Abstract
Brain tumors arise from abnormal cell growth in the brain. Glioblastoma, the most common and aggressive type, poses significant challenges for identification during surgery. The primary goal of this study is to identify and differentiate normal brain tissue from glioblastoma tissue using optical coherence tomography (OCT) and OCT angiography (OCTA). These techniques offer a non-invasive way to analyze the morphological and microvascular alternations associated with glioblastoma in an animal model. To monitor the changes in morphology and vascular distribution of brain tissue as glioblastoma tumors grow, time-series OCT and OCTA results were collected for comparison. Texture analysis of OCT images was proposed using the gray-level co-occurrence matrix (GLCM), from which homogeneity and variance were calculated as discriminative parameters. Additionally, OCTA was used to assess microvascular characteristics, including vessel diameter, density, and fractal dimension. The findings demonstrate that the proposed methods can effectively distinguish between normal and cancerous brain tissue in vivo.
1. Introduction
Brain tumors are masses of abnormal cells within the brain. They are classified by cell type, location, grade, and distinct features. Brain tumors may be primary, originating from the brain itself, or metastatic, where cancer cells spread from other parts of the body to the brain through the bloodstream. Metastatic tumors are generally more prevalent than primary brain tumors [1,2]. Depending on the severity and behavior of the tumor, it can be categorized as benign or malignant. Tumors are graded on a scale from 1 to 4, ranging from slow-growing to aggressively reproducing. Benign tumors, although slow-growing and non-invasive, can still cause long-term problems if they become large enough to exert pressure on surrounding tissues. In contrast, malignant tumors grow rapidly and invade nearby cells, disrupting the normal function of the brain regions they infiltrate [3,4]. Glioblastoma (GBM), classified as Grade 4, falls under the category of gliomas, glioneuronal, and neuronal tumors within the family of adult-type diffuse gliomas. The GBM subtype is particularly aggressive due to its rapid and uncontrolled proliferation and high resistance to therapies [5]. These tumors often disrupt the vascular network of the brain, leading to increased blood supply demands and capillary leakage, which may cause fluid accumulation in the brain. Such invasive growth can damage brain function, causing symptoms, like vision loss, seizures, and cognitive issues [6].
Brain tumors present as a diverse and complex array of diseases with variations in type, location, and severity, which significantly challenges detection. In the past few decades, advancements in neuroimaging, molecular diagnostics, and biotechnology have provided a sophisticated range of techniques for the diagnosis of brain tumors. The most commonly used imaging modalities at the preoperative stage are computed tomography (CT) and magnetic resonance imaging (MRI) [7,8]. These non-invasive techniques yield detailed structural images of the brain, facilitating the identification of a tumor’s presence, location, and size to allow diagnosis and operation planning. Furthermore, advanced imaging methods, such as functional MRI, positron emission tomography (PET), and magnetic resonance spectroscopy (MRS), can offer additional insights into a tumor’s metabolic activity, blood flow, and chemical composition—factors that are crucial in the diagnostic process [9,10,11]. During surgery, the brain may shift unpredictably due to certain factors, such as the opening of the skull, tissue removal, or gravity, leading to significant positional changes [12]. Hence, intraoperative techniques are crucial. For instance, intraoperative MRI (iMRI) provides live imaging to aid surgeons during procedures, but the setup is costly and prolongs both preparation and operation times [13]. Intraoperative ultrasound (ioUS) offers a more affordable option; however, it must overcome artifact issues caused by the skull–soft tissue interface and the presence of gas bubbles in brain fluid [14]. Intraoperative computed tomography (iCT) can provide real-time feedback, verify tumor resection, and detect bleeding, but it carries the risk of radiation exposure [15]. Fluorescent-guided techniques use fluorescent agents to visualize the tumor area. While this method can address the challenges posed by brain shift, it cannot detect low-grade gliomas, and tumor tissues may exist outside the fluorescent areas [16]. In general, these techniques are constrained by imaging resolution limits, making it challenging to accurately delineate small tumor regions or define the margins between normal and cancerous tissues.
Various methods have been proposed for the treatment of brain tumors, such as resection, radiotherapy, and chemotherapy. Radiotherapy utilizes high-energy particles, often from nuclear decay reactions, to damage the DNA of tumor cells, which helps to shrink tumors and prevent their recurrence [17]. Chemotherapy employs drugs to kill or debilitate cancer cells and can be used in conjunction with radiotherapy [18]. Resection surgery is the most common treatment to entail the physical excision of the brain tumor, either partially or completely [19]. Since glial tumors are infiltrative, accurately determining tumor margins during surgery is particularly challenging [20]. Incomplete tumor removal can lead to recurrence, while inaccurate differentiation risks damaging the neurological functions of healthy brain tissues. Moreover, post-treatment survival and recovery rates are inversely related to the extent of resection and residual tumor volume [21]. Therefore, a non-invasive imaging method is required for real-time differentiation between normal and tumorous brain tissue during resection surgery.
