Investigation of an Improved Angular Spectrum Method Based on Holography
Abstract
:1. Introduction
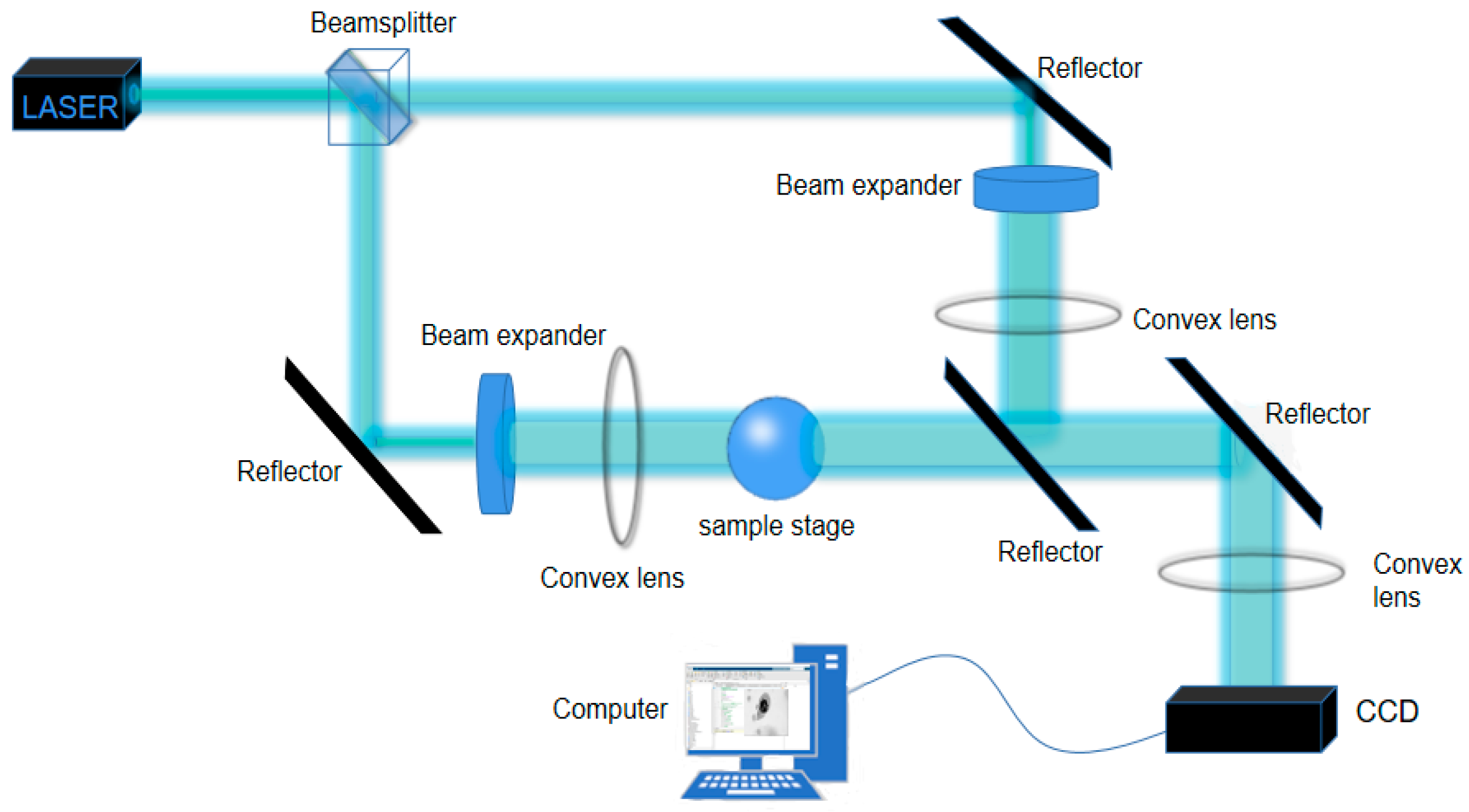
2. Holographic Principle
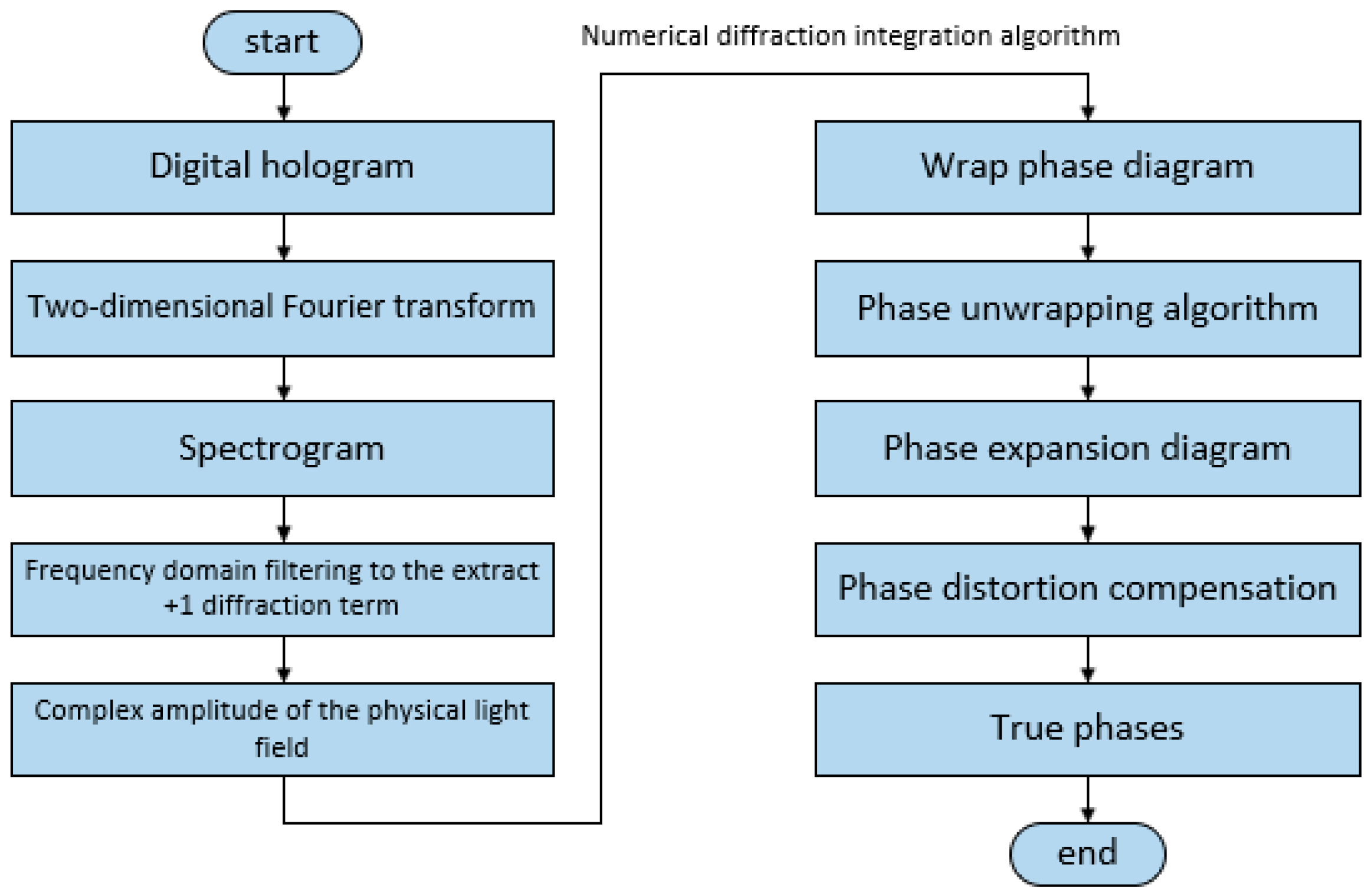
3. Method
- (1)
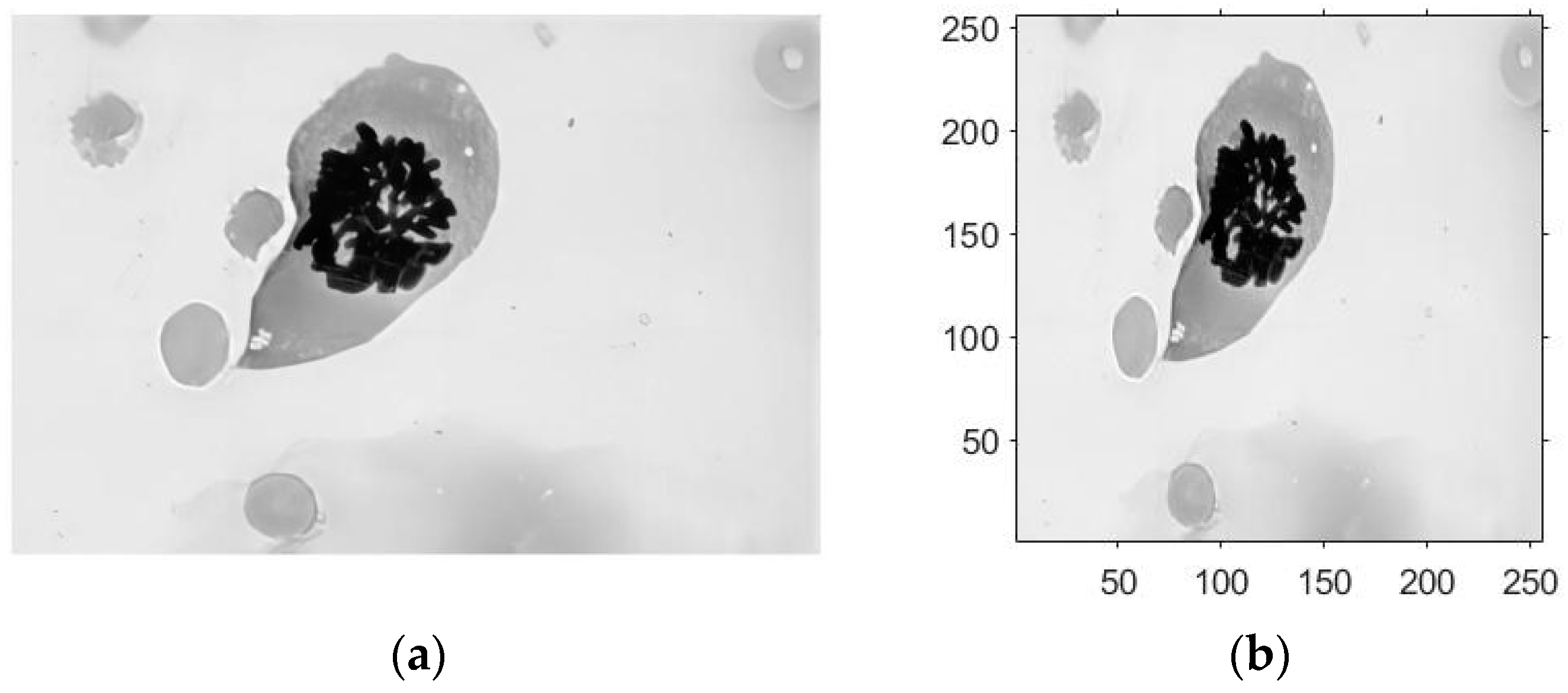
- The modeling process begins with input data, where an RGB cell image with M × N pixels is preprocessed and processed. The original image is then resized to 