1. Introduction
Precise and high-throughput analysis of DNA sequences is fundamental to numerous applications in molecular diagnostics, genotyping, and nucleic acid thermodynamics [
1,
2,
3,
4,
5,
6]. Advances in molecular diagnostics increasingly demand analytical platforms capable of performing a highly parallel, sensitive, and rapid detection of biomolecular interactions. Traditional fluorescence-based DNA analysis techniques, such as microarrays and quantitative PCR, have provided powerful means for identifying and quantifying nucleic acid sequences [
7,
8,
9,
10,
11,
12,
13]. However, these methods are often limited by low throughput due to serial signal acquisition and require precise and complex optical measurement setup. Conventional melting curve assays, such as fluorescence-based quantitative PCR and microarray hybridization for example, provide valuable information about sequence composition and hybridization stability, but they are often limited by the need for bulky optical systems, slow thermal cycling, and complex signal processing. These constraints hinder the rapid, parallel analysis of multiple DNA targets, which is increasingly demanded in clinical diagnostics and large-scale genomic studies.
To overcome these challenges, optical microcavity-based sensing and lasing approaches have emerged as promising alternatives, offering ultrahigh sensitivity and the potential for multiplexed biochemical analysis [
14,
15]. Optical microcavities offer an attractive platform for next-generation biomolecular sensing because of their ability to confine light, enhance light–matter interactions, and transduce subtle molecular changes into large measurable optical signals [
16,
17,
18,
19,
20,
21,
22]. Among various cavity designs, Fabry–Perot (FP) resonators provide straightforward fabrication and precise spectral control, enabling compact and scalable architectures for bioassays. In particular, plano-concave FP (PC-FP) microcavity is capable of sustaining stable lasing modes with high-quality (Q) factors and small mode volumes, facilitating the sensitive detection of DNA hybridization.
In this work, we demonstrate a parallel DNA sequence analysis platform based on an array of PC-FP microcavity lasers. By functionalizing the cavity surfaces with sequence-specific DNA probes and monitoring the lasing emission spectra under controlled temperature gradients, our system enables the simultaneous detection of multiple targets with high throughput. This approach combines the benefits of cavity-enhanced light–matter interaction and arrayed multiplexing, offering a compact and scalable solution for rapid nucleic acid characterization. While existing laser-based detection systems [