Optical coherence tomography (OCT) is a non-invasive imaging technique that reconstructs the structure of biological tissue according to the backscattered signals received from the sample [22]. As it solely relies on these optical backscattered signals, no additional contrast agents or fluorescent dyes are necessary for imaging. OCT can deliver imaging resolutions at a micrometer scale along both depth and transverse directions with an imaging depth range of 1–2 mm [23]. Thanks to advancements in Fourier-domain OCT, it is possible to capture the 3D microstructure of samples in vivo, and its use has been demonstrated in various biomedical applications, such as retina, cornea, skin, and cardiac diseases [24,25,26,27]. In addition to the non-invasive reconstruction of 3D sample structures, a multitude of OCT functionalities, such as elastography, polarization, and angiography, have been extensively developed [28,29,30,31]. For vascular imaging with OCT, different methods have been proposed, including speckle variance, correlation mapping, optical microangiography, and intensity variation [32,33,34,35]. For brain tumor diagnosis, previous reports have demonstrated OCT’s ability to differentiate between normal tissues, tumor tissues, and necrosis, thus enabling the detection of residual tumor tissues [36,37,38]. Furthermore, polarization-sensitive OCT, a functional extension, allows the retrieval of not only the backscattered intensity but also the polarization properties of biological tissues [39,40,41]. Previous studies have also confirmed that high-resolution OCT, which includes full-field OCT and μ-OCT, can further identify tumor types and grades [42,43]. As an alternative strategy to direct tumor identification over normal tissues, some research focuses on studying tumor-associated macrophages that are generated during cancer progression. Benjamin J. Vakoc et al. showcase a pioneering study using OCTA for tumor analysis. With OCTA, the tumor can be quantitatively evaluated in terms of tumor volume, vessel length, vessel diameter, tortuosity, and other metrics [44]. Yingting Pan et al. proposed the use of contrast-enhanced optical Doppler tomography with intralipid to visualize 3D images of capillary networks within a brain tumor. Additionally, the cerebral blood flow velocity wase quantitatively assessed [45]. Hubert Dolezyczek et al. utilized optical coherence microscopy (OCM) to study glioblastoma development in a mouse model. In their study, brain tumors were scanned with OCM on Days 1, 3, 11, and 14 following tumor cell injection, and several vascular parameters were also estimated to compare the control and tumor groups, including vessel area, total vessel length total number of junctions, and total number of endpoints. Until Day 11 after tumor cell injection, the control and tumor groups could be distinguished based on these four parameters [46]. Furthermore, Nitesh Katta et al. employed OCT as a guiding tool for laser surgery on brain tumors. After laser coagulation or ablation, the coagulated or ablated area could be identified from OCTA images [47].
In this study, we employed a swept-source OCT system to examine changes in optical properties and vascular patterns during brain tumor progression. We conducted serial scans of the brain tumor, comparing the optical homogeneity and intensity variations between normal and cancerous tissues. Moreover, tumor-induced angiogenesis is investigated, assessing parameters, such as vessel diameter, vessel density, and fractal dimension. These optical and vascular assessments allowed for the mapping of tumor distribution. Additionally, the criteria established for recognizing cancerous tissue can potentially aid in the precise targeting and effectiveness assessment of treatments applied to the brain tumor.
2. Materials and Methods
2.1. Setup of Swept-Source OCT
To non-invasively investigate the features of a brain tumor, a swept-source OCT system was developed with a center wavelength of 1060 nm, as shown in Figure 1. This OCT system, based on a Michelson interferometer, is comprised of a fiber coupler with a 50/50 coupling ratio (TW1064R5A2B, Thorlabs, Newton, NJ, USA). The optical beam of the light source (Axsun, EXCELITAS Technologies, Waltham, MA, USA) is divided into the reference and sample arms. Within the sample arm, the light beam is collimated on a two-axis galvanometer, then focused on the sample by a scan lens (LSM02-BB, Thorlabs, Newton, NJ, USA). The signal, which is backscattered or reflected from both arms, is detected by a balanced detector (PDB481C-AC, Thorlabs, Newton, NJ, USA) with the other 50/50 fiber coupler. The axial and transverse resolutions are 7 and 10 μm, respectively. Due to the scan rate of the light source reaching 100 kHz, the frame rate can achieve 100 frames/s. A 3D scan covers a physical area of 4 mm × 4 mm.
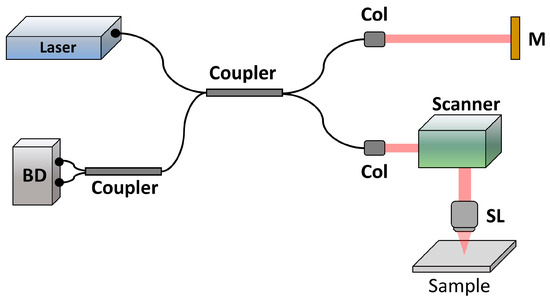
Figure 1.
Setup of swept-source OCT for animal experiments. Col: collimator, M: mirror, SL: scan lens, and BD: balanced detector.
2.2. Animal Preparation
In this study, 6-week-old C57BL/6 mice were used, and a chronic cranial window procedure was performed to facilitate long-term imaging of brain tissue. Under anesthesia, a small portion of the skull was carefully removed using a dental drill, and Glioma261 cells were subsequently introduced into the brain tissue using a fine needle attached to a microsyringe. After the injection, the skull opening was sealed to maintain structural integrity. To promote cell growth, the integrity of the skull was preserved for seven days following the tumor cells’ implantation. On the seventh day post-implantation, a transparent window with a 4 mm diameter was created using a dental drill to enable precise OCT scanning. The opening was then sealed with a coverslip to preserve the imaging environment. This protocol was approved by the Institutional Animal Care and Use Committee of Chang Gung University (IACUC Approval No.: CGU108-140). The cranial window technique allowed for real-time monitoring of tumor evolution. During imaging sessions, mice were immobilized using a custom-designed, 3D-printed restraint system to minimize motion artifacts due to respiration. This methodological approach represents a significant advancement in preclinical glioma studies, providing deeper insights into the complex dynamics of this aggressive cancer.
In this study, a control group (N = 3) and a tumor group (N = 7) are included. In the tumor group, two mice were repeatedly scanned with OCT for 7 days, and the remaining five were scanned for 14 days. For the control group, animals were fitted with a transparent window, but no tumor cells were injected. The window region was then scanned. In our experiments, multiple 3D images of the same window region for each measurement were recorded to ensure comprehensive data collection. Each 3D volume comprised 1000 × 1000 pixels, covering an area of 4 mm × 4 mm, which means that each B-scan consisted of 1000 A-scans, and a 3D volume contained 1000 B-scans. For further analysis, 500 B-scans from the central region of each 3D dataset were used to analyze homogeneity and variance. For angiographic analysis, projection-view OCTA results were employed to evaluate vessel diameter, vessel density, and fractal dimension. To investigate changes in these vascular parameters, regions near the tumor implantation and adjacent areas were selected for comparative analysis. Finally, the results from five mice were averaged to calculate the mean value and standard deviation.