256 × 256 pixels, subjected to grayscale processing, and the object data are then generated by sampling the 256 × 256 pixel image.
- (2)
- Suppose we have a two-dimensional image f(x,y), in which the two dimensions x and y represent the horizontal and vertical directions of the image; its corresponding complex function can then be defined as F(u,v). When we perform the Fourier transform of the function F(u,v), we can obtain its frequency domain information G(u,v), and finally, we can obtain the spatial domain information of the original image via the inverse Fourier transform of G(u,v), which is the basic formula behind the reasoning of angular spectral theory.
- (3)
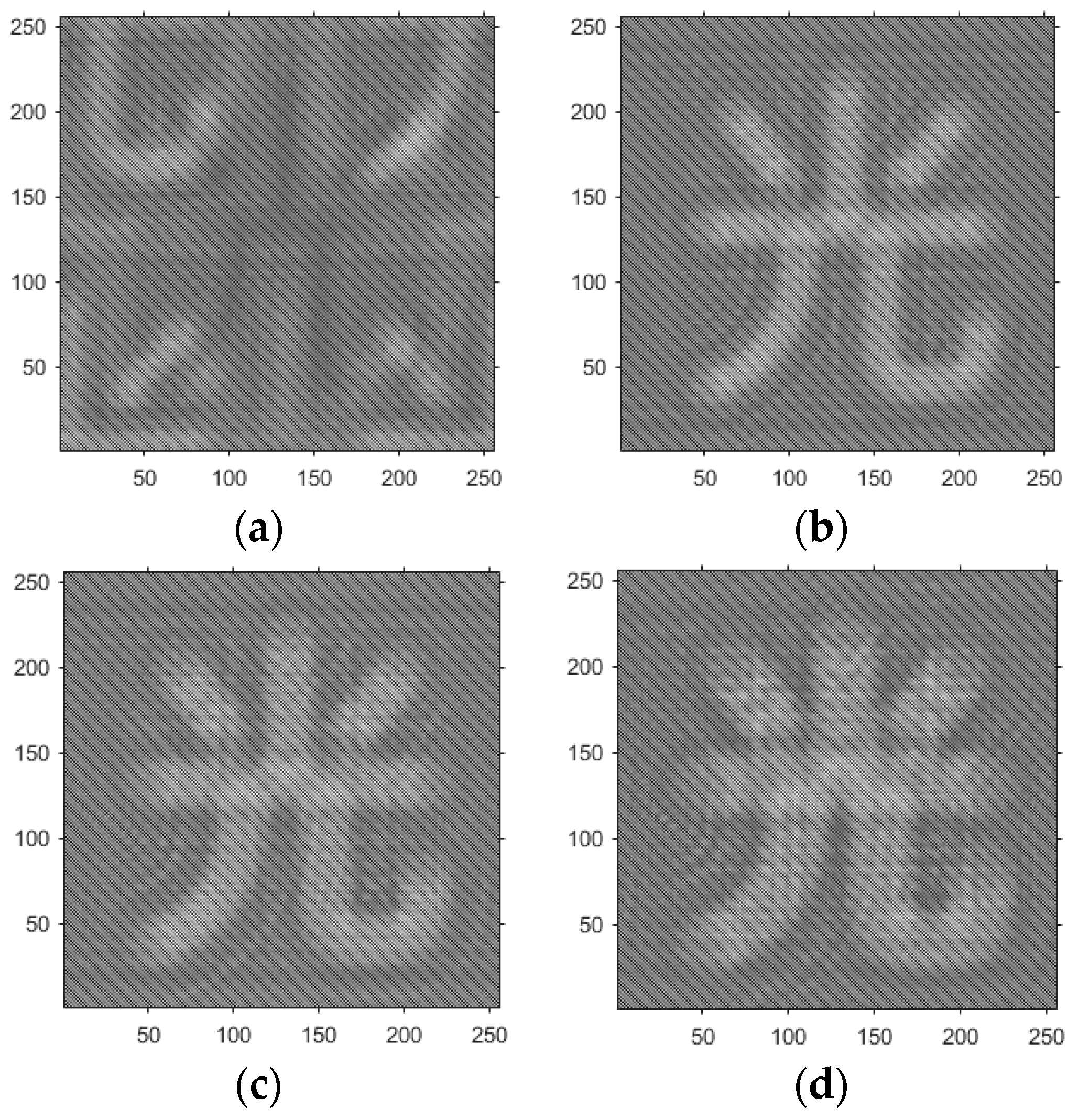
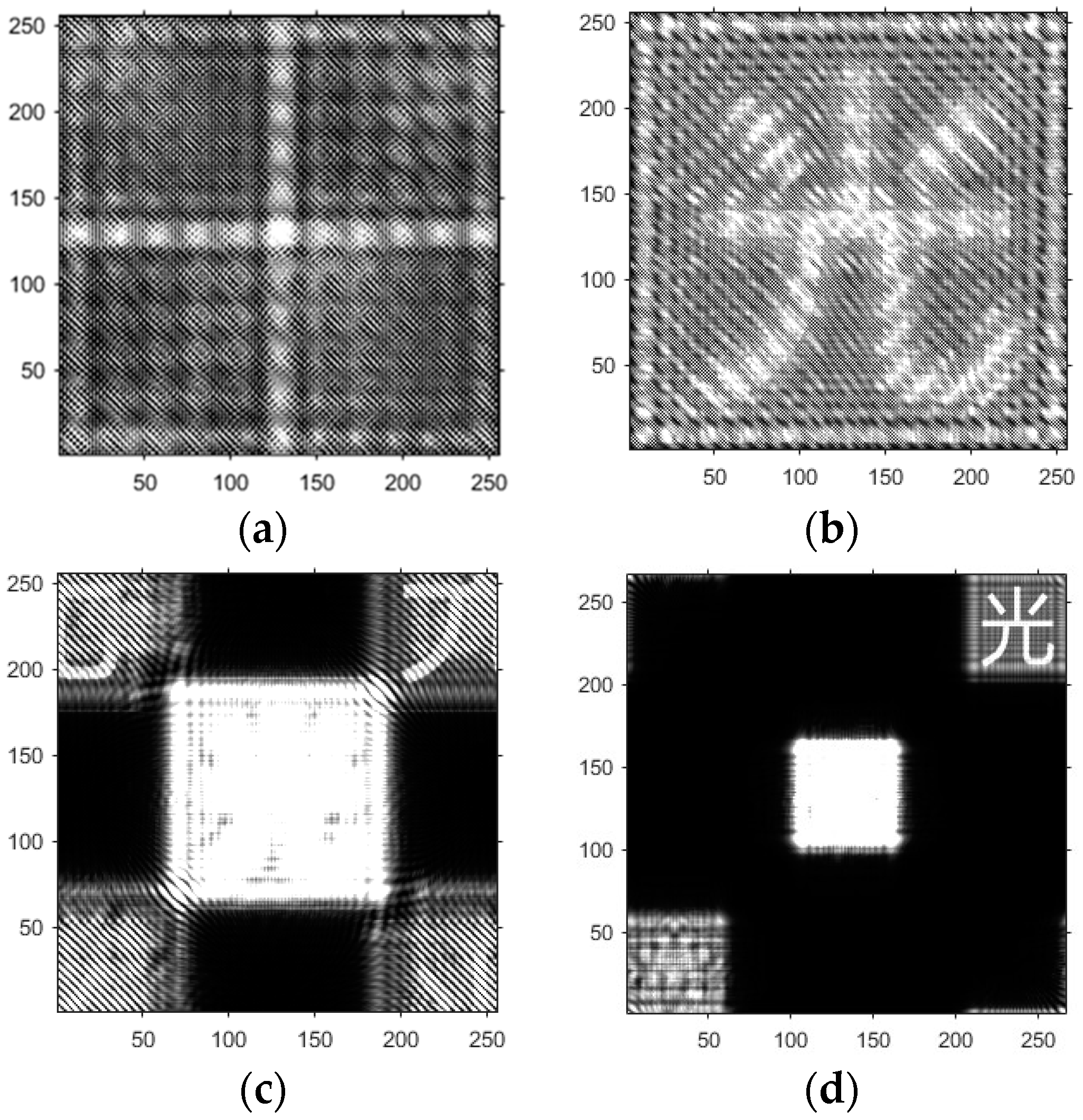
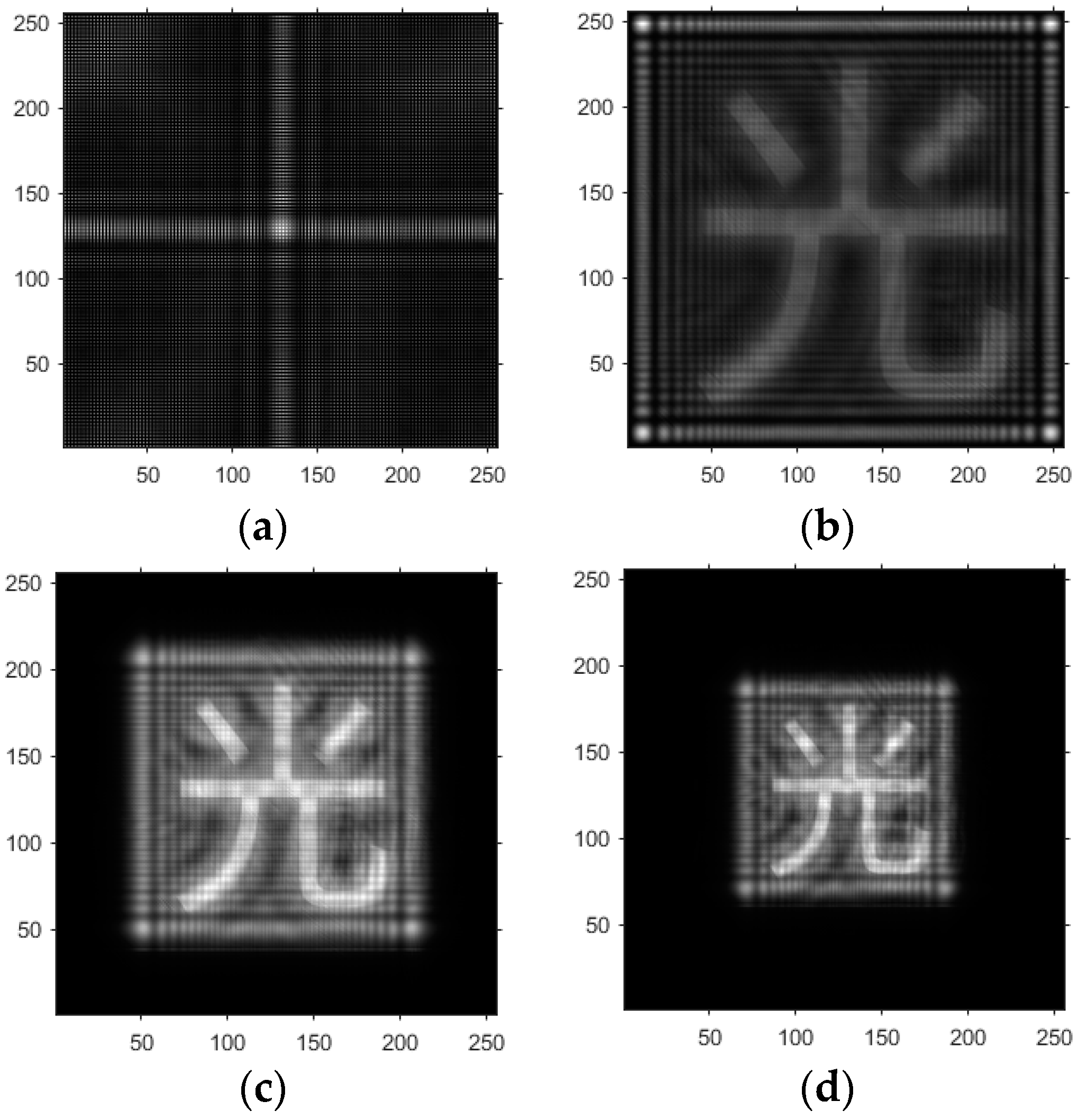
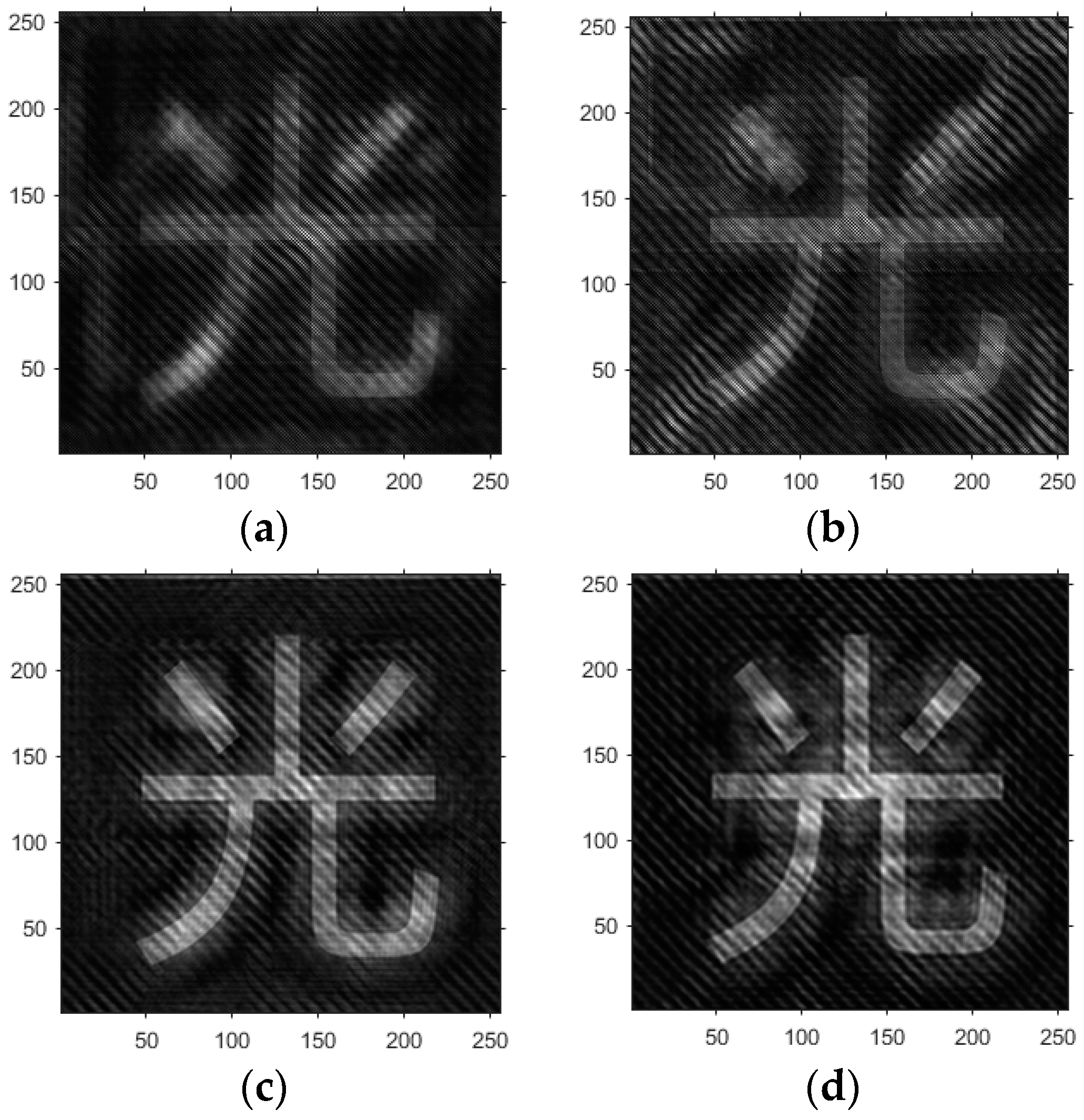