16,
17,
19,
20,
21,
22] revolutionized analysis time by completing DNA detection within mere milliseconds, they had limitations in simultaneous analysis/detection (e.g., utilizing a single-ring resonator laser cavity). In contrast, our novel platform further enables array-based multiplexed analysis. The results demonstrate that microcavity-based laser arrays are capable of providing a powerful alternative to conventional fluorescence assays, paving the way toward integrated, high-throughput genetic analysis and real-time molecular diagnostics.
2. Materials and Methods
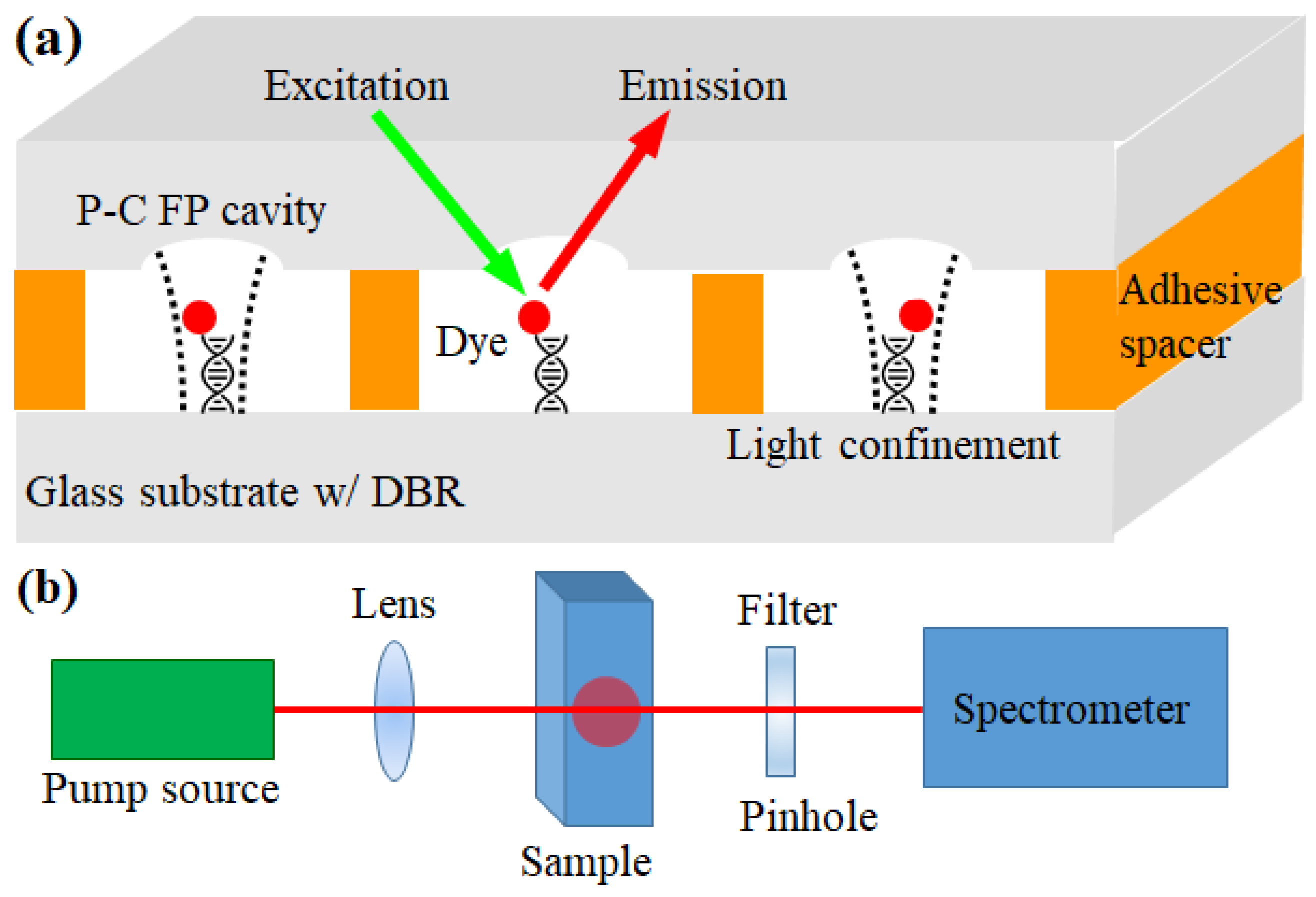
The PC-FP microcavity was fabricated on a glass substrate based on a previously studied microwell fabrication technique [
23,
24]. Using the CO
2 laser (RF-pumped, Firestar vi30, Synrad, Mukilteo, WA, USA, pulse repetition rate = 20 kHz) ablation method, a series of microwells was produced on the polished surface of fused silica glass. The previously characterized power and pulse duration (power of ~0.9 W and 30 ms pulse duration time) of the CO
2 laser ablation results in a microwell that is 3 µm in depth, 40 µm in diameter, and 200 µm in curvature radius. Three different microwells were produced and they were separated by 5 mm on the glass substrate.
A dielectric mirror stack was then deposited on both substrates (w/ and w/o microwells) to form the PC-FP cavity. The mirror consisted of a high-reflectivity distributed Bragg reflector (DBR) composed of alternating SiO2/TiO2 layers (15 pairs, reflectivity > 99% at the center wavelength of 610 nm) deposited on the substrate. The resulting mirror has relatively low reflectance to the light outside of the band to make sure labeling dyes can be optically pumped from outside. The top and bottom (flat) substrates were then assembled using a 5-micron-thick adhesive spacer. Silica beads with a diameter of 5 µm were well dispersed in the NOA 81 adhesive (Norland, Cranbury, NJ, USA), and the adhesive was cured with UV light. The spacer was used to not only assemble the two glass substrates but also simultaneously seal a fluid channel in the middle, enabling the injection and withdrawal of liquid samples.