2.3. Analytic Method
OCT is predicated on the capture of backscattered signals from a sample, which facilitates the reconstruction of its depth-resolved structure. This backscattered signal is intrinsically linked to the tissue architecture, which allows for differentiation between normal and pathological states. To enhance the specificity of this differentiation, texture features from OCT images are analyzed using the gray-level co-occurrence matrix (GLCM) [48,49]. GLCM is a statistical tool used for examining the texture of an image. It analyzes the spatial relationship of pixels at specific distances and angles and records how frequently pairs of pixels with specific values (gray-levels) occur in an image. A GLCM is a matrix where the number of rows and columns is equal to the number of gray levels in the image. Using GLCMs, several texture features can be calculated, such as contrast, correlation, energy, and homogeneity. These features help in various applications, such as image analysis, pattern recognition, and machine vision to describe the texture of the underlying image. Within this study, two significant parameters have been focused on, namely homogeneity and variance, which serve to quantify the distinctions in backscattering properties that emerge during the progression of a brain tumor. The GLCM method analyzes patterns of spatial relationships of pixel intensities, effectively capturing the texture by examining the distribution of recurring local variations in intensity across a specified neighborhood. In the context of OCT images, these patterns or textures can be indicative of the organizational structure of the tissue, which changes in disease states, such as tumorigenesis. The equations for the estimation of homogeneity (HOM) and variance (VAR) can be expressed as follows:
where N is the number of gray-levels in each B-scan. i and j are the gray levels in GLCM, G(i,j). Homogeneity reflects the closeness of the distribution of elements in the GLCM to the GLCM diagonal and is a measure of the textural uniformity. Variance measures the dispersion of the GLCM elements and is indicative of the textural contrast. The mathematical estimation of these parameters provides a quantitative basis for assessing tissue structure at the microscopic level, which can be crucial for the accurate identification of cancerous changes within the brain. Additionally, tumor growth is accompanied by angiogenesis to provide oxygen and nutrition for tumor cells. Thus, tumor angiography can be beneficial to investigate the vascular changes due to tumors. In this study, OCTA is performed according to the estimated speckle variance. Moving particles in biological tissue, such as red blood cells, result in a larger variation in backscattered intensity. In contrast, the variation in backscattered intensity of the static structure of biological tissue is much smaller. Therefore, the vessels can be acquired by estimation of the speckle variance [50]. The estimation of the speckle variance, SV, can be expressed as follows:
where m is the total number of B-scans for the estimation of the speckle variance. Ijik is the spatial distribution of intensity. The principle underlying this technique is that the backscattered intensity of light from moving particles exhibits greater temporal variance compared to the more uniform scattering from stationary tissues. By quantifying this variance, the OCTA can generate detailed images of the vascular network within the biological tissue. High speckle variance corresponds to areas with active blood flow, thereby enabling the mapping of the tumor vasculature. Such imaging is critical for understanding the angiogenic process within tumors and may offer insights into the efficacy of anti-angiogenic therapies. In this study, the algorithm for estimating vessel diameter is similar to that described in the previous report, which includes filtering, binarization, and skeletonization [51]. To reduce the background noise for further analysis, an enhancement filter was employed, utilizing multi-scale Gaussian filtering in conjunction with Hessian matrix analysis. This technique effectively highlights tubular structures, thereby enhancing vessel detection against the noisy background [52]. Vessel density refers to the quantification of blood vessel density within a particular area of tissue. In this study, vessel density is expressed as a percentage and represents the proportion of the selected area occupied by blood vessels. It can be estimated after the process of skeletonization. This quantitative measure can be useful in assessing vascular health, monitoring disease progression, and evaluating the effectiveness of treatments in various ocular and systemic conditions. The fractal dimension is a quantitative measurement used to characterize the complexity and irregularity of vascular networks within biological tissues. Blood vessels form intricate patterns that often exhibit self-similar or repeating structures across different spatial scales. The fractal dimension is a mathematical parameter that quantifies the degree of complexity and self-similarity of these vascular networks. A higher fractal dimension indicates a more complex and irregular vascular pattern, while a lower fractal dimension suggests a simpler and more ordered arrangement of blood vessels.
3. Results
Figure 2 delineates the progression of tissue changes from Day 1 to Day 5 after craniectomy, as captured by OCT and OCTA. The red arrow in Figure 2a represents the surface of the coverslip to provide a transparent window for imaging. On Days 1 and 2, the backscattered intensity across the brain tissue appeared uniform, with no discernible differences between regions. From Day 3 to Day 5, an increase in the heterogeneity of the backscattered intensity was observed, probably indicative of the contrast between normal and tumor cells. The tumor tissue presented with a heightened backscattered intensity compared to normal tissue, suggesting a potential biomarker for distinguishing healthy versus tumorous regions. Some intensity clots, as indicated by the white arrows, can be found, as shown, in Figure 2c,d. It can be noted that tumor tissue caused stronger backscattered intensity than that of normal tissue. Additionally, corresponding OCT angiography results are shown in Figure 2f–j. On Day 1, the vasculature is be well-defined, with clear vessel boundaries and branching patterns evident. On Day 3, a blurring of vessel boundaries can be noted, alongside an increase in vessel diameter. By Days 4 and 5, the vascular architecture began to exhibit signs of angiogenesis, with the emergence of a fine and mesh-like vessel network, as highlighted by the yellow arrows in Figure 2j.

Figure 2.
(a–e) 2D OCT images of tumorous brain obtained on Day 1 to Day 5 after removal of the skull section. (f–j) Corresponding OCT angiography obtained on Day 1 to Day 5 after removal of the skull section. The red arrow indicates the surface of the coverslip, and the white arrows represent the intensity clots. The yellow arrows show the emerged mesh-like vessel network. The red lines in (f–j) indicate the corresponding locations of (a–e).
