection accuracy and reduce the likelihood of false measurements. Further, Race et al. [59] developed an automated platform to detect human papillomavirus (HPV16) antibodies, which is linked to oropharyngeal cancer, using just a single drop of serum. In this user-friendly, single-use diagnostic tool, a droplet of serum is placed into a cartridge that is illuminated with a custom laser scanning instrument, allowing optimal light coupling into the GMR sensor for fluorescence detection. The results demonstrated enhanced excitation under “on-resonance conditions”, even without a powerful laser. It led to increased spot intensities above the LOD and enabled the identification of spots that would otherwise be undetectable as depicted in Figure 5c. However, the study also detected anti-E7 antibodies in the serum of healthy controls and thus suggested the biomarker being not reliable for identifying HPV-related oropharyngeal cancer due to the observed enhanced fluorescence.
In this study, they reported photonic crystals featuring a 40 nm grating depth, a 360 nm period, and a 36% duty cycle. The device comprised a 150 nm thick high-refractive-index TiO2 layer deposited atop an 840 nm thick low-refractive-index SiO2 layer. It had a resonant wavelength near 628 nm with an approximate 2 nm bandwidth and about 80% reflection efficiency. This configuration achieved an optimal specificity of 0.71, and the PCEF platform exhibited an LOD lower than the clinically relevant serum anti-E7 antibody concentration (in the pM to nM range as determined by ELISA), which made it well suited for detecting cancer biomarkers in blood at low concentrations.
Collectively, these studies demonstrate that GMR-based sensors enable real-time, cross-scale analysis. This argument is supported by Chan et al.’s monitoring of drug-induced apoptosis and proliferation in breast cancer cell populations, Magnusson et al.’s quantification of antibody–antigen molecular interactions, and Race et al.’s detection of cancer-linked biomarkers like HPV antibodies. These reports reflect that GMR-based biosensing systems help in understanding cancer-related processes such as apoptosis evasion, biomarker dysregulation, and therapeutic response.
By integrating surface functionalization techniques and microfluidic systems, GMR sensors efficiently detect a wide range of biomarkers at ultra-low concentrations, offering a label-free, non-invasive solution for early cancer diagnosis. Xiao et al. reported that after the integration of the microfluidic modules, the GMR system’s functions and parameters improved significantly. The tunability was enhanced by achieving a tuning bandwidth of 13.181 nm (optofluidics) in comparison with 25.39 nm (optically tuned) along with an improved spectral response characterized by a reduction in FWHM from 2.504 nm to 1.090 nm, while maintaining high reflectance levels (>85%) [74]. These improvements demonstrated that integrating sub-microfluidic modules not only enabled dynamic tuning of the filter but also enhanced its overall optical performance. In addition to microfluidic integration, GMR was combined with up-conversion nanoparticles (UCNPs) for detecting low-abundance biomarkers such as cardiac troponin I (cTnI). Their ultra-low concentrations make detection difficult as normal levels are near 1 pg/Ml in serum. To address this limitation, Tseng et al. proposed an immunosensor by integrating GMR with UCNPs [75]. Prior to this innovation, up-conversion-based immunoassays were offering a limit of detection (LOD) of 130 pg/mL across a range (10–10,000,000 pg/mL). The GMR-based UCNP hybrid system, however, improved detection capabilities, achieving an LOD of 0.00024 pg/mL. Furthermore, the system’s detection range was optimized to 0.001–100 pg/mL, specifically targeting clinically low-abundance concentrations. This advancement represented an increase in LOD and a detection range compared with the single-molecule array (Simoa) [76], setting a new benchmark for cTnI analysis in patients undergoing chemotherapy [77]. Based on the above discussions, it can be concluded that the advancements in GMR-based biosensors have significantly enhanced their versatility, sensitivity, and precision in cancer biomarker detection. GMR technology has proven valuable in capturing subtle cellular changes and improving diagnostic accuracy, with applications in drug discovery and personalized medicine. The use of colorimetric sensors has further contributed to its reliability, positioning GMR sensors as powerful tools in cancer diagnostics.
4. Challenges and Future Perspectives
GMR-based biosensors have advanced significantly in terms of precision and versatility for cancer biomarker detection, leveraging surface functionalization, microfluidic systems, and integration with photonic crystals and fluorescence enhancement that improves precision and versatility. It is important to note that as GMR sensors are usually built using dielectric materials, these have minimal loss compared with metal-based SPR sensors and therefore are ideal for wavelength filtering responses. Although GMR sensors can theoretically achieve high quality (Q) factors, practical limitations arise from fabrication defects such as surface roughness, structural irregularities, and non-uniformity. As a result it enhances radiative losses or coupling losses [78]. These losses can be reduced by lowering the refractive index contrast as reported by Li et al. [79], which causes the guided mode to be more effectively confined within the structure. This enhanced confinement sharpens the resonance, resulting in a narrower FWHM. However, at a very low refractive index contrast (Δn < 0.01), as reported by Triggs et al., the GMR spatial resolution deteriorates [80]. The effect is due to weakened confinement of the resonant mode caused by insufficient contrast between the grating and the surrounding medium. To address this limitation, materials such as graphene, which enable strong refractive index modulation and rapid response, have been proposed [81]. Further studies have demonstrated that GMR condition can be achieved in low-index waveguide materials such as SU8, which is placed on top of a high-index lithium niobate through a hybrid configuration [28]. Therefore, the flexibility in choosing GMR materials, whether high or low index, provides multiple material options enabling possible integration with metal gratings for enhanced light manipulation which is favorable for applications like sensing, nonlinear optics, and electro-optics [82,83]. Moreover, tunability of GMR sensors is possible through grating design, dielectric layer thickness [84], and angle of incidence [85]. Additionally, GMR structures have spectroscopic advantages like non-destructive analysis, multiple sensing [86], real-time monitoring [87], and label-free sensing capabilities [38]. Notably, the chirped GMR approach allows spectral data to be captured and processed as spatial information, eliminating the need for an expensive spectrometer. This can lead to cost savings in system design as it integrates the roles of both sensing and spectral analysis into one element [88]. Another key advantage of GMR structures is their ability to couple light with out-of-plane illumination, allowing for better tolerance to variations in the incident angle [89]. This is especially useful in situations where precise light alignment is difficult, such as end-fire coupling, waveguide coupling in SPR sensors [90]. Their low cost, miniaturization, and portability make them ideal for POC diagnostics, enabling sensitive, label-free detection at low concentrations for non-invasive early cancer diagnosis.