Prior to assembling the PC-FP cavities, a patterning process was performed to functionalize each PC-FP cavity with distinct DNA probes for the parallel analysis of different DNA samples. First, one-dimensional microchannels with open tops were formed from polydimethylsiloxane (PDMS) on a flat-bottom substrate using photolithography followed by lift-off technology. Note that the flat side was functionalized, not the microwell side, to ensure the uniform immobilization of the DNA probes during the process. The PDMS fluidic channel has a width of 200 µm and a height of 50 µm. To immobilize DNA probe molecules on the bottom surface of each channel, the glass substrate surface was functionalized with bis(sulfosuccinimidyl)-succinimidyl ester (BS3), a homofunctional amine–amine crosslinker. BS3 was dissolved in phosphate-buffered saline (PBS) at a concentration of 0.1 mg/mL, and the substrate with PDMS channels was immersed in the solution for 30 min. The substrate was washed with PBS. The functionalized substrate surface was then incubated with streptavidin by immersing it in a 1 mg/mL streptavidin/PBS solution for 30 min. The streptavidin-functionalized glass substrate was then washed sequentially with PBS, DI water, and Tris-acetate-EDTA (TAE)/12.5 mM MgCl2 buffer solution.
Single-stranded DNA (ssDNA) molecules serving as the probe DNA were 21 nucleotides long. Three different randomized sequences were prepared as shown in
Table 1, and they were biotinylated at the 5′ end. All ssDNA molecules used in the experiment were purchased from Integrated DNA Technologies, and the probe DNA was dissolved in TAE/MgCl
2 buffer at a concentration of 1 μM. Three different solutions were carefully introduced into each PDMS channels. Through this process, the probe DNA molecules were immobilized at their respective positions via streptavidin–biotin bonds, after which the PDMS channels were removed. Channels carrying different probe DNAs were assembled in precise alignment with the upper concave substrate, completing each PC-FP cavity as described earlier. The complete device allowed for simultaneous temperature control and optical monitoring across multiple channels.
The cavity array was pumped using a pump laser light source (Continuum, Santa Clara, CA, USA, optical parametric oscillator, 518 nm wavelength, 5 ns pulse width, and 20 Hz repetition rate) through an optical setup, as illustrated in
Figure 1b. The emitted light was collected through a filter and pinhole, rejecting the optical pump light, and directed to a spectrometer (HR550i, Horiba Jobin Yvon) equipped with a cooled CCD detector.
3. Results
First, we evaluated the performance of our system in detecting target DNA molecules using only a single PC-FP cavity. We labeled a single-stranded DNA molecule that had a complementary base pair to the DNA 1 probe sequence in
Table 1 with Cy3 dye. The target DNA was prepared in TAE/MgCl