To further delve into the structural and vascular evolution associated with brain tumor growth, OCT imaging was performed daily for a period of 14 days following the craniectomy. The series of 2D OCT images charted in Figure 3 illustrates the progressive changes from Day 1 to Day 14 after the removal of the skull section. On Day 1, the scattering intensity throughout the brain region was largely homogenous. Bone meal used for the fixation of the coverslip is indicated by the red arrow, which marks the start of the observational period. In Figure 3b–e, a gradual shift towards an ununiform scattering pattern was observed, likely signaling the growth and increasing density of the tumor tissue. On Day 6 (as shown in Figure 3f,n), marked by yellow arrows in Figure 3f, surface deformation of the brain tissue became apparent, presumably due to the expanding mass of the tumor. In parallel, as indicated by the white arrows, the backscattered intensity grew stronger and more heterogeneous in the tumor region.
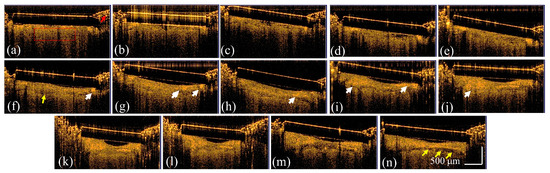
Figure 3.
2D OCT images of the tumorous brain obtained on (a–n) Day 1–Day 14 after removal of the skull section. The red arrow indicates the bone meal for fixation of the coverslip. The white arrows represent the stronger backscattered intensity resulting from tumorous tissue, and the yellow arrows indicate the deformation of brain tissue. The red rectangle in (a) indicates the region selected for the further analysis of homogeneity and variance.
For a more quantitative assessment of brain tissue changes, texture analysis was applied to the OCT images using two specific parameters: homogeneity and variance. The analysis spanned a 14-day period post-skull removal, drawing on a dataset of 500 images per day to ensure comprehensive coverage. The selected region for the analysis of homogeneity and variance in each B-scan is indicated by the red rectangle in Figure 3a. Figure 4a,b plot the analytic results of homogeneity and variance evaluated from five mice and the same brain areas were repeatedly scanned with OCT from Day 1 to Day 14 after removing the skull. In Figure 4a, the analytic data across five mice indicates a significant decline in homogeneity over time, particularly after Day 4. This decline in homogeneity suggests an increase in tissue complexity, as tumor growth tends to disrupt the previously uniform structure of healthy brain tissue. In parallel, the variance showed an upward trend with the tumor’s growth time, peaking before Day 12, as shown in Figure 4b. After Day 12, even though the mean variance decreased, it remained higher than that on Day 1, reflecting the ongoing presence of the tumor. The results presented in Figure 4 underscore the ability of OCT to detect textural changes indicative of tumor presence. Tumor cells, varying in size and irregularly dispersed, contribute to the inhomogeneity and variance observed in the backscattered intensity. These changes in tissue texture are measurable and provide insight into the physical alterations occurring within the brain as a tumor progresses.

Figure 4.
Analytic results of (a) homogeneity and (b) variance estimated from the tumorous brains of 5 mice obtained on Day 1 to Day 14 after removal of the skull sections.
Figure 5a–n shows the corresponding projection-view OCTA results of Figure 3 and Figure 5o, which represent the photo taken on Day 14 after removing the skull. The red circle in Figure 5a that marks the injection region and subsequent images captures the progression and spread of tumor-induced vascular alterations. Initially, the vasculature in the brain tissue is clearly defined, with distinct vascular boundaries that can be easily discerned, as shown in Figure 5a. The injection site is marked, providing a reference for subsequent changes. Over time, as the tumor cells proliferate, they induce the formation of new blood vessels (angiogenesis). This is visible in the changing vascular patterns, where boundaries become less distinct, as shown in Figure 5b–n. Interestingly, some of the main vessels visible on Day 1 seem to disappear or become obscured as the tumor progresses. This could be due to the vessels being compressed or remodeled by the growing tumor, or the formation of a denser network of smaller vessels that are harder to distinguish individually.
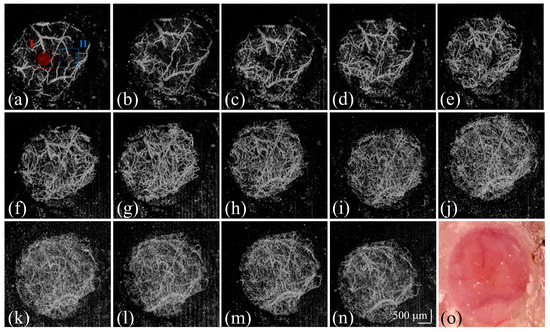
Figure 5.
(a–n) Corresponding projection-view OCTA results of Figure 3 and (o) the photo taken on Day 14 after removing the skull. The red-circle region indicates the implantation region of the tumor cells. The regions marked by the (I) red and (II) blue squares are used for the following analysis of the OCTA results.
The quantitative analysis of the vasculature in the context of brain tumor growth is essential for understanding how the tumor affects blood vessel formation and structure. OCTA is particularly useful in this regard because it can measure microvascular changes non-invasively. Figure 6 plots the quantitative analysis of OCTA results of five mice, including vessel diameter, vessel density, and fractal dimension. Here, two regions were chosen for the evaluation of the vascular parameters. The first region of interest (ROI) was directly at the site of tumor cell implantation. The second ROI was deliberately chosen to be 1 mm away from the tumor implantation site. The first and second regions are indicated by the red and blue squares in Figure 5a, respectively. The central region, where tumor cells were implanted, probably shows direct effects of the tumor on the vascular parameters. In contrast, the peripheral region may provide insights into the indirect or systemic effects. Analyzing these two regions and comparing their vascular parameters over time can provide a detailed view of the spatial dynamics of tumor-induced angiogenesis. In Figure 6a, the mean values of the vessel diameter in ROI I and II gradually increase until Day 7, and then they decrease. This pattern likely reflects an initial increase in blood flow to support tumor growth before Day 7, followed by the formation of new vessels with smaller diameters, which reduce the average vessel diameter. Figure 6b plots the analytic results of the vessel density. It can be noted that the vessel density significantly increased at the beginning of tumor growth and became slightly varied after Day 5. In Figure 6c, the mean values of fractal dimension also increased with growth time in both ROIs. Vascular networks can be considered fractal-like because of their branching nature. A fractal dimension analysis allows for the quantification of the complexity and density of the vessel branching pattern. In tumor angiogenesis, the fractal dimension might increase as the network becomes more intricate and disorganized, a common feature in tumorous tissues.