On the other side, there are some existing challenges associated with GMR-based sensors that need to be carefully addressed. It includes sensitivity to environmental changes like temperature fluctuations [91] and mechanical vibrations [92], which could introduce noise and reduce accuracy in real-world applications. A promising advancement is the introduction of a bowtie-shaped sensor that leverages GMRs in a chirped modality [93]. This design offers high sensitivity to refractive index changes while being robust against mechanical and thermal noise. These sensors have effectively measured parameters like salinity and bacterial growth in noisy environments, highlighting their potential for real-world applications. Another real-world challenge faced by the nanofabrication-based sensors is the reproducibility of results due to fabrication inconsistency and requirement of nanoscale precision for optimal performance. Recent investigations by Sahoo et al. [25] provide useful guidelines for fabricating nanostructures more reliably and achieving a higher success rate in exploratory experiments. Furthermore, detecting low-abundance biomarkers in early-stage cancer is challenging due to limited sensitivity. GMR sensors need to be functionalized with specific molecules to capture target biomarkers; however, this process is complex and time-consuming and may limit multi-biomarker detection. Biocompatibility issues with some materials used in GMR sensors can also restrict their use in clinical or in vivo applications. For example, gold (Au) is generally biocompatible; however, it can induce foreign body reactions when shaped into nanoparticles [94]. Silver used in biosensors can release silver ions (Ag+), which can lead to cytotoxicity and oxidative stress; therefore, limiting its usefulness in prolonged in vivo applications [95]. Additionally, non-specific binding of other molecules can interfere with signals, reducing the sensor’s selectivity [96]. By implementing protective coatings and suitable receptors, the challenges associated with non-specific binding can be effectively mitigated [97]. Moreover, advanced algorithms are needed to accurately interpret signals and filter out noise, which otherwise can make real-time analysis more challenging [98]. Improving sensitivity of GMR sensor can further enhance its usability [99]. This requires a multifaceted approach involving structural optimization (e.g., particle swarm optimization) [100], material selection [101], surface functionalization [102], light optimization [103,104], and advanced measurement techniques [105].
Now, we compare other photonic devices and sensors relevant to cancer biomarker detection, with a focus on enhancing the limit of detection and real-time monitoring [106]. SPR biosensors offer high sensitivity, specificity, and real-time monitoring for cancer biomarker detection. They effectively detect HER2 in breast cancer [107] and biomarkers like PSA (LOD: 0.15 ng/mL), IL-8 (LOD: 2.5 pM), CEA (LOD: 0.5 ng/mL), and fibronectin (LOD: 2.5 µg/mL) [108]. A plasmonic sensor with split-ring resonators achieved an LOD of 0.016 RIU for brain cancer cells [109]. Bladder cancer-related miRNA biomarkers were detected with an LOD of 26.3 fM using a biochip with time-gated luminescent probes and photonic crystals [110]. Other than SPR sensors, interferometric and photonic crystal biosensors offer high sensitivity for detecting low-abundance cancer biomarkers [111]. CYFRA 21-1, a lung cancer biomarker, is quantified with an LOQ (limit of quantification) of 81 pg/mL with enhanced diagnostic accuracy using compensated interferometry [112].
Given the aforementioned facts, GMR biosensors present a promising and cost-effective solution for POC diagnostics. They facilitate highly sensitive, label-free detection of biomarkers by utilizing economical components such as filtered LEDs and CMOS cameras [86]. This simple, label-free GMR approach has achieved sensitivity in pg/mL levels comparable to those of commercial enzyme-linked immunoassays (ELISAs), while eliminating the need for labels, enzymatic amplification, or trained laboratory personnel. The integration of chirped gratings further simplifies the optical setup by encoding spectral resonance information into the spatial domain, effectively combining sensing and spectrometer functions within a single element. Since the technology omits the requirement for expensive dispersive optics or external spectrometers, it allows operation at a single wavelength, optimized for the camera’s signal-to-noise ratio [88]. GMR sensors also support multiplexed detection of clinically relevant biomarkers, such as troponin, procalcitonin, and C-reactive protein in complex samples like undiluted urine, using antibody immobilization to achieve ultra-low detection limits and minimize non-specific binding [86]. However, the critical aspect for POC deployment is the fabrication reproducibility since nanometer-scale variations can detune resonance behavior. This issue can be minimized by tight control of electron-beam exposure and development processes that ensures consistency in resonance wavelength, amplitude, and sensitivity [25]. In light of these considerations, the integrating of a GMR sensor with basic optical components and a low-cost camera results in a portable, compact, and fault-tolerant system that is ideal for practical applications. With component costs under USD 10, it is highly suitable for affordable POC devices [43]. Importantly, the clinical utility of these tools relies on collaborations with healthcare professionals, ensuring that technical capabilities are effectively aligned with practical needs. Despite its simplicity, the GMR platform does not compromise performance, positioning it as a robust, scalable solution for accurate and decentralized testing [113].
The future of GMR sensors in cancer biomarker detection depends on advancements in multiplexing, artificial intelligence (AI) integration, miniaturization, and real-time data processing. Multiplexing enables simultaneous biomarker detection, whereas AI improves data analysis. Shen et al. have applied machine learning to the GMR filter design by training a forward deep neural network on simulation data to rapidly generate millions of structures. They have used Fano resonance-based inverse matching to optimize asymmetric spectral line shapes, achieving narrow linewidths (6.8–8.7 nm) in the visible spectrum. This approach combines data-driven designs with Fano models to efficiently engineer high-performance optical filters, bypassing traditional iterative optimization [114,115]. Cloud-based connectivity and the Internet of Medical Things (IoMT) will further enable remote monitoring, personalized treatments, and telemedicine applications. In the future, hybrid plasmonic–GMR structures along with nanomaterials could enhance sensitivity to ultra-low biomarker levels. A hybrid device combining GMR sensors for rapid screening with other optical technologies for detailed analysis could enhance diagnostics.