2 buffer solution at a concentration of 1 μM and injected it into the fluid channel of the PC-FP cavity via capillary action. After a 5 min hybridization process, buffer solution was injected into the fluid channel using external pressure. Sufficient volume was injected to remove unbound DNA and dye samples, which was confirmed via fluorescence. Subsequently, the PC-FP cavity was optically pumped using an external optical pump, following the optical experimental procedure described earlier.
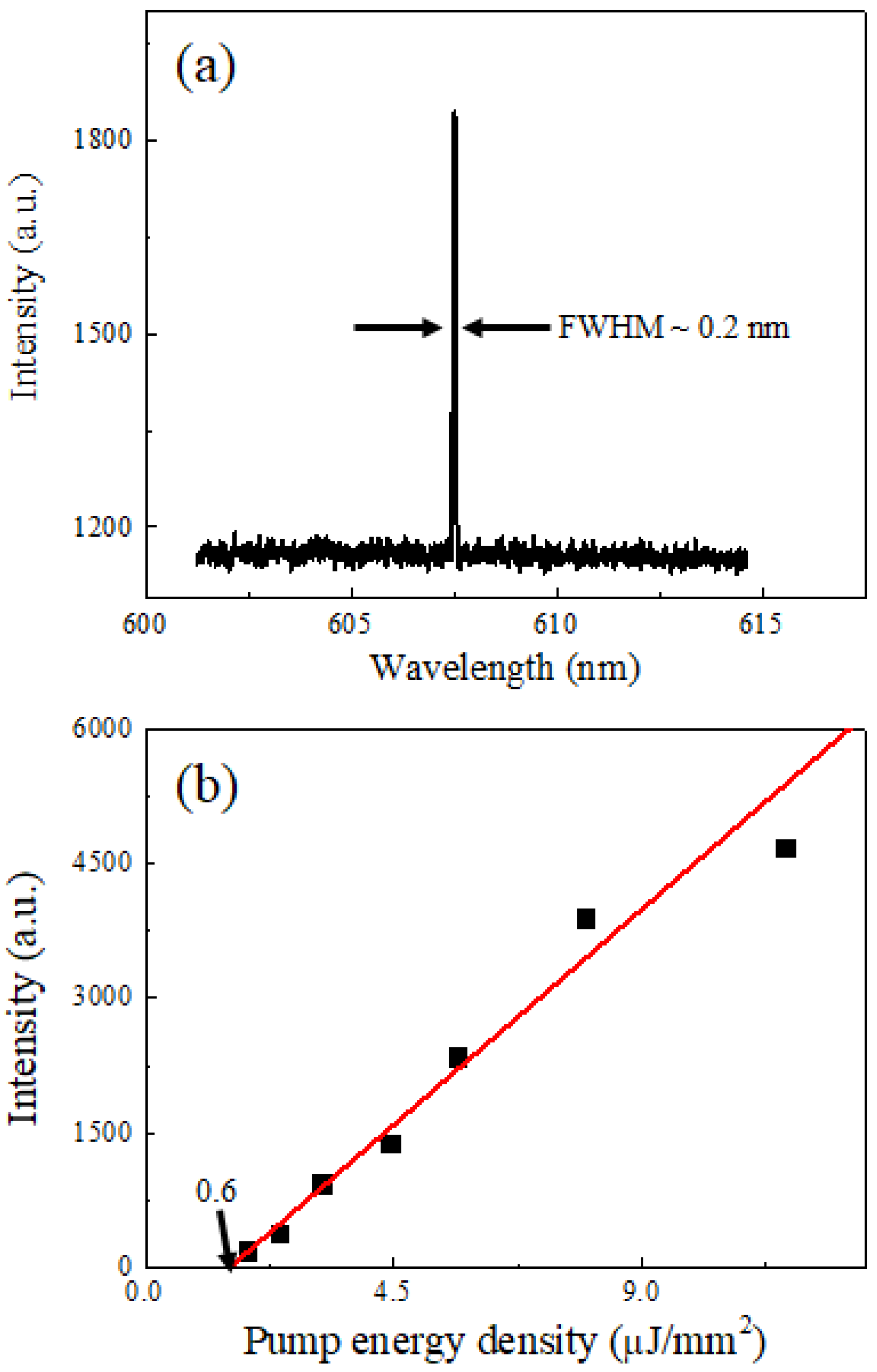
Figure 2 reveals the lasing characteristics of the PC-FP cavity with hybridized target DNA molecules under optical pumping. When the microcavity was excited with a pump energy density of 4.5 µJ/mm
2, a distinct and narrow emission peak appeared at a wavelength of approximately 607 nm, as shown in
Figure 2a. The sharp linewidth and high intensity of the peak indicate the onset of stimulated emission within the PC-FP microcavity. Below the lasing threshold, the emission spectrum was broad and weak, corresponding to spontaneous fluorescence from the Cy3 dye molecules. As the pump energy increased beyond the threshold, the emission intensity rose sharply, and the spectral linewidth collapsed to less than 0.2 nm, confirming laser oscillation.
The spectrally integrated emission intensity as a function of pump energy density is plotted in
Figure 2b. The data exhibit a clear nonlinear increase in intensity, typical of lasing behavior. A threshold pump energy density of approximately 0.6 µJ/mm
2 was determined from the intersection of linear fits to the spontaneous and stimulated emission regions. Above this threshold, the emission intensity increased linearly with pump energy density, following the trend shown by the red fitting line. These results confirm the formation of a coherent laser emission originating from the Cy3-labeled target DNA within the microcavity structure.
The target DNA hybridized with the probe and exhibited the distinct laser emission pattern shown above. The same experiment was repeated using a sample labeled with Cy-3 dye that shared the same 21-base sequence but contained a single-base mismatch in the middle. In this case, under identical conditions and pump density, virtually zero optical signals were observed, differing from
Figure 2a. This confirms that our DNA detection system using the PC-FP cavity laser was capable of detecting the target DNA molecules with single-base mismatch precision only with the single-laser oscillation. In this experiment, to demonstrate DNA detection at the fastest possible speed, we analyzed DNA molecules with 21-base sequences that could be analyzed/detected at room temperature. However, if we investigate laser emission at higher temperatures, we can also analyze longer DNA molecules with more base sequences (previous studies have shown that DNA with up to 100 base sequences can be detected with single-base mismatch accuracy).