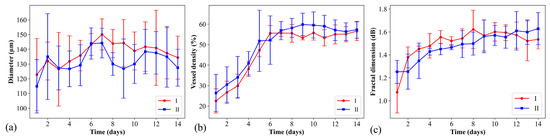
Figure 6.
Quantitative analysis of OCTA results of 5 mice including (a) vessel diameter, (b) vessel density, and (c) fractal dimension. The red and blue lines represent the analytic results evaluated from ROI 1 and 2, as indicated by the red and blue squares in Figure 5a.
4. Discussion
Through the careful evaluation of homogeneity and variance in OCT image textures, the impact of tumor growth on brain tissue structure can be monitored and documented. These metrics offer a non-invasive means to observe and potentially predict tumor behavior, potentially aiding in the assessment of treatment efficacy and planning of therapeutic interventions. Additionally, SV-OCT results underscore the dynamic nature of tumor progression and its impact on both tissue architecture and vasculature. The enhanced backscattered intensity and the vascular changes observable through OCT and OCTA offer valuable insights into tumor development and may serve as critical indicators for the differentiation of tissue status.
In this study, our objective was to investigate morphological changes in brain tissue related to tissue scattering and vessels. An OCT system with resolutions of 7 and 10 μm along the axial and transverse directions, respectively, was employed, which was sufficient to identify these morphological changes. However, higher cellular resolutions could provide more detailed insights, especially in the early stages of tumor growth. Previous reports have demonstrated that full-field OCT, with a high transverse resolution of approximately 1 μm, can be used for ex vivo imaging of brain tumors, enabling the identification of neuronal cell bodies, myelinated axon bundles, and vasculature. However, due to limitations in the imaging speed and depth, the use of full-field OCT for in vivo imaging remains challenging [42]. Furthermore, µOCT, offering axial and transverse resolutions of 1.8 and 2.0 µm, respectively, has been proposed for ex vivo brain tumor imaging and grade discrimination. This method has shown satisfactory accuracy in discriminating brain tumor grades when compared with histological results [43]. Based on these findings, employing OCT with higher imaging resolutions appears to be effective for investigating brain tumors in their early stages.
In addition to identifying brain tissue morphology, the polarization properties of brain tissue have been employed to further differentiate between normal and tumorous tissues. Yashin et al. proposed using cross-polarization OCT for an ex vivo study involving 30 glioma patients [40,41]. Their analyses of cross-polarization OCT data yielded significant diagnostic accuracy. They also discovered that certain parameters, such as attenuation and forward cross-scattering, are effective in distinguishing between normal and tumorous brain tissues. Although previous reports have shown that OCT microstructural results can differentiate tumorous from normal brain tissue, the diagnostic accuracy can be further enhanced by utilizing the functional capabilities of OCT [53].
Tumor growth is invariably associated with angiogenesis, which alters the vasculature to enhance nutrient and oxygen transport, thereby changing microvascular characteristics. Consequently, identifying microvasculature in brain tumors can effectively differentiate tumor distribution. Previous research has advocated using OCTA to distinguish tumor microvasculature from normal brain vessels in the animal model [54]. This study employed a 1310-nm OCT system for in vivo investigations, analyzing vascular parameters including fractional blood volume, vessel tortuosity, diameter, orientation, and directionality. The significant findings reveal that the average fractal blood volume in tumor tissue is approximately 60%, compared to 85% in non-tumorous tissue. Additionally, vessel tortuosity in tumor tissues is higher than in normal tissues. These results suggest that OCTA, along with detailed vascular parameters, holds potential for diagnostic and therapeutic studies of brain tumors. Based on the previous research, our study utilized both OCT and OCTA to further explore brain tumor progression, focusing on daily changes in morphology and vascular structures. Moreover, we propose analytical methods for quantitative evaluation of these changes, including scattering homogeneity and variation in OCT images, as well as vessel diameter, density, and fractal dimension in OCTA results.
As tumors grow, tumor often stimulate the formation of new blood vessels to supply the necessary nutrients for their growth. An increase in vessel density within the tumor region would typically be expected in comparison to normal brain tissue. Tumors can also influence the diameter of blood vessels, causing existing vessels to dilate or newly formed vessels to have different sizes compared to normal tissue. Tracking changes in vessel diameter over time can help distinguish between healthy and tumorous tissue, as tumors might cause vessels to become enlarged in the early stage to support the increased blood flow required by the growing tumor mass. The results demonstrate the potential of texture analysis and OCTA as valuable tools for the non-invasive monitoring of tumor-related angiogenesis. It could be particularly useful in preclinical studies, drug development, and the evaluation of anti-angiogenic therapies that aim to disrupt the blood supply to tumors.
5. Conclusions
In this study, we propose the use of OCT and OCTA to investigate the growth of brain tumors. OCT can identify morphological changes and detect tumor-induced variations in backscattered light intensity. Furthermore, the homogeneity and variance of the OCT backscattered intensity are evaluated, which indicates that tumor growth alters tissue homogeneity and increases intensity variation. Additionally, OCTA is utilized to assess angiogenesis associated with tumor growth, and quantitative evaluations are performed to analyze vessel diameter, vessel density, and fractal dimension. The angiographic results demonstrate an increase in vessel density and an increase in the mean vessel diameter as the tumor grows in the early tumor stage. Moreover, the fractal dimension of the vessels also increases with tumor progression. These findings suggest that OCT and OCTA could be effective, non-invasive tools for evaluating brain tumor growth and distinguishing between normal and tumorous brain tissue.