5. Conclusions
This review highlights the fundamentals and recent advancements of GMR-based biosensors for cancer biomarker detection. Its significance is explained in detail, including the operational mechanisms and the applications in drug discovery and cancer diagnostics. Their potential for developing cost-effective and cutting-edge technology-driven POC sensing devices is highlighted. These sensors offer highly sensitive, label-free, and non-invasive methods for early cancer diagnosis and monitoring. Integrating GMR sensors with technologies like microfluidics, photonic crystals, and fluorescence has greatly improved detection accuracy and sensitivity. The challenges like sensitivity to environmental noise and non-specific binding need to be carefully addressed; nevertheless, improvements in sensor design, materials, biorecognition elements, and algorithms have helped to overcome many of these problems. Addressing broader challenges like late-stage cancer detection, limited biomarkers, psychosocial impacts, and the effectiveness of current treatments requires collaboration between scientists, healthcare providers, policymakers, and society. Ongoing and future developments in AI could enhance their performance and accessibility, which supports early detection of cancer, particularly in collecting and analyzing raw data. The integration of GMR devices with mobile applications significantly enhances the usability of health monitoring and early warning systems. This advancement offers affordable and efficient healthcare solutions, particularly in remote or underserved regions. Furthermore, these POC devices have potential to reduce hospital burden and minimize the necessity for specialist consultations in specific cases. In summary, this review provides a guide for future cancer research focused on developing highly sensitive GMR-based biosensors to improve diagnostic abilities, especially in resource-limited areas.
Funding
P.K.S., A.A.B. and M.S. acknowledge financial support by the SERB-SURE, DST, (ANRF), India (Grant No. SUR/2022/004910), and JKST&IC, J&K, India (Grant No. JKST&IC/SRE/941-45). K.L. acknowledges the support of the University of York.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- International Agency for Research on Cancer. Global Cancer Burden Growing, amidst Mounting Need for Services. Saudi Med. J. 2024, 45, 326–327. [Google Scholar]
- Bray, F.; Laversanne, M.; Sung, H.; Ferlay, J.; Siegel, R.L.; Soerjomataram, I.; Jemal, A. Global Cancer Statistics 2022: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2024, 74, 229–263. [Google Scholar] [CrossRef] [PubMed]
- Larkins, M.C.; Thombare, A. Point-of-Care Testing. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2025. Available online: https://www.ncbi.nlm.nih.gov/books/NBK592387/ (accessed on 25 April 2025).
- Tantiwanichapan, K.; Jolivot, R.; Jomphoak, A.; Srisuai, N.; Chananonnawathorn, C.; Lertvanithpol, T.; Horprathum, M.; Boonruang, S. Demonstration of Cross Reaction in Hybrid Graphene Oxide/Tantalum Dioxide Guided Mode Resonance Sensor for Selective Volatile Organic Compound. Sci. Rep. 2023, 13, 10799. [Google Scholar] [CrossRef]
- Qu, X.; Hu, Y.; Xu, C.; Li, Y.; Zhang, L.; Huang, Q.; Moshirian-Farahi, S.S.; Zhang, J.; Xu, X.; Liao, M. Optical Sensors of Volatile Organic Compounds for Non-Invasive Diagnosis of Diseases. Chem. Eng. J. 2024, 485, 149804. [Google Scholar] [CrossRef]
- Pulumati, A.; Pulumati, A.; Dwarakanath, B.S.; Verma, A.; Papineni, R.V.L. Technological Advancements in Cancer Diagnostics: Improvements and Limitations. Cancer Rep. 2023, 6, e1764. [Google Scholar] [CrossRef]
- Masson, J.-F. Surface Plasmon Resonance Clinical Biosensors for Medical Diagnostics. ACS Sens. 2017, 2, 16–30. [Google Scholar] [CrossRef]
- Chakraborty, B.; Das, S.; Gupta, A.; Xiong, Y.; T-V, V.; Kizer, M.E.; Duan, J.; Chandrasekaran, A.R.; Wang, X. Aptamers for Viral Detection and Inhibition. ACS Infect. Dis. 2022, 8, 667–692. [Google Scholar] [CrossRef]
- Zaytseva, N.; Miller, W.; Goral, V.; Hepburn, J.; Fang, Y. Microfluidic Resonant Waveguide Grating Biosensor System for Whole Cell Sensing. Appl. Phys. Lett. 2011, 98, 163703. [Google Scholar] [CrossRef]
- Zourob, M.; Elwary, S.; Fan, X.; Mohr, S.; Goddard, N.J. Label-Free Detection with the Resonant Mirror Biosensor. In Biosensors and Biodetection; Rasooly, A., Herold, K.E., Eds.; Methods in Molecular Biology; Humana Press: Totowa, NJ, USA, 2009; Volume 503, pp. 89–138. ISBN 978-1-60327-566-8. [Google Scholar] [CrossRef]
- Quaranta, G.; Basset, G.; Martin, O.J.F.; Gallinet, B. Recent Advances in Resonant Waveguide Gratings. Laser Photonics Rev. 2018, 12, 1800017. [Google Scholar] [CrossRef]