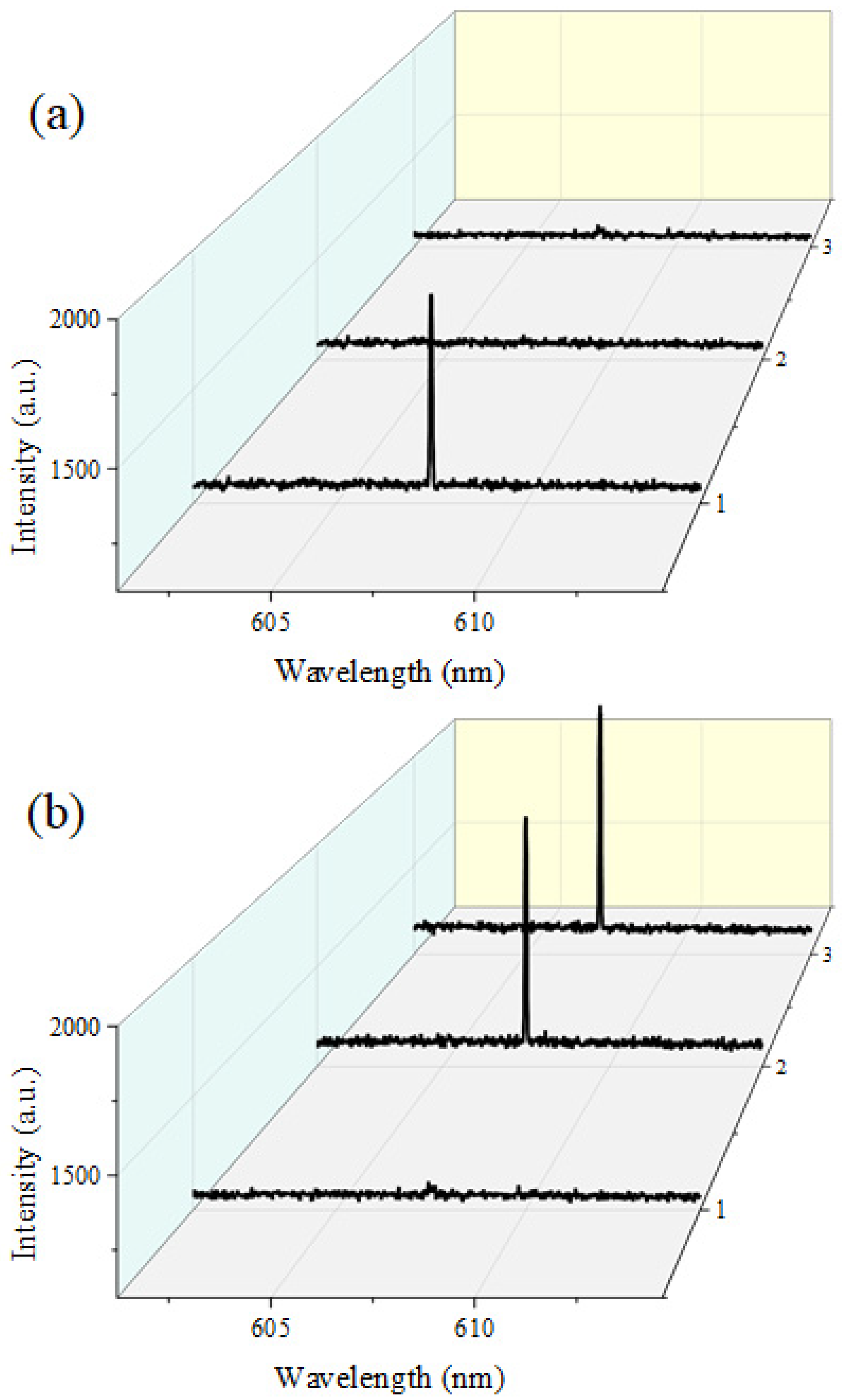
Figure 3 depicts the simultaneous laser emission spectra obtained from three independent PC-FP microcavities integrated within our parallel DNA analysis platform. Each cavity was functionalized with a different probe DNA sequence (DNA 1–3) as described earlier, allowing selective optical response depending on the hybridization state of the corresponding target DNA.