Author Contributions
Conceptualization, T.N.-H. and M.-T.T.; methodology, M.-T.T.; software, C.-H.W.; validation, T.N.-H., T.-A.W., C.-H.W. and M.-T.T.; formal analysis, T.-A.W. and C.-H.W.; investigation, T.N.-H.; data curation, T.-A.W.; writing—original draft preparation, T.N.-H., T.-A.W. and M.-T.T.; writing—review and editing, T.N.-H., T.-A.W. and M.-T.T.; visualization, M.-T.T.; supervision, M.-T.T.; project administration, M.-T.T.; funding acquisition, M.-T.T. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Chang Gung Memorial Hospital, grant numbers (CMRPD2H0243, CMRPD2H0242, CMRPD2H0241), and the National Science and Technology Council, grant number MOST 110-2223-E-182-001-MY3.
Institutional Review Board Statement
The animal protocol was approved by Animal Care and Use Committee of Chang Gung University (IACUC Approval No.: CGU108-140).
Informed Consent Statement
Not applicable.
Data Availability Statement
The data of this article are available from the corresponding author upon reasonable request.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- McFaline-Figueroa, J.R.; Lee, E.Q. Brain Tumors. Am. J. Med. 2018, 131, 874–882. [Google Scholar] [CrossRef] [PubMed]
- Gavrilovic, I.T.; Posner, J.B. Brain metastases: Epidemiology and pathophysiology. J. Neuro-Oncol. 2005, 75, 5–14. [Google Scholar] [CrossRef] [PubMed]
- Ogasawara, C.; Philbrick, B.D.; Adamson, D.C. Meningioma: A Review of Epidemiology, Pathology, Diagnosis, Treatment, and Future Directions. Biomedicines 2021, 9, 319. [Google Scholar] [CrossRef] [PubMed]
- Sever, R.; Brugge, J.S. Signal transduction in cancer. Cold Spring Harb. Perspect. Med. 2015, 5, a006098. [Google Scholar] [CrossRef] [PubMed]
- Louis, D.N.; Perry, A.; Wesseling, P.; Brat, D.J.; Cree, I.A.; Figarella-Branger, D.; Hawkins, C.; Ng, H.K.; Pfister, S.M.; Reifenberger, G.; et al. The 2021 WHO Classification of Tumors of the Central Nervous System: A summary. Neuro Oncol. 2021, 23, 1231–1251. [Google Scholar] [CrossRef] [PubMed]
- Louis, D.N.; Ohgaki, H.; Wiestler, O.D.; Cavenee, W.K.; Burger, P.C.; Jouvet, A.; Scheithauer, B.W.; Kleihues, P. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 2007, 114, 97–109. [Google Scholar] [CrossRef] [PubMed]
- Jang, S.J.; Gardner, J.M.; Ro, J.Y. Diagnostic approach and prognostic factors of cancers. Adv. Anat. Pathol. 2011, 18, 165–172. [Google Scholar] [CrossRef] [PubMed]
- Mabray, M.C.; Barajas, R.F., Jr.; Cha, S. Modern brain tumor imaging. Brain Tumor Res. Treat. 2015, 3, 8–23. [Google Scholar] [CrossRef] [PubMed]
- Gao, H.; Jiang, X. Progress on the diagnosis and evaluation of brain tumors. Cancer Imaging 2013, 13, 466–481. [Google Scholar] [CrossRef]
- Fuss, T.L.; Cheng, L.L. Metabolic Imaging in Humans. Top. Magn. Reson. Imaging 2016, 25, 223–235. [Google Scholar] [CrossRef][Green Version]
- Overcast, W.B.; Davis, K.M.; Ho, C.Y.; Hutchins, G.D.; Green, M.A.; Graner, B.D.; Veronesi, M.C. Advanced imaging techniques for neuro-oncologic tumor diagnosis, with an emphasis on PET-MRI imaging of malignant brain tumors. Curr. Oncol. Rep. 2021, 23, 34. [Google Scholar] [CrossRef]
- Luo, M.; Larson, P.S.; Martin, A.J.; Miga, M.I. Accounting for Deformation in Deep Brain Stimulation Surgery With Models: Comparison to Interventional Magnetic Resonance Imaging. IEEE Trans. Biomed. Eng. 2020, 67, 2934–2944. [Google Scholar] [CrossRef] [PubMed]
- Rogers, C.M.; Jones, P.S.; Weinberg, J.S. Intraoperative MRI for Brain Tumors. J. Neuro-Oncol. 2021, 151, 479–490. [Google Scholar] [CrossRef]
- Dixon, L.; Lim, A.; Grech-Sollars, M.; Nandi, D.; Camp, S. Intraoperative ultrasound in brain tumor surgery: A review and implementation guide. Neurosurg. Rev. 2022, 45, 2503–2515. [Google Scholar] [CrossRef] [PubMed]
- Verburg, N.; de Witt Hamer, P.C. State-of-the-art imaging for glioma surgery. Neurosurg. Rev. 2021, 44, 1331–1343. [Google Scholar] [CrossRef]
- Bin-Alamer, O.; Abou-Al-Shaar, H.; Gersey, Z.C.; Huq, S.; Kallos, J.A.; McCarthy, D.J.; Head, J.R.; Andrews, E.; Zhang, X.; Hadjipanayis, C.G. Intraoperative Imaging and Optical Visualization Techniques for Brain Tumor Resection: A Narrative Review. Cancers 2023, 15, 4890. [Google Scholar] [CrossRef]
- Baskar, R.; Lee, K.A.; Yeo, R.; Yeoh, K.W. Cancer and radiation therapy: Current advances and future directions. Int. J. Med. Sci. 2012, 9, 193–199. [Google Scholar] [CrossRef]
- Liu, Y.P.; Zheng, C.C.; Huang, Y.N.; He, M.L.; Xu, W.W.; Li, B. Molecular mechanisms of chemo- and radiotherapy resistance and the potential implications for cancer treatment. MedComm 2021, 2, 315–340. [Google Scholar] [CrossRef] [PubMed]