- Han, Q.; Pang, J.; Li, Y.; Sun, B.; Ibarlucea, B.; Liu, X.; Gemming, T.; Cheng, Q.; Zhang, S.; Liu, H.; et al. Graphene Biodevices for Early Disease Diagnosis Based on Biomarker Detection. ACS Sens. 2021, 6, 3841–3881. [Google Scholar] [CrossRef]
- Bhalla, N.; Jolly, P.; Formisano, N.; Estrela, P. Introduction to Biosensors. Essays Biochem. 2016, 60, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Chiang, C.C.; Tseng, W.-C.; Tsai, W.-T.; Huang, C.-S. Handheld Biosensor System Based on a Gradient Grating Period Guided-Mode Resonance Device. Biosensors 2023, 14, 21. [Google Scholar] [CrossRef] [PubMed]
- Schmid, J.H.; Sinclair, W.; García, J.; Janz, S.; Lapointe, J.; Poitras, D.; Li, Y.; Mischki, T.; Lopinski, G.; Cheben, P.; et al. Silicon-on-Insulator Guided Mode Resonant Grating for Evanescent Field Molecular Sensing. Opt. Express 2009, 17, 18371–18380. [Google Scholar] [CrossRef]
- Yadav, A.; Mishra, M.; Tripathy, S.K.; Kumar, A.; Singh, O.P.; Sharan, P. Improved Surface Plasmon Effect in Ag-Based SPR Biosensor with Graphene and WS2: An Approach Towards Low Cost Urine-Glucose Detection. Plasmonics 2023, 18, 2273–2283. [Google Scholar] [CrossRef]
- Pandiaraj, S.; Muthuramamoorthy, M.; Alanazi, N.; Alodhayb, A.N. Enhanced Sensitivity of SPR-Based Biosensor for Waterborne Pathogen Monitoring: A Numerical Analysis. Plasmonics 2024, 19, 2913–2921. [Google Scholar] [CrossRef]
- Goodrum, R.; Li, H. Advances in Three Dimensional Metal Enhanced Fluorescence Based Biosensors Using Metal Nanomaterial and Nano-Patterned Surfaces. Biotechnol. J. 2024, 19, 2300519. [Google Scholar] [CrossRef]
- Sohrabi, H.; Mahmoudi-Maleki, R.; Majidi, M.R.; Oroojalian, F.; Mokhtarzadeh, A.A.; de la Guardia, M. Recent Advances in Nanostructure-Enhanced Optical Assays Focused on Luminescence-Based Biosensors for Detection of Cancer Biomarkers. TrAC Trends Anal. Chem. 2024, 176, 117753. [Google Scholar] [CrossRef]
- Boschi, A.; Iachetta, G.; Buonocore, S.; Hubarevich, A.; Hurtaud, J.; Moreddu, R.; d’Amora, M.; Formoso, M.B.; Tantussi, F.; Dipalo, M.; et al. Interferometric Biosensor for High Sensitive Label-Free Recording of HiPS Cardiomyocytes Contraction in Vitro. Nano Lett. 2024, 24, 6451–6458. [Google Scholar] [CrossRef]
- Fallahi, V.; Kordrostami, Z.; Hosseini, M. Cancer Detection by Photonic Crystal Optical Biosensors: Effect of Hexagonal Micro Ring Resonator Design. Mater. Sci. Semicond. Process. 2024, 174, 108188. [Google Scholar] [CrossRef]
- Gowdhami, D.; Balaji, V.R.; Murugan, M.; Robinson, S.; Hegde, G. Photonic Crystal Based Biosensors: An Overview. ISSS J. Micro Smart Syst. 2022, 11, 147–167. [Google Scholar] [CrossRef]
- Myersa, F.B.; Lee, L.P. Innovations in Optical Microfluidic Technologies for Point-of-Care Diagnostics. Lab Chip 2008, 8, 2015–2031. [Google Scholar] [CrossRef] [PubMed]
- Dobbs, D.W.; Gershkovich, I.; Cunningham, B.T. Fabrication of a Graded-Wavelength Guided-Mode Resonance Filter Photonic Crystal. Appl. Phys. Lett. 2006, 89, 123113. [Google Scholar] [CrossRef]
- Sahoo, P.K.; Coates, E.; Silver, C.D.; Li, K.; Krauss, T.F. On the Reproducibility of Electron-Beam Lithographic Fabrication of Photonic Nanostructures. Sci. Rep. 2024, 14, 8703. [Google Scholar] [CrossRef] [PubMed]
- Magnusson, R. The Guided-Mode Resonance Biosensor: Principles, Models, and Applications. In Proceedings of the 2020 IEEE Photonics Conference (IPC), Virtual, 28 September–1 October 2020; IEEE: Piscataway, NJ, USA, 2020; pp. 1–2. [Google Scholar] [CrossRef]
- Foland, S.; Choi, K.H.; Lee, J.B. Pressure-Tunable Guided-Mode Resonance Sensor for Single-Wavelength Characterization. Opt. Lett. 2010, 35, 3871–3873. [Google Scholar] [CrossRef]
- Guo, T.; Evans, J.; Wang, N.; Jin, Y.; He, J.; Sun, Y. Guided Mode Resonance in a Low-Index Waveguide Layer. Appl. Sci. 2021, 11, 3312. [Google Scholar] [CrossRef]
- Koussi, E.-K. Micro Patterning of Complex Waveguide Resonant Gratings (WRG). Available online: https://www.researchgate.net/publication/351105934_Micro_patterning_of_complex_Waveguide_Resonant_Gratings_WRG (accessed on 23 April 2025).
- Magnusson, R.; Lee, K.J.; Hemmati, H.; Ko, Y.H.; Wenner, B.R.; Allen, J.W.; Allen, M.S.; Gimlin, S.; Weidanz, D.W. The Guided-Mode Resonance Biosensor: Principles, Technology, and Implementation. In Proceedings of the Frontiers in Biological Detection: From Nanosensors to Systems X, San Francisco, CA, USA, 28–29 January 2018. [Google Scholar] [CrossRef]
- Tibuleac, S.; Magnusson, R. Reflection and Transmission Guided-Mode Resonance Filters. J. Opt. Soc. Am. A 1997, 14, 1617–1626. [Google Scholar] [CrossRef]
- Sahoo, P.K.; Sarkar, S.; Joseph, J. High Sensitivity Guided-Mode-Resonance Optical Sensor Employing Phase Detection. Sci. Rep. 2017, 7, 7607. [Google Scholar] [CrossRef]
- Hsu, H.-Y.; Lan, Y.-H.; Huang, C.-S. A Gradient Grating Period Guided-Mode Resonance Spectrometer. IEEE Photonics J. 2018, 10, 1–9. [Google Scholar] [CrossRef]