In
Figure 3a, only the target DNA complementary to probe DNA 1 was introduced into the channel. As a result, a lasing emission was observed exclusively from PC-FP cavity 1, while cavities 2 and 3 revealed virtually zero optical signals without any spectral peaks. The single narrow emission peak near 607 nm from PC-FP cavity 1 confirms that lasing occurs solely in the cavity where hybridization between the probe and target DNA took place, demonstrating the molecular selectivity of the detection scheme.
In contrast,
Figure 3b illustrates the spectral response when target DNA 2 and 3 were simultaneously introduced into the same microfluidic channel. In this case, laser peaks emerged from PC-FP cavities 2 and 3, corresponding to the binding of their respective targets, whereas cavity 1 revealed no lasing. The clear one-to-one correspondence between the specific DNA targets and the lasing cavities verifies that each microcavity operates as an independent molecular detection unit. The laser wavelengths measured from each cavity and different target all exhibited deviations within 0.5 nm of 607 nm and showed laser output intensities within the error margin at the same pump density. The same statistical values were measured in PC-FP arrays fabricated under identical conditions on other substrates, enabling reliable laser characteristics to be obtained under the same experimental conditions. Although our system detects DNA targets based solely on the presence or absence of laser oscillation, independent of characteristics like laser wavelength, this consistency within the laser array demonstrates that it can guarantee the reliability of our detection system.
These results confirm that the proposed PC-FP laser array enables parallel, sequence-specific DNA analysis within a single optical platform. The ability to resolve discrete lasing spectra from multiple cavities provides a direct and unambiguous digital readout for multiplexed biomolecular identification. However, at this point, not only our PC-FP cavity but also the flat mirror-to-mirror FP cavity can function as a laser microcavity, potentially causing mutual interference between laser oscillation signals. To clarify that this is not problematic to our platform, we performed the same laser oscillation experiments using an FP cavity without microwells. In this case, compared to the PC-FP cavity, it exhibited a laser threshold difference exceeding the order of magnitude. This demonstrated that under appropriately controlled experimental conditions, our PC-FP laser array-based DNA detection platform does not need to worry about interference from laser signals in flat mirror parts of our system. Moreover, our laser utilizing a PC-FP cavity exhibits tolerance to misalignment, unlike flat FPs where even minor substrate misalignment adversely affects the Q-factor, causing a rapid deterioration in laser characteristics. This prevents optical misalignment failures that commonly accompany detection systems using fluidic channels, thereby providing a significantly more reliable DNA detection platform. Therefore, we demonstrated that high-throughput DNA molecular recognition can be performed in parallel by simultaneously pumping as many PC-FP lasers as the external optical pump allows and identifying the cavities within the array where lasers oscillate.
4. Conclusions
The experimental results demonstrate that the PC-FP microcavity provides a robust optical environment for highly sensitive DNA detection and multiplexed analysis. The clear lasing threshold and narrow linewidth observed from the Cy3-labeled complementary targets confirm that hybridization events can be transduced into binary optical responses, functioning effectively as a “digital” readout analogous to previous optofluidic DNA lasers. The tenfold reduction in threshold compared to a flat FP cavity highlights the importance of geometric mode confinement, which minimizes diffraction loss and enhances intracavity photon density.
The parallel lasing operation from three independent cavities (
Figure 3) confirms that the platform can perform multi-target analysis within a single microfluidic domain without spectral interference. Such spatial multiplexing could be further scaled by the lithographic patterning of larger PC-FP arrays or by combining it with temperature-gradient control to perform melting analysis in parallel. The single-base mismatch discrimination further verifies the cavity’s sensitivity to molecular hybridization energetics, making it suitable for SNP genotyping or kinetic melting studies.
Compared with fluorescence microarrays and qPCR, this solid-state PC-FP cavity array eliminates the need for bulky optics and scanning systems while providing real-time, threshold-based molecular identification. Future improvements—such as integration with CMOS photodiodes, microheaters, and wavelength-multiplexed pump sources—could extend the platform toward fully chip-scale, high-throughput nucleic acid diagnostics.