- Rivera, M.; Norman, S.; Sehgal, R.; Juthani, R. Updates on Surgical Management and Advances for Brain Tumors. Curr. Oncol. Rep. 2021, 23, 35. [Google Scholar] [CrossRef]
- Vanderweyen, D.C.; Theaud, G.; Sidhu, J.; Rheault, F.; Sarubbo, S.; Descoteaux, M.; Fortin, D. The role of diffusion tractography in refining glial tumor resection. Brain Struct. Funct. 2020, 225, 1413–1436. [Google Scholar] [CrossRef]
- Ercan, S.; Ataizi, Z.S.; Ataizi, F.; Bulduk, E.B. Effects of total and subtotal glial tumor resection on survival. Eur. J. Med. Investig. 2020, 4, 73–77. [Google Scholar] [CrossRef]
- Huang, D.; Swanson, E.A.; Lin, C.P.; Schuman, J.S.; Stinson, W.G.; Chang, W.; Hee, M.R.; Flotte, T.; Gregory, K.; Puliafito, C.A.; et al. Optical coherence tomography. Science 1991, 254, 1178–1181. [Google Scholar] [CrossRef]
- Venkateswaran, N.; Galor, A.; Wang, J.; Karp, C.L. Optical coherence tomography for ocular surface and corneal diseases: A review. Eye Vis. 2018, 5, 13. [Google Scholar] [CrossRef]
- Lim, L.S.; Aung, H.T.; Aung, T.; Tan, D.T. Corneal imaging with anterior segment optical coherence tomography for lamellar keratoplasty procedures. Am. J. Ophthalmol. 2008, 145, 81–90. [Google Scholar] [CrossRef] [PubMed]
- Hee, M.R.; Izatt, J.A.; Swanson, E.A.; Huang, D.; Schuman, J.S.; Lin, C.P.; Puliafito, C.A.; Fujimoto, J.G. Optical coherence tomography of the human retina. Arch. Ophthalmol. 1995, 113, 325–332. [Google Scholar] [CrossRef]
- Sattler, E.; Kästle, R.; Welzel, J. Optical coherence tomography in dermatology. J. Biomed. Opt. 2013, 18, 061224. [Google Scholar] [CrossRef]
- Vignali, L.; Solinas, E.; Emanuele, E. Research and clinical applications of optical coherence tomography in invasive cardiology: A review. Curr. Cardiol. Rev. 2014, 10, 369–376. [Google Scholar] [CrossRef] [PubMed]
- Kennedy, B.F.; Kennedy, K.M.; Oldenburg, A.L.; Adie, S.G.; Boppart, S.A.; Sampson, D.D. Optical Coherence Elastography. In Optical Coherence Tomography: Technology and Applications; Drexler, W., Fujimoto, J.G., Eds.; Springer International Publishing: Cham, Switzerland, 2015; pp. 1007–1054. [Google Scholar]
- de Boer, J.F.; Hitzenberger, C.K.; Yasuno, Y. Polarization sensitive optical coherence tomography—A review [Invited]. Biomed. Opt. Express 2017, 8, 1838–1873. [Google Scholar] [CrossRef] [PubMed]
- de Carlo, T.E.; Romano, A.; Waheed, N.K.; Duker, J.S. A review of optical coherence tomography angiography (OCTA). Int. J. Retin. Vitr. 2015, 1, 5. [Google Scholar] [CrossRef]
- Tsai, M.T.; Chang, F.Y.; Lee, C.K.; Gong, C.S.; Lin, Y.X.; Lee, J.D.; Yang, C.H.; Liu, H.L. Investigation of temporal vascular effects induced by focused ultrasound treatment with speckle-variance optical coherence tomography. Biomed. Opt. Express 2014, 5, 2009–2022. [Google Scholar] [CrossRef][Green Version]
- Enfield, J.; Jonathan, E.; Leahy, M. In vivo imaging of the microcirculation of the volar forearm using correlation mapping optical coherence tomography (cmOCT). Biomed. Opt. Express 2011, 2, 1184–1193. [Google Scholar] [CrossRef] [PubMed]
- Jonathan, E.; Enfield, J.; Leahy, M.J. Correlation mapping method for generating microcirculation morphology from optical coherence tomography (OCT) intensity images. J. Biophotonics 2011, 4, 583–587. [Google Scholar] [CrossRef] [PubMed]
- An, L.; Qin, J.; Wang, R.K. Ultrahigh sensitive optical microangiography for in vivo imaging of microcirculations within human skin tissue beds. Opt. Express 2010, 18, 8220–8228. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.K.; An, L.; Francis, P.; Wilson, D.J. Depth-resolved imaging of capillary networks in retina and choroid using ultrahigh sensitive optical microangiography. Opt. Lett. 2010, 35, 1467–1469. [Google Scholar] [CrossRef] [PubMed]
- Strenge, P.; Lange, B.; Draxinger, W.; Grill, C.; Danicke, V.; Theisen-Kunde, D.; Hagel, C.; Spahr-Hess, S.; Bonsanto, M.M.; Handels, H.; et al. Differentiation of different stages of brain tumor infiltration using optical coherence tomography: Comparison of two systems and histology. Front. Oncol. 2022, 12, 896060. [Google Scholar] [CrossRef] [PubMed]
- Giese, A.; Böhringer, H.; Leppert, J.; Kantelhardt, S.; Lankenau, E.; Koch, P.; Birngruber, R.; Hüttmann, G. Non-Invasive Intraoperative Optical Coherence Tomography of the Resection Cavity during Surgery of Intrinsic Brain Tumors; SPIE: San Jose, CA, USA, 2006; Volume 6078. [Google Scholar]