- Tabassum, S.; Kumar, R.; Dong, L. Nanopatterned Optical Fiber Tip for Guided Mode Resonance and Application to Gas Sensing. IEEE Sens. J. 2017, 17, 7262–7272. [Google Scholar] [CrossRef]
- You, L.; Zha, D.; Anslyn, E.V. Recent Advances in Supramolecular Analytical Chemistry Using Optical Sensing. Chem. Rev. 2015, 115, 7840–7892. [Google Scholar] [CrossRef]
- Luchansky, M.S.; Bailey, R.C. High-Q Optical Sensors for Chemical and Biological Analysis. Anal. Chem. 2011, 84, 793–821. [Google Scholar] [CrossRef] [PubMed]
- Barth, I.; Conteduca, D.; Reardon, C.; Johnson, S.; Krauss, T.F. Common-Path Interferometric Label-Free Protein Sensing with Resonant Dielectric Nanostructures. Light Sci. Appl. 2020, 9, 96. [Google Scholar] [CrossRef] [PubMed]
- Joseph, S.; Rajpal, S.; Kar, D.; Devinder, S.; Pandey, S.; Mishra, P.; Joseph, J. Guided Mode Resonance Immunosensor for Label-Free Detection of Pathogenic Bacteria Pseudomonas Aeruginosa. Biosens. Bioelectron. 2023, 241, 115695. [Google Scholar] [CrossRef]
- Wawro, D.; Ding, Y.; Gimlin, S.; Zimmerman, S.; Kearney, C.; Pawlowski, K.; Magnusson, R. Guided-Mode Resonance Sensors for Rapid Medical Diagnostic Testing Applications. In Proceedings of the Optical Fibers and Sensors for Medical Diagnostics and Treatment Applications IX, San Jose, CA, USA, 24–25 January 2009; SPIE: Bellingham, WA, USA, 2009; Volume 7173, pp. 18–26. [Google Scholar] [CrossRef]
- Fang, Y.; Ferrie, A.M.; Fontaine, N.H.; Mauro, J.; Balakrishnan, J. Resonant Waveguide Grating Biosensor for Living Cell Sensing. Biophys. J. 2006, 91, 1925–1940. [Google Scholar] [CrossRef]
- Fang, Y.; Li, G.; Ferrie, A.M. Non-Invasive Optical Biosensor for Assaying Endogenous G Protein-Coupled Receptors in Adherent Cells. J. Pharmacol. Toxicol. Methods 2007, 55, 314–322. [Google Scholar] [CrossRef]
- Nezhadbadeh, S.; Neumann, A.; Zarkesh-Ha, P.; Brueck, S.R.J. Chirped-Grating Spectrometer-on-a-Chip. Opt. Express 2020, 28, 24501–24510. [Google Scholar] [CrossRef]
- Triggs, G.J.; Wang, Y.; Reardon, C.P.; Fischer, M.; Evans, G.J.O.; Krauss, T.F. Chirped Guided-Mode Resonance Biosensor. Optica 2017, 4, 229–234. [Google Scholar] [CrossRef]
- Morales, M.A.; Halpern, J.M. Guide to Selecting a Biorecognition Element for Biosensors. Bioconjug. Chem. 2018, 29, 3231–3239. [Google Scholar] [CrossRef]
- Zhou, Y.; Wang, B.; Guo, Z.; Wu, X. Guided Mode Resonance Sensors with Optimized Figure of Merit. Nanomaterials 2019, 9, 837. [Google Scholar] [CrossRef]
- Conteduca, D.; Arruda, G.S.; Barth, I.; Wang, Y.; Krauss, T.F.; Martins, E.R. Beyond Q: The Importance of the Resonance Amplitude for Photonic Sensors. ACS Photonics 2022, 9, 1757–1763. [Google Scholar] [CrossRef]
- Molaei-Yeznabad, A.; Bahador, H. Refractive Index-Based Optimized Ternary Photonic Crystal Biosensor for Ultra Precise Detection of Cancer Cells. Sens. Imaging 2025, 26, 36. [Google Scholar] [CrossRef]
- Juan-Colas, J.; Conteduca, D.; Barth, I.; Krauss, T.F. Guided Mode Resonances for Sensing and Imaging. EPJ Web Conf. 2019, 215, 11001. [Google Scholar] [CrossRef]
- Morris, K.; de Arruda, G.S.; Martins, A.; Coupe, S.T.; Martins, E.R.; Krauss, T.F.; Quinn, S.D. BPS2025-Ultrasensitive detection of Alzheimer’s disease biomarkers using a nanoparticle-enhanced resonant biosensor. Biophys. J. 2025, 124, 500a. [Google Scholar] [CrossRef]
- Kim, W.-J.; Kim, B.K.; Kim, A.; Huh, C.; Ah, C.S.; Kim, K.-H.; Hong, J.; Park, S.H.; Song, S.; Song, J.; et al. Response to Cardiac Markers in Human Serum Analyzed by Guided-Mode Resonance Biosensor. Anal. Chem. 2010, 82, 9686–9693. [Google Scholar] [CrossRef]
- Abdallah, M.G.; Buchanan-Vega, J.A.; Lee, K.J.; Wenner, B.R.; Allen, J.W.; Allen, M.S.; Gimlin, S.; Wawro Weidanz, D.; Magnusson, R. Quantification of Neuropeptide Y with Picomolar Sensitivity Enabled by Guided-Mode Resonance Biosensors. Sensors 2019, 20, 126. [Google Scholar] [CrossRef]
- Wawro, D.; Koulen, P.; Ding, Y.; Zimmerman, S.; Magnusson, R. Guided-Mode Resonance Sensor System for Early Detection of Ovarian Cancer. In Proceedings of the Optical Diagnostics and Sensing X: Toward Point-of-Care Diagnostics, San Francisco, CA, USA, 25–26 January 2010; SPIE: Bellingham, WA, USA, 2010; Volume 7572, pp. 85–90. [Google Scholar] [CrossRef]
- Tu, Y.-K.; Tsai, M.-Z.; Lee, I.-C.; Hsu, H.-Y.; Huang, C.-S. Integration of a Guided-Mode Resonance Filter with Microposts for in-Cell Protein Detection. Analyst 2016, 141, 4189–4195. [Google Scholar] [CrossRef]
- Lin, S.-F.; Ding, T.-J.; Liu, J.-T.; Lee, C.-C.; Yang, T.-H.; Chen, W.-Y.; Chang, J.-Y. A Guided Mode Resonance Aptasensor for Thrombin Detection. Sensors 2011, 11, 8953–8965. [Google Scholar] [CrossRef]
- Bakshi, S.; Sahoo, P.K.; Li, K.; Johnson, S.; Raxworthy, M.J.; Krauss, T.F. Nanophotonic and Hydrogel-Based Diagnostic System for the Monitoring of Chronic Wounds. Biosens. Bioelectron. 2023, 242, 115743. [Google Scholar] [CrossRef]