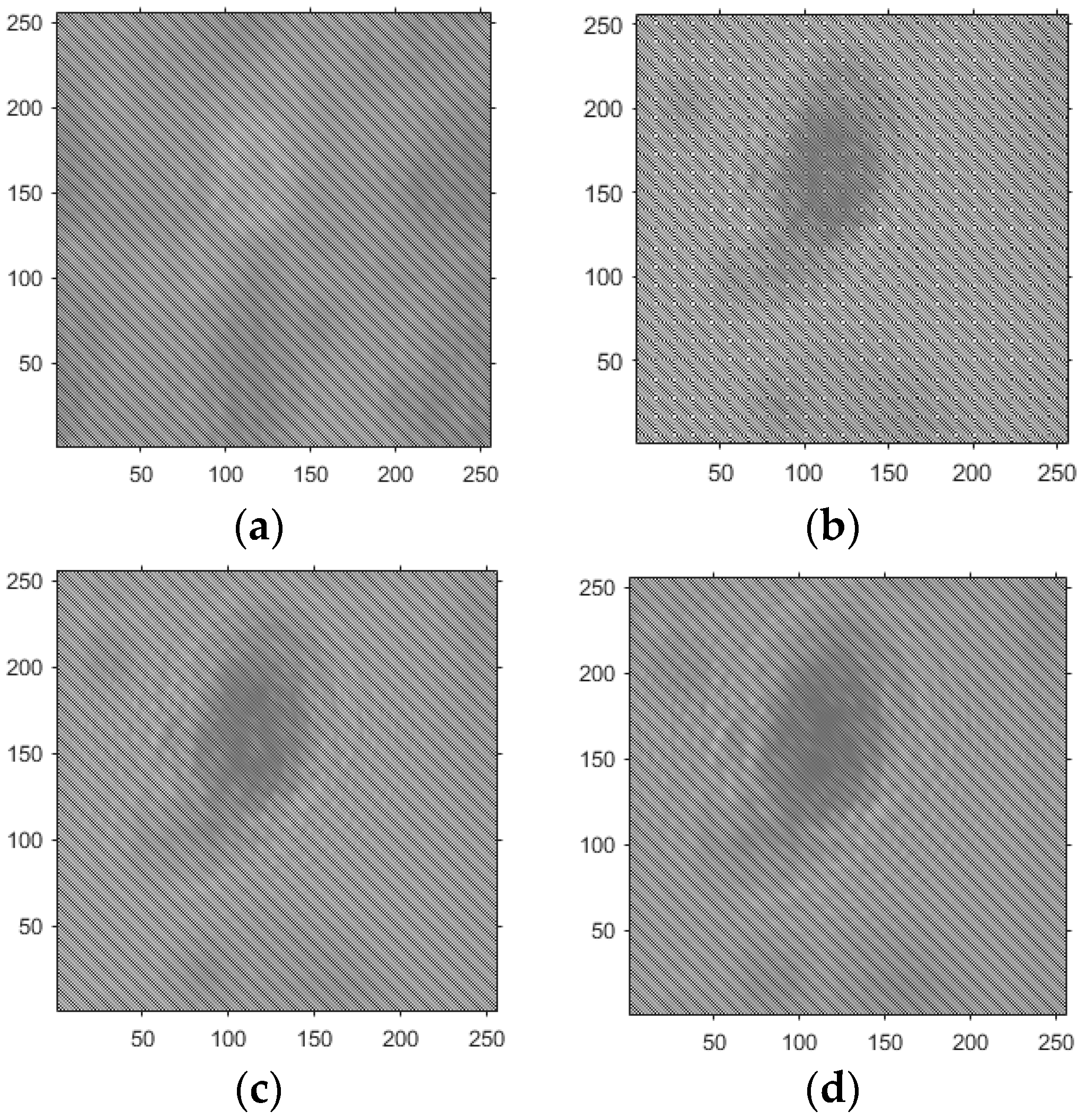
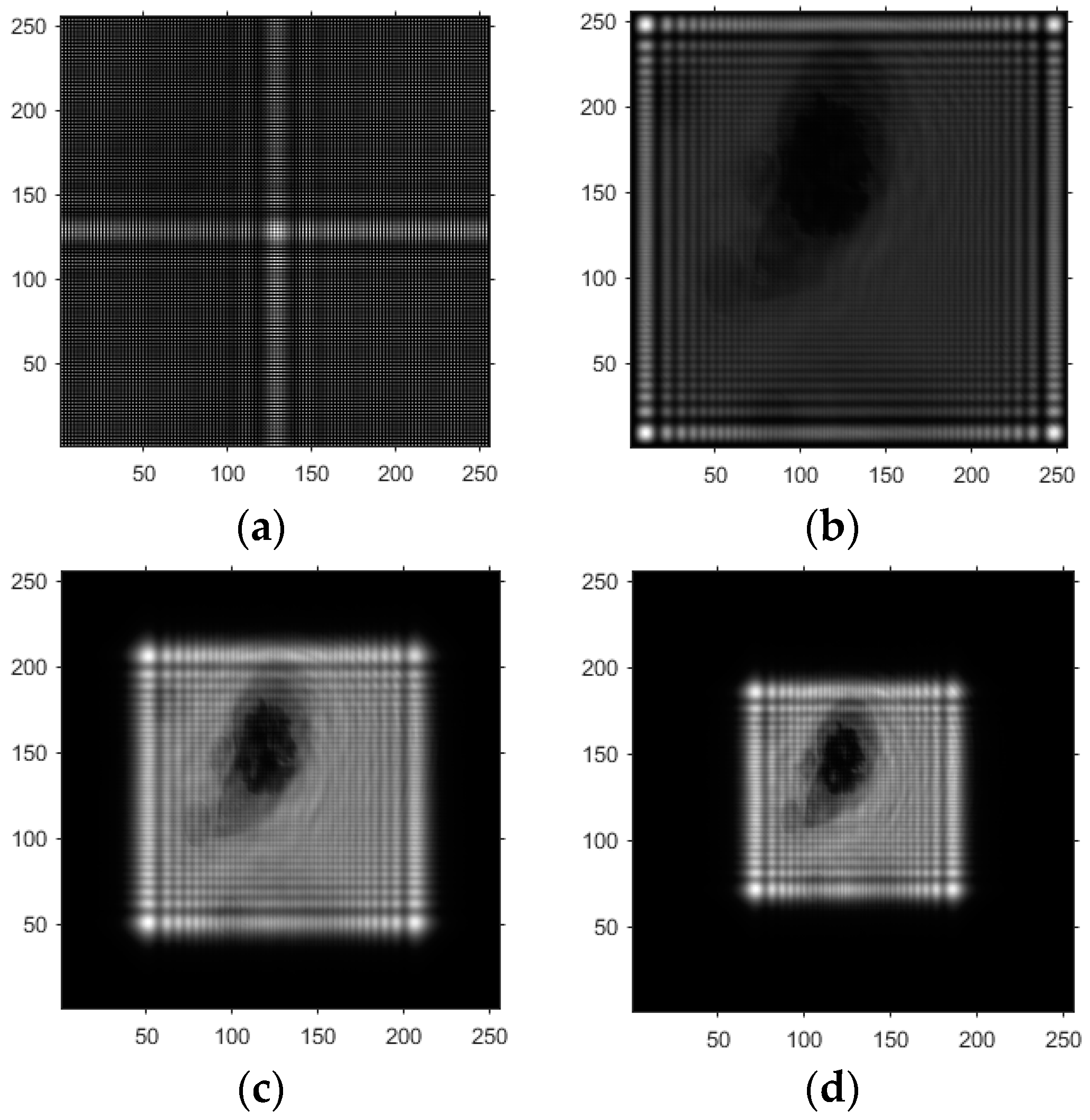
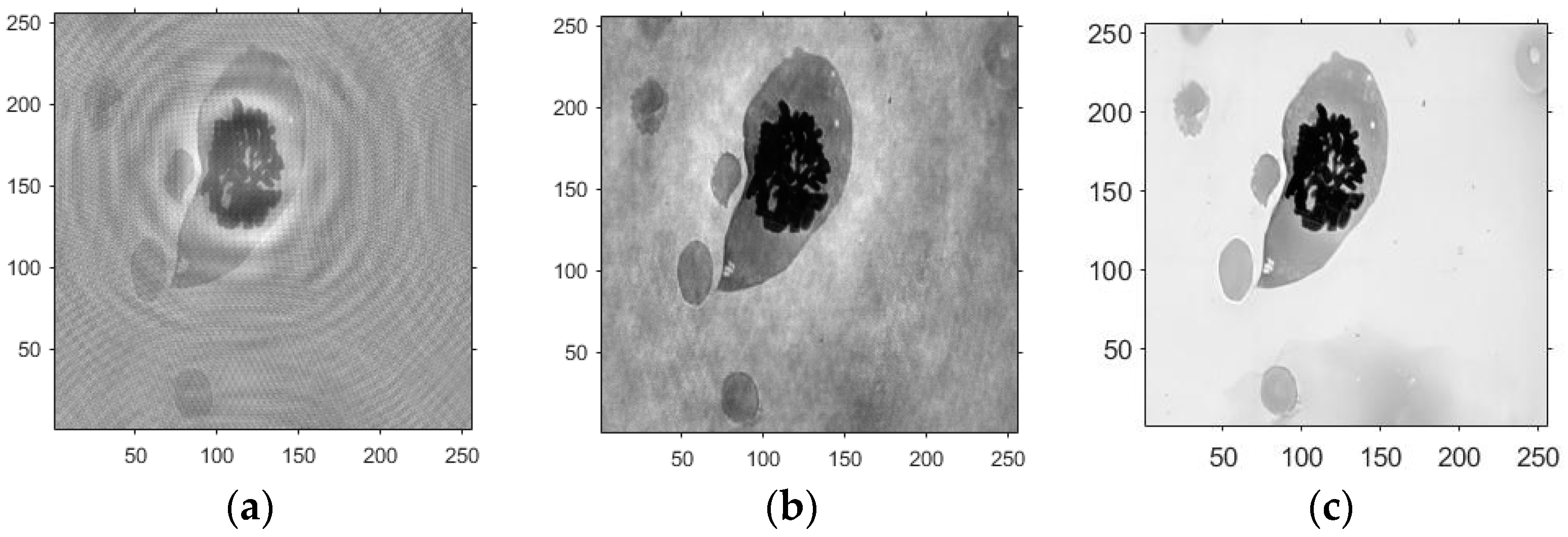
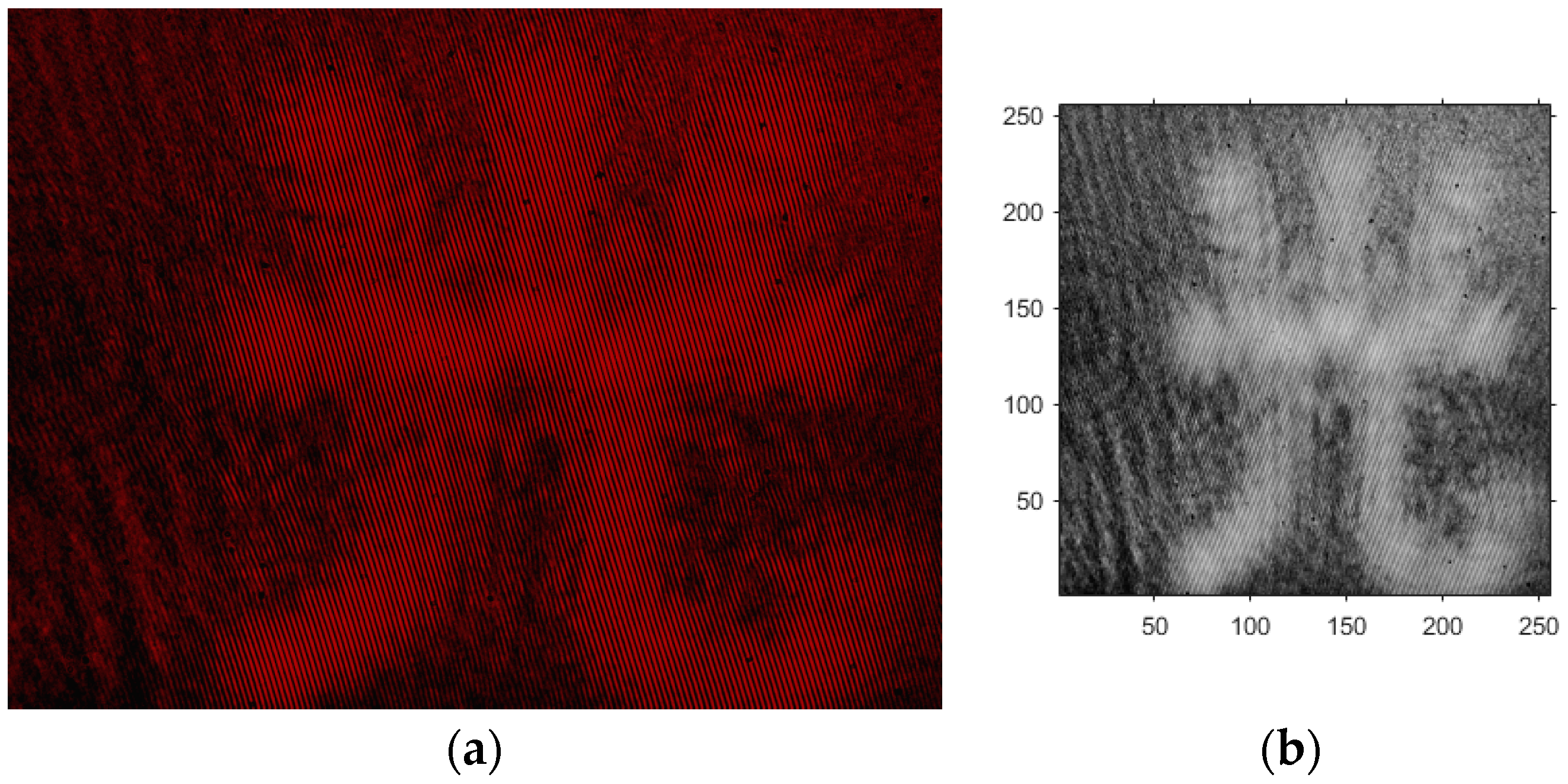
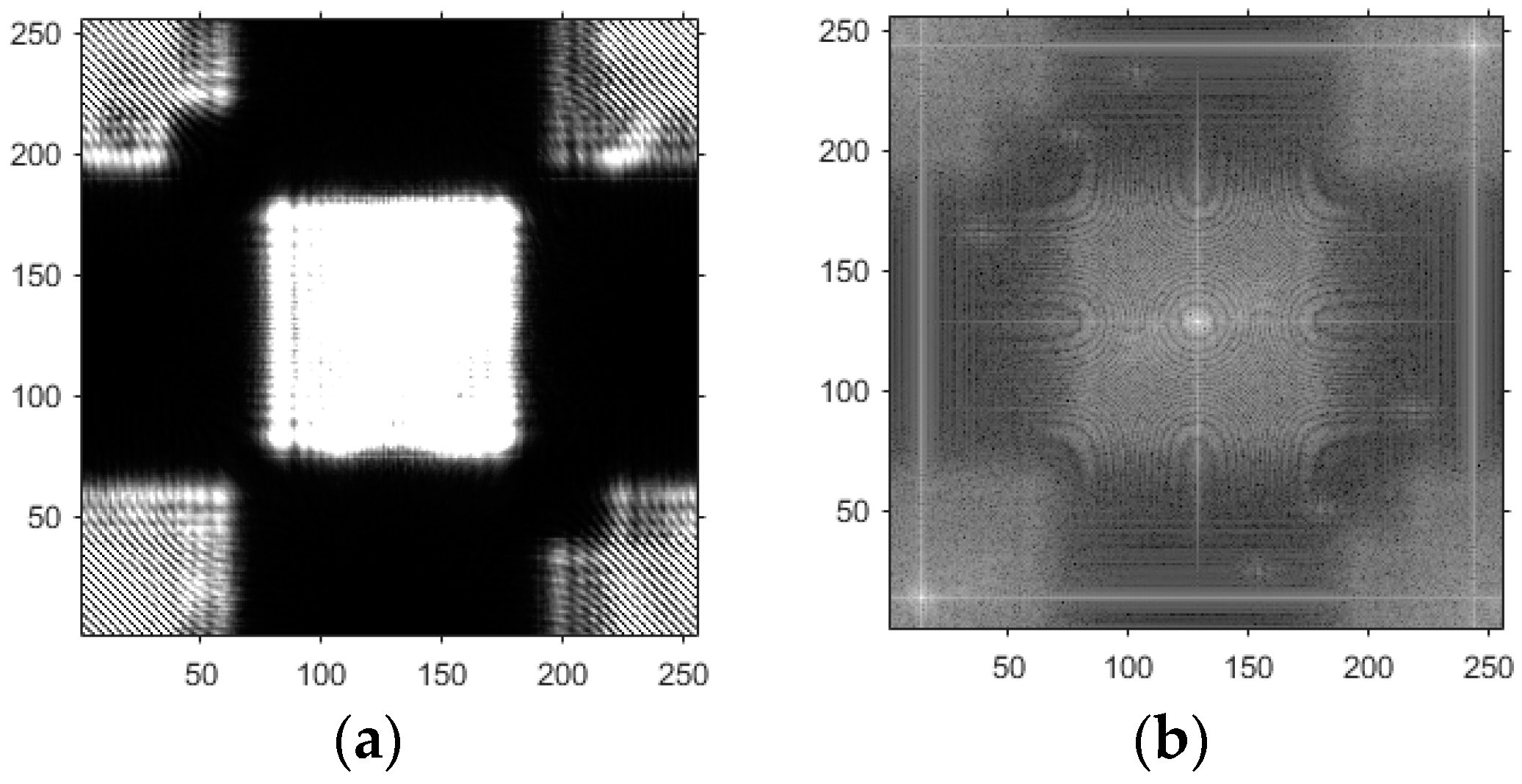
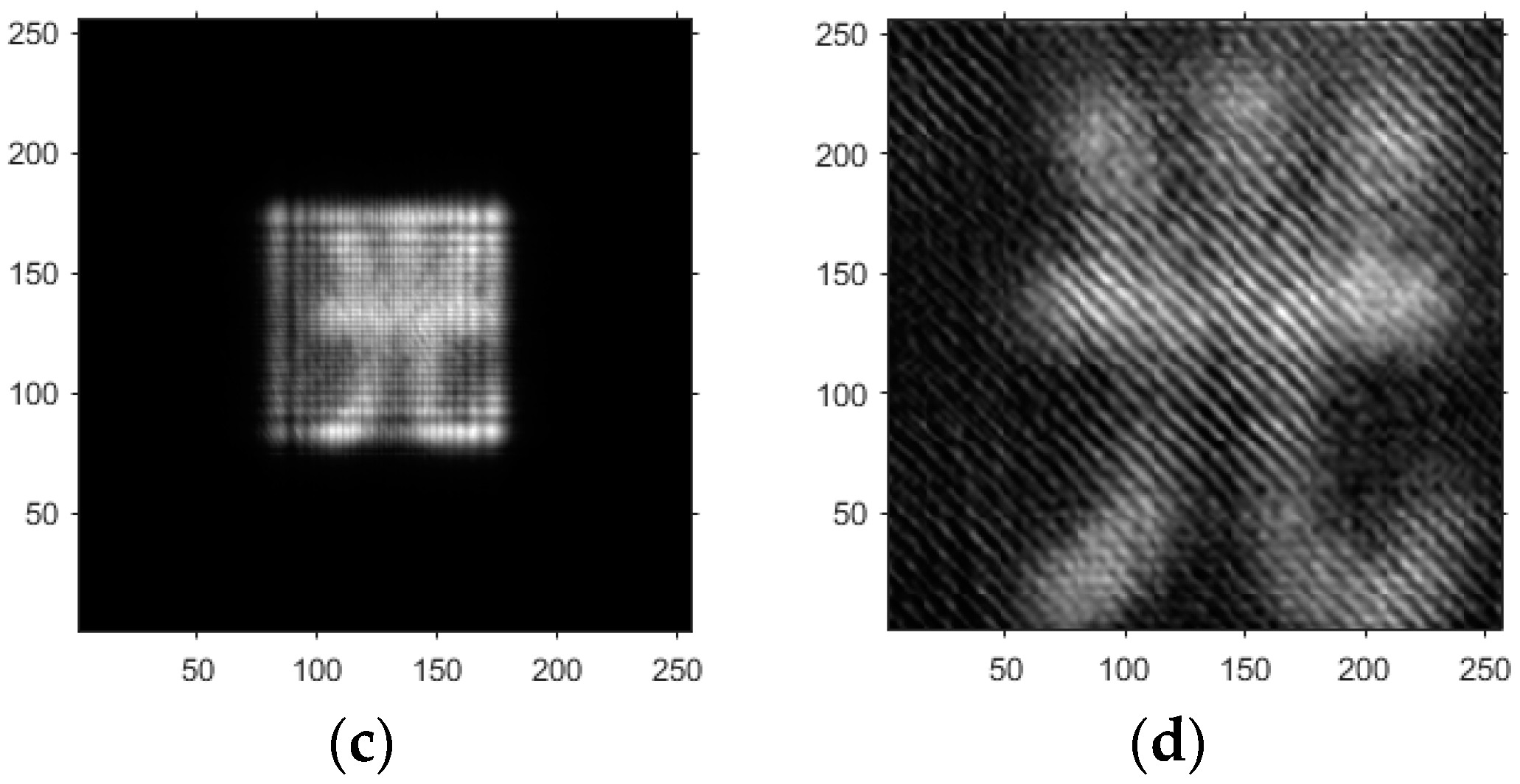
- Based on the above principles, five models were established and the superiority of the improved angular spectrum algorithm for the phase reconstruction of cell images was experimentally verified. Model 1, in digital holography experiments, based on the reproduction method of the improved angular spectrum method, created diffraction images of the object reference 1: 1 light field with diffraction distances of 10, 20, 30, and 40 cm; Model 2, in digital holography experiments, based on the reproduction method of the improved angular spectrum method, set up the zero-level image, the original image, and the conjugate image with the diffraction distances of 10, 20, 30, and 40 cm, respectively; Model 3, in the digital holography experiment, set up holographic spectral images with diffraction distances of 10, 20, 30, and 40 cm, based on the improved angular spectrum reproduction method; Model 4, in the digital holography experiment, set up CCD screen interference patterns with diffraction distances of 10, 20, 30, and 40 cm, based on the improved angular spectrum reproduction method; Model 5, in the digital holography experiment, set up diffraction distances of 10, 20, 30, and 40 cm for the restored image according to the improved angular spectrum reproduction method. The five models were processed accordingly using light word images and chromosome cell images, respectively, and the results are shown in Models 1–9, respectively.