- SoRelle, E.D.; Yecies, D.W.; Liba, O.; Bennett, F.C.; Graef, C.M.; Dutta, R.; Mitra, S.; Joubert, L.M.; Cheshier, S.; Grant, G.A.; et al. Spatiotemporal Tracking of Brain-Tumor-Associated Myeloid Cells in Vivo through Optical Coherence Tomography with Plasmonic Labeling and Speckle Modulation. ACS Nano 2019, 13, 7985–7995. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.Q.; Chiu, K.S.; Liu, X.R.; Hsiao, T.Y.; Zhao, G.; Li, S.J.; Lin, C.P.; Sun, C.W. Polarization-Sensitive Optical Coherence Tomography for Brain Tumor Characterization. IEEE J. Sel. Top. Quantum Electron. 2019, 25, 1–7. [Google Scholar] [CrossRef]
- Yashin, K.S.; Kiseleva, E.B.; Gubarkova, E.V.; Moiseev, A.A.; Kuznetsov, S.S.; Shilyagin, P.A.; Gelikonov, G.V.; Medyanik, I.A.; Kravets, L.Y.; Potapov, A.A.; et al. Cross-Polarization Optical Coherence Tomography for Brain Tumor Imaging. Front. Oncol. 2019, 9, 201. [Google Scholar] [CrossRef]
- Yashin, K.S.; Kiseleva, E.B.; Moiseev, A.A.; Kuznetsov, S.S.; Timofeeva, L.B.; Pavlova, N.P.; Gelikonov, G.V.; Medyanik, I.A.; Kravets, L.Y.; Zagaynova, E.V.; et al. Quantitative nontumorous and tumorous human brain tissue assessment using microstructural co- and cross-polarized optical coherence tomography. Sci. Rep. 2019, 9, 2024. [Google Scholar] [CrossRef]
- Assayag, O.; Grieve, K.; Devaux, B.; Harms, F.; Pallud, J.; Chretien, F.; Boccara, C.; Varlet, P. Imaging of non-tumorous and tumorous human brain tissues with full-field optical coherence tomography. Neuroimage Clin. 2013, 2, 549–557. [Google Scholar] [CrossRef]
- Yu, X.; Hu, C.; Zhang, W.; Zhou, J.; Ding, Q.; Sadiq, M.T.; Fan, Z.; Yuan, Z.; Liu, L. Feasibility evaluation of micro-optical coherence tomography (μOCT) for rapid brain tumor type and grade discriminations: μOCT images versus pathology. BMC Med. Imaging 2019, 19, 102. [Google Scholar] [CrossRef]
- Vakoc, B.J.; Lanning, R.M.; Tyrrell, J.A.; Padera, T.P.; Bartlett, L.A.; Stylianopoulos, T.; Munn, L.L.; Tearney, G.J.; Fukumura, D.; Jain, R.K.; et al. Three-dimensional microscopy of the tumor microenvironment in vivo using optical frequency domain imaging. Nat. Med. 2009, 15, 1219–1223. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.; You, J.; Volkow, N.D.; Park, K.; Du, C. Ultrasensitive detection of 3D cerebral microvascular network dynamics in vivo. Neuroimage 2014, 103, 492–501. [Google Scholar] [CrossRef] [PubMed]
- Dolezyczek, H.; Rapolu, M.; Niedzwiedziuk, P.; Karnowski, K.; Borycki, D.; Dzwonek, J.; Wilczynski, G.; Malinowska, M.; Wojtkowski, M. Longitudinal in-vivo OCM imaging of glioblastoma development in the mouse brain. Biomed. Opt. Express 2020, 11, 5003–5016. [Google Scholar] [CrossRef]
- Katta, N.; Estrada, A.D.; McElroy, A.B.; Gruslova, A.; Oglesby, M.; Cabe, A.G.; Feldman, M.D.; Fleming, R.D.; Brenner, A.J.; Milner, T.E. Laser brain cancer surgery in a xenograft model guided by optical coherence tomography. Theranostics 2019, 9, 3555–3564. [Google Scholar] [CrossRef] [PubMed]
- Haralick, R.M.; Shanmugam, K.; Dinstein, I. Textural Features for Image Classification. IEEE Trans. Syst. Man Cybern. 1973, SMC-3, 610–621. [Google Scholar] [CrossRef]
- Sawyer, T.W.; Chandra, S.; Rice, P.F.S.; Koevary, J.W.; Bartaon, J.K. Three-dimensional texture analysis of optical coherence tomography images of ovarian tissue. Phys. Med. Biol. 2018, 63, 235020. [Google Scholar] [CrossRef]
- Mariampillai, A.; Leung, M.K.; Jarvi, M.; Standish, B.A.; Lee, K.; Wilson, B.C.; Vitkin, A.; Yang, V.X. Optimized speckle variance OCT imaging of microvasculature. Opt. Lett. 2010, 35, 1257–1259. [Google Scholar] [CrossRef]
- Untracht, G.R.; Matos, R.S.; Dikaios, N.; Bapir, M.; Durrani, A.K.; Butsabong, T.; Campagnolo, P.; Sampson, D.D.; Heiss, C.; Sampson, D.M. OCTAVA: An open-source toolbox for quantitative analysis of optical coherence tomography angiography images. PLoS ONE 2021, 16, e0261052. [Google Scholar] [CrossRef]
- Jerman, T.; Pernuš, F.; Lkar, B.; Špiclin, Ž. Blob Enhancement and Visualization for Improved Intracranial Aneurysm Detection. IEEE Trans. Vis. Comput. Graph. 2016, 22, 1705–1717. [Google Scholar] [CrossRef]
- Achkasova, K.A.; Moiseev, A.A.; Yashin, K.S.; Kiseleva, E.B.; Bederina, E.L.; Loginova1, M.M.; Medyanik, I.A.; Gelikonov, G.V.; Zagaynova, E.V.; Gladkova, N.D. Nondestructive label-free detection of peritumoral white matter damage using crosspolarization optical coherence tomography. Front. Oncol. 2023, 13, 1133074. [Google Scholar] [CrossRef] [PubMed]
- Andleeb, F.; Katta, N.; Gruslova, A.; Muralidharan, B.; Estrada, A.; McElroy, A.B.; Ullah, H.; Brenner, A.J.; Milner, T.E. Differentiation of Brain Tumor Microvasculature From Normal Vessels Using Optical Coherence Angiography. Laser Surg. Med. 2021, 53, 1386–1394. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).