- Yang, J.-M.; Yang, N.-Z.; Chen, C.-H.; Huang, C.-S. Gradient Waveguide Thickness Guided-Mode Resonance Biosensor. Sensors 2021, 21, 376. [Google Scholar] [CrossRef]
- Lin, Y.-C.; Hsieh, W.-H.; Chau, L.-K.; Chang, G.-E. Intensity-Detection-Based Guided-Mode-Resonance Optofluidic Biosensing System for Rapid, Low-Cost, Label-Free Detection. Sens. Actuators B Chem. 2017, 250, 659–666. [Google Scholar] [CrossRef]
- Yeh, C.-T.; Barshilia, D.; Hsieh, C.-J.; Li, H.-Y.; Hsieh, W.-H.; Chang, G.-E. Rapid and Highly Sensitive Detection of C-Reaction Protein Using Robust Self-Compensated Guided-Mode Resonance Biosensing System for Point-of-Care Applications. Biosensors 2021, 11, 523. [Google Scholar] [CrossRef] [PubMed]
- Race, C.M.; Kwon, L.E.; Foreman, M.T.; Huang, Q.; Inan, H.; Kesiraju, S.; Le, P.; Lim, S.J.; Smith, A.M.; Zangar, R.C. An Automated Microfluidic Assay for Photonic Crystal Enhanced Detection and Analysis of an Antiviral Antibody Cancer Biomarker in Serum. IEEE Sens. J. 2017, 18, 1464–1473. [Google Scholar] [CrossRef] [PubMed]
- Du, Y.; Li, Z.; Li, L.; Chen, Z.G.; Sun, S.-Y.; Chen, P.; Shin, D.M.; Khuri, F.R.; Fu, H. Distinct Growth Factor-Induced Dynamic Mass Redistribution (DMR) Profiles for Monitoring Oncogenic Signaling Pathways in Various Cancer Cells. J. Recept. Signal Transduct. 2009, 29, 182–194. [Google Scholar] [CrossRef]
- Huang, C.-S.; George, S.; Lu, M.; Chaudhery, V.; Tan, R.; Zangar, R.C.; Cunningham, B.T. Application of Photonic Crystal Enhanced Fluorescence to Cancer Biomarker Microarrays. Anal. Chem. 2011, 83, 1425–1430. [Google Scholar] [CrossRef] [PubMed]
- George, S.; Chaudhery, V.; Lu, M.; Takagi, M.; Amro, N.; Pokhriyal, A.; Tan, Y.; Ferreira, P.; Cunningham, B.T. Sensitive Detection of Protein and miRNA Cancer Biomarkers Using Silicon-Based Photonic Crystals and a Resonance Coupling Laser Scanning Platform. Lab. Chip 2013, 13, 4053–4064. [Google Scholar] [CrossRef]
- Karawdeniya, B.I.; Damry, A.M.; Murugappan, K.; Manjunath, S.; Bandara, Y.M.N.D.Y.; Jackson, C.J.; Tricoli, A.; Neshev, D. Surface Functionalization and Texturing of Optical Metasurfaces for Sensing Applications. Chem. Rev. 2022, 122, 14990–15030. [Google Scholar] [CrossRef]
- Hosseini, S.; Ibrahim, F.; Djordjevic, I.; Koole, L.H. Recent Advances in Surface Functionalization Techniques on Polymethacrylate Materials for Optical Biosensor Applications. Analyst 2014, 139, 2933–2943. [Google Scholar] [CrossRef]
- Kaja, S.; Hilgenberg, J.D.; Collins, J.L.; Shah, A.A.; Wawro, D.; Zimmerman, S.; Magnusson, R.; Koulen, P. Detection of Novel Biomarkers for Ovarian Cancer with an Optical Nanotechnology Detection System Enabling Label-Free Diagnostics. J. Biomed. Opt. 2012, 17, 081412. [Google Scholar] [CrossRef]
- Cruceriu, D.; Baldasici, O.; Balacescu, O.; Berindan-Neagoe, I. The Dual Role of Tumor Necrosis Factor-Alpha (TNF-α) in Breast Cancer: Molecular Insights and Therapeutic Approaches. Cell. Oncol. 2020, 43, 1–18. [Google Scholar] [CrossRef]
- Knüpfer, H.; Preiß, R. Significance of Interleukin-6 (IL-6) in Breast Cancer (Review). Breast Cancer Res. Treat. 2007, 102, 129–135. [Google Scholar] [CrossRef]
- Chen, W.; Long, K.D.; Lu, M.; Chaudhery, V.; Yu, H.; Choi, J.S.; Polans, J.; Zhuo, Y.; Harley, B.A.; Cunningham, B.T. Photonic Crystal Enhanced Microscopy for Imaging of Live Cell Adhesion. Analyst 2013, 138, 5886–5894. [Google Scholar] [CrossRef] [PubMed]
- Poulikakos, L.V.; Lawrence, M.; Barton, D.R.; Jeffrey, S.S.; Dionne, J.A. Guided-Mode-Resonant Dielectric Metasurfaces for Colorimetric Imaging of Material Anisotropy in Fibrous Biological Tissue. ACS Photonics 2020, 7, 3216–3227. [Google Scholar] [CrossRef]
- Fang, Y.; Ferrie, A.M.; Fontaine, N.H.; Yuen, P.K. Characteristics of Dynamic Mass Redistribution of Epidermal Growth Factor Receptor Signaling in Living Cells Measured with Label-Free Optical Biosensors. Anal. Chem. 2005, 77, 5720–5725. [Google Scholar] [CrossRef]
- Zaytseva, N.; Lynn, J.G.; Wu, Q.; Mudaliar, D.J.; Sun, H.; Kuang, P.Q.; Fang, Y. Resonant Waveguide Grating Biosensor-Enabled Label-Free and Fluorescence Detection of Cell Adhesion. Sens. Actuators B Chem. 2013, 188, 1064–1072. [Google Scholar] [CrossRef]
- Chan, L.L.; Gosangari, S.L.; Watkin, K.L.; Cunningham, B.T. A Label-Free Photonic Crystal Biosensor Imaging Method for Detection of Cancer Cell Cytotoxicity and Proliferation. Apoptosis 2007, 12, 1061–1068. [Google Scholar] [CrossRef]
- Magnusson, R.; Wawro, D.; Zimmerman, S.; Ding, Y. Resonant Photonic Biosensors with Polarization-Based Multiparametric Discrimination in Each Channel. Sensors 2011, 11, 1476–1488. [Google Scholar] [CrossRef]