- (4)
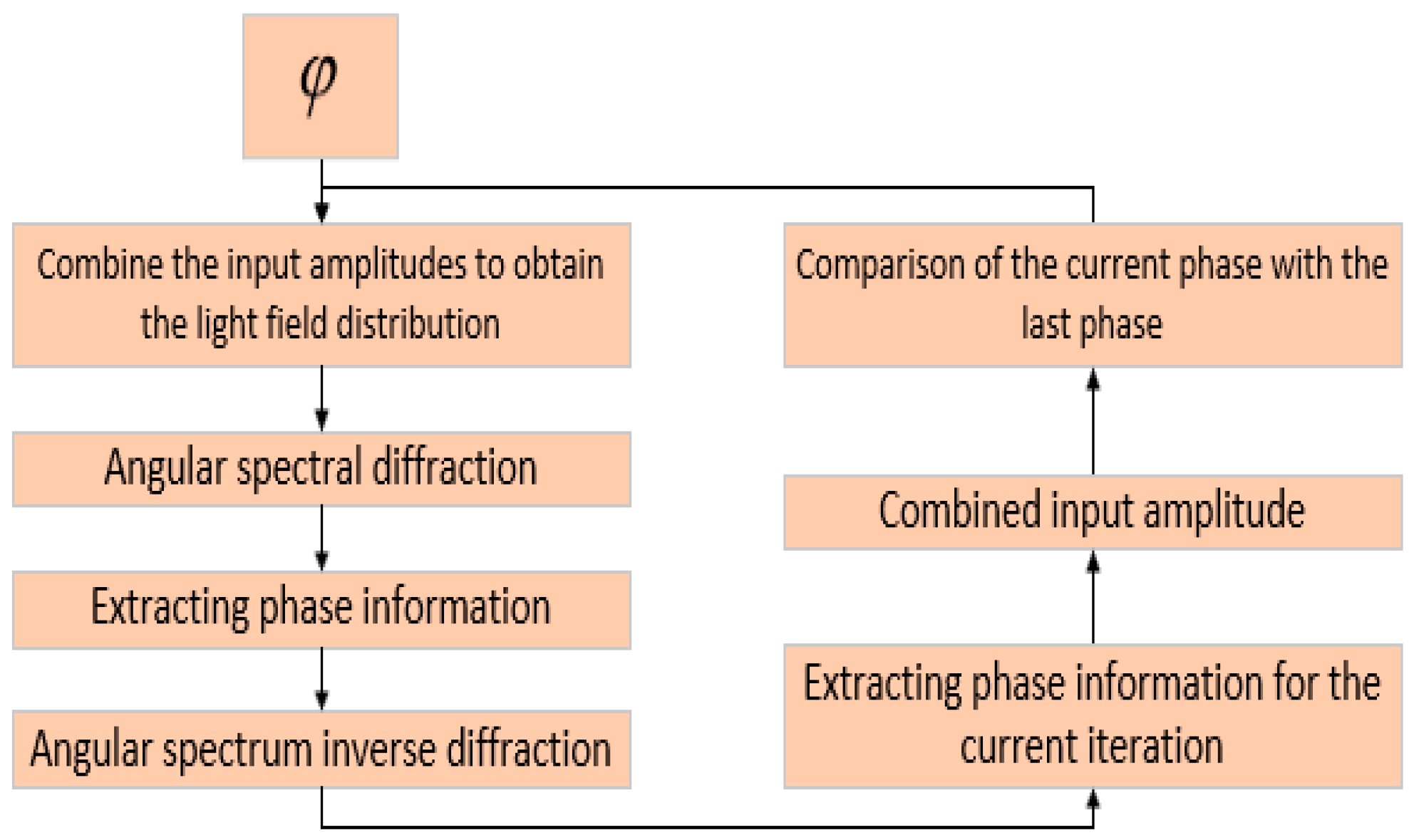
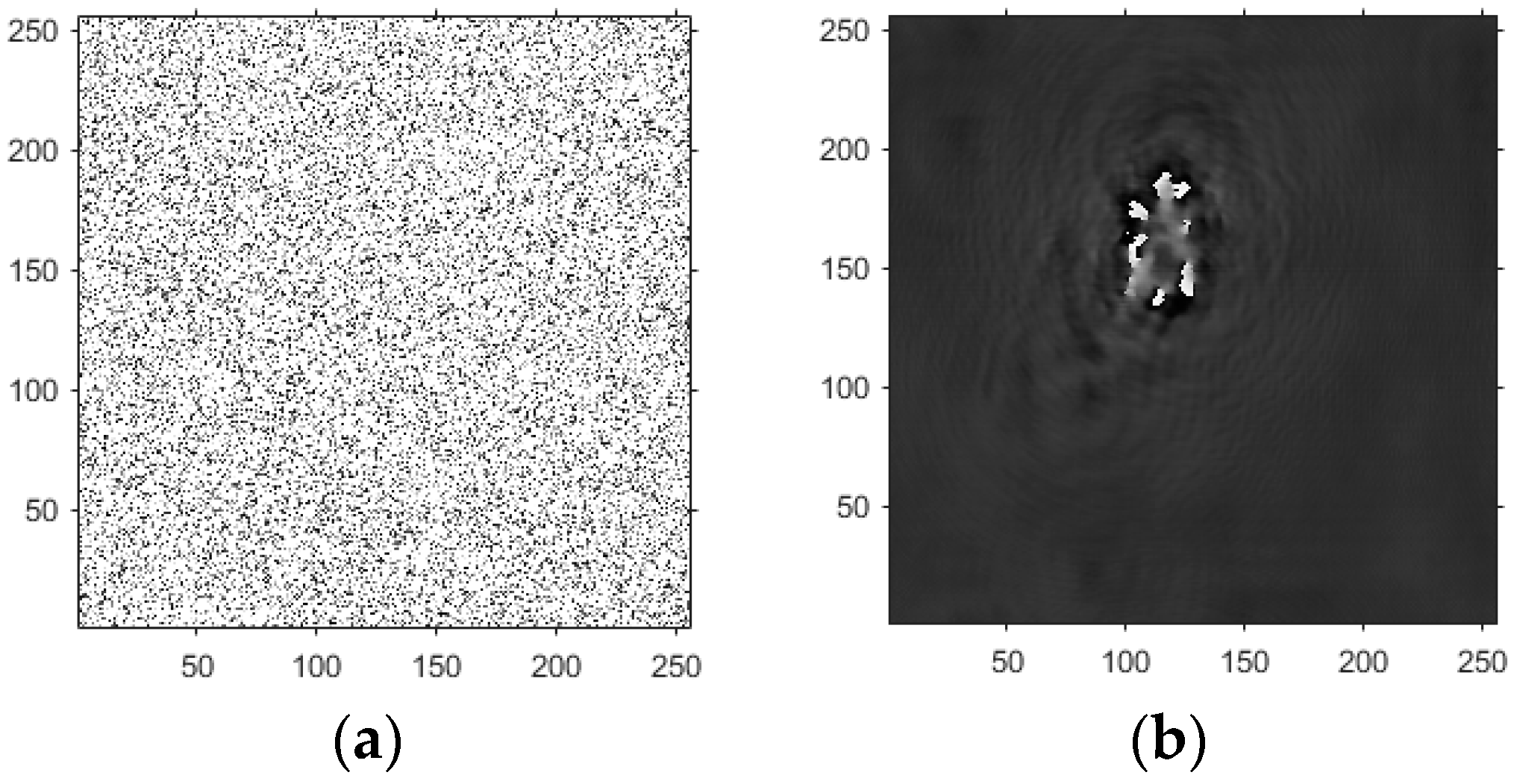