- Xiao, G.; Zhu, Q.; Shen, Y.; Li, K.; Liu, M.; Zhuang, Q.; Jin, C. A Tunable Submicro-Optofluidic Polymer Filter Based on Guided-Mode Resonance. Nanoscale 2015, 7, 3429–3434. [Google Scholar] [CrossRef]
- Tseng, Y.-T.; Chiu, Y.-C.; Pham, V.-D.; Wu, W.-H.; Le-Vu, T.T.; Wang, C.-H.; Kuo, S.-W.; Chan, M.W.Y.; Lin, C.-H.; Li, S.-C.; et al. Ultrasensitive Upconversion Nanoparticle Immunoassay for Human Serum Cardiac Troponin I Detection Achieved with Resonant Waveguide Grating. ACS Sens. 2024, 9, 455–463. [Google Scholar] [CrossRef]
- Wilson, D.H.; Rissin, D.M.; Kan, C.W.; Fournier, D.R.; Piech, T.; Campbell, T.G.; Meyer, R.E.; Fishburn, M.W.; Cabrera, C.; Patel, P.P.; et al. The Simoa HD-1 Analyzer: A Novel Fully Automated Digital Immunoassay Analyzer with Single-Molecule Sensitivity and Multiplexing. SLAS Technol. 2016, 21, 533–547. [Google Scholar] [CrossRef]
- Empana, J.-P.; Lerner, I.; Perier, M.-C.; Guibout, C.; Jabre, P.; Bailly, K.; Andrieu, M.; Climie, R.; van Sloten, T.; Vedie, B.; et al. Ultrasensitive Troponin I and Incident Cardiovascular Disease. Arterioscler. Thromb. Vasc. Biol. 2022, 42, 1471–1481. [Google Scholar] [CrossRef]
- Huang, L.; Jin, R.; Zhou, C.; Li, G.; Xu, L.; Overvig, A.; Deng, F.; Chen, X.; Lu, W.; Alù, A.; et al. Ultrahigh-Q Guided Mode Resonances in an All-Dielectric Metasurface. Nat. Commun. 2023, 14, 3433. [Google Scholar] [CrossRef] [PubMed]
- Li, C.-H.; Chang, C.-Y.; Chen, Y.-R.; Huang, C.-S. Blood Biomarker Detection Using Integrated Microfluidics with Optical Label-Free Biosensor. Sensors 2024, 24, 6756. [Google Scholar] [CrossRef] [PubMed]
- Triggs, G.J.; Fischer, M.; Stellinga, D.; Scullion, M.G.; Evans, G.J.O.; Krauss, T.F. Spatial Resolution and Refractive Index Contrast of Resonant Photonic Crystal Surfaces for Biosensing. IEEE Photonics J. 2015, 7, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.-S.; Kwak, J.Y.; Seong, T.-Y.; Hwang, G.W.; Kim, W.M.; Kim, I.; Lee, K.-S. Optimization of Tunable Guided-Mode Resonance Filter Based on Refractive Index Modulation of Graphene. Sci. Rep. 2019, 9, 19951. [Google Scholar] [CrossRef]
- Fallah, H.; Hakim, L.; Boonruang, S.; Mohammed, W.S.; Hsu, S.H. Polymer-Based Guided-Mode Resonance Sensors: From Optical Theories to Sensing Applications. ACS Appl. Polym. Mater. 2023, 5, 9700–9713. [Google Scholar] [CrossRef]
- Alam, M.Z.; Sun, X.; Mojahedi, M.; Aitchison, J.S. Augmented Low Index Waveguide for Confining Light in Low Index Media. Laser Photonics Rev. 2017, 11, 1500224. [Google Scholar] [CrossRef]
- Cu, D.T.; Wu, H.W.; Chen, H.P.; Su, L.C.; Kuo, C.C. Exploiting Thin-Film Properties and Guided-Mode Resonance for Designing Ultrahigh-Figure-of-Merit Refractive Index Sensors. Sensors 2024, 24, 960. [Google Scholar] [CrossRef]
- Ura, S.; Tsuji, R.; Inoue, J.; Kintaka, K. Multidimensional Angle Sensing Method Using Guided-Mode Resonance. Opt. Rev. 2021, 28, 650–654. [Google Scholar] [CrossRef]
- Kenaan, A.; Li, K.; Barth, I.; Johnson, S.; Song, J.; Krauss, T.F. Guided Mode Resonance Sensor for the Parallel Detection of Multiple Protein Biomarkers in Human Urine with High Sensitivity. Biosens. Bioelectron. 2020, 153, 112047. [Google Scholar] [CrossRef]
- Lue, J.-H.; Ding, T.-J.; Yang, T.-H.; Huang, H.-C.; Hsu, C.-L.; Liu, J.-T.; Chen, W.-Y.; Chang, J.-Y. Real-Time Monitoring DNA Hybridization by Guided Resonant Mode Biosensor. In Proceedings of the 2011 6th IEEE International Conference on Nano/Micro Engineered and Molecular Systems, Kaohsiung, Taiwan, 20–23 February 2011; IEEE: Piscataway, NJ, USA, 2011; pp. 1208–1211. [Google Scholar] [CrossRef]
- Drayton, A.; Li, K.; Simmons, M.; Reardon, C.; Krauss, T.F. Performance Limitations of Resonant Refractive Index Sensors with Low-Cost Components. Opt. Express 2020, 28, 32239–32248. [Google Scholar] [CrossRef]
- Juan-Colás, J.; Hitchcock, I.S.; Coles, M.; Johnson, S.; Krauss, T.F. Quantifying Single-Cell Secretion in Real Time Using Resonant Hyperspectral Imaging. Proc. Natl. Acad. Sci. USA 2018, 115, 13204–13209. [Google Scholar] [CrossRef] [PubMed]
- Gupta, G.; Kondoh, J. Tuning and Sensitivity Enhancement of Surface Plasmon Resonance Sensor. Sens. Actuators B Chem. 2007, 122, 381–388. [Google Scholar] [CrossRef]
- Özdemir, Ş.K.; Turhan-Sayan, G. Temperature Effects on Surface Plasmon Resonance: Design Considerations for an Optical Temperature Sensor. J. Light. Technol. 2003, 21, 805. [Google Scholar] [CrossRef]
- Tamayo, J.; Kosaka, P.M.; Ruz, J.J.; Paulo, Á.S.; Calleja, M. Biosensors Based on Nanomechanical Systems. Chem. Soc. Rev. 2013, 42, 1287–1311. [Google Scholar] [CrossRef]
- Li, K.; Suliali, N.J.; Sahoo, P.K.; Silver, C.D.; Davrandi, M.; Wright, K.; Reardon, C.; Johnson, S.D.; Krauss, T.F. Noise Tolerant Photonic Bowtie Grating Environmental Sensor. ACS Sens. 2024, 9, 1857–1865. [Google Scholar] [CrossRef]
- Kus-Liśkiewicz, M.; Fickers, P.; Ben Tahar, I. Biocompatibility and Cytotoxicity of Gold Nanoparticles: Recent Advances in Methodologies and Regulations. Int. J. Mol. Sci. 2021, 22, 10952. [Google Scholar] [CrossRef]
- Greulich, C.; Kittler, S.; Epple, M.; Muhr, G.; Köller, M. Studies on the Biocompatibility and the Interaction of Silver Nanoparticles with Human Mesenchymal Stem Cells (hMSCs). Langenbecks Arch. Surg. 2009, 394, 495–502. [Google Scholar] [CrossRef]
- Lichtenberg, J.Y.; Ling, Y.; Kim, S. Non-Specific Adsorption Reduction Methods in Biosensing. Sensors 2019, 19, 2488. [Google Scholar] [CrossRef]
- Salaluk, S.; Jiang, S.; Viyanit, E.; Rohwerder, M.; Landfester, K.; Crespy, D. Design of Nanostructured Protective Coatings with a Sensing Function. ACS Appl. Mater. Interfaces 2021, 13, 53046–53054. [Google Scholar] [CrossRef]
- Gambhir, M.; Gupta, S. Advanced Optimization Algorithms for Grating Based Sensors: A Comparative Analysis. Optik 2018, 164, 567–574. [Google Scholar] [CrossRef]
- Moghaddas, S.A.J.; Shahabadi, M.; Mohammad-Taheri, M. Guided Mode Resonance Sensor With Enhanced Surface Sensitivity Using Coupled Cross-Stacked Gratings. IEEE Sens. J. 2014, 14, 1216–1222. [Google Scholar] [CrossRef]
- AlAameri, H.M.H.; Shokooh-Saremi, M. Guided-Mode Resonance Sensors with High Sensitivity and Asymmetric Structures. J. Nanophotonics 2024, 18, 016005. [Google Scholar] [CrossRef]
- Finco, G.; Bideskan, M.Z.; Vertchenko, L.; Beliaev, L.Y.; Malureanu, R.; Lindvold, L.R.; Takayama, O.; Andersen, P.E.; Lavrinenko, A.V. Guided-Mode Resonance on Pedestal and Half-Buried High-Contrast Gratings for Biosensing Applications. Nanophotonics 2021, 10, 4289–4296. [Google Scholar] [CrossRef]
- Antoniou, M.; Tsounidi, D.; Petrou, P.S.; Beltsios, K.G.; Kakabakos, S.E. Functionalization of Silicon Dioxide and Silicon Nitride Surfaces with Aminosilanes for Optical Biosensing Applications. Med. Devices Sens. 2020, 3, e10072. [Google Scholar] [CrossRef]
- Hsiung, C.-T.; Huang, C.-S. Refractive Index Sensor Based on Gradient Waveguide Thickness Guided-Mode Resonance Filter. IEEE Sens. Lett. 2018, 2, 1–4. [Google Scholar] [CrossRef]
- Abdulhalim, I. Optimized Guided Mode Resonant Structure as Thermooptic Sensor and Liquid Crystal Tunable Filter. Chin. Opt. Lett. 2009, 7, 667–670. [Google Scholar] [CrossRef]
- Pandey, V.; Pal, S. Investigating the Performance of Metal-Assisted Guided Mode Resonance Based Structures for Biosensing Applications. IEEE Sens. J. 2019, 19, 4412–4418. [Google Scholar] [CrossRef]
- Chen, C.; Wang, J. Optical Biosensors: An Exhaustive and Comprehensive Review. Analyst 2020, 145, 1605–1628. [Google Scholar] [CrossRef]
- Chen, W.; Li, Z.; Cheng, W.; Wu, T.; Li, J.; Li, X.; Liu, L.; Bai, H.; Ding, S.; Li, X.; et al. Surface Plasmon Resonance Biosensor for Exosome Detection Based on Reformative Tyramine Signal Amplification Activated by Molecular Aptamer Beacon. J. Nanobiotechnol. 2021, 19, 450. [Google Scholar] [CrossRef]
- Homola, J. Surface Plasmon Resonance Sensors for Detection of Chemical and Biological Species. Chem. Rev. 2008, 108, 462–493. [Google Scholar] [CrossRef]
- Khodaie, A.; Heidarzadeh, H.; Harzand, F.V. Development of an Advanced Multimode Refractive Index Plasmonic Optical Sensor Utilizing Split Ring Resonators for Brain Cancer Cell Detection. Sci. Rep. 2025, 15, 433. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Li, Z.; Lin, Q.; Wei, Y.; Wang, J.; Li, Y.; Yang, R.; Yuan, Q. Highly Sensitive Detection of Bladder Cancer-Related miRNA in Urine Using Time-Gated Luminescent Biochip. ACS Sens. 2019, 4, 2124–2130. [Google Scholar] [CrossRef] [PubMed]
- Calvo-Lozano, O.; García-Aparicio, P.; Raduly, L.-Z.; Estévez, M.C.; Berindan-Neagoe, I.; Ferracin, M.; Lechuga, L.M. One-Step and Real-Time Detection of microRNA-21 in Human Samples for Lung Cancer Biosensing Diagnosis. Anal. Chem. 2022, 94, 14659–14665. [Google Scholar] [CrossRef]
- Kammer, M.N.; Kussrow, A.K.; Webster, R.L.; Chen, H.; Hoeksema, M.; Christenson, R.; Massion, P.P.; Bornhop, D.J. Compensated Interferometry Measures of CYFRA 21–1 Improve Diagnosis of Lung Cancer. ACS Comb. Sci. 2019, 21, 465–472. [Google Scholar] [CrossRef]
- Krauss, T.F.; Miller, L.; Wälti, C.; Johnson, S. Photonic and Electrochemical Biosensors for Near-Patient Tests–a Critical Comparison. Optica 2024, 11, 1408–1418. [Google Scholar] [CrossRef]
- Shen, R.; He, R.; Chen, L.; Guo, J. Inverse Design of Hybrid Metal-Dielectric Guided Mode Resonance Optical Filters with a Deep Learning Neural Network and Fano Function Matching. Opt. Mater. Express 2022, 12, 3600–3613. [Google Scholar] [CrossRef]
- Araki, K.; Zhang, R.Z. Resonant-Mode Metasurface Thermal Super Mirror by Deep Learning-Assisted Optimization Algorithms. J. Quant. Spectrosc. Radiat. Transf. 2024, 329, 109195. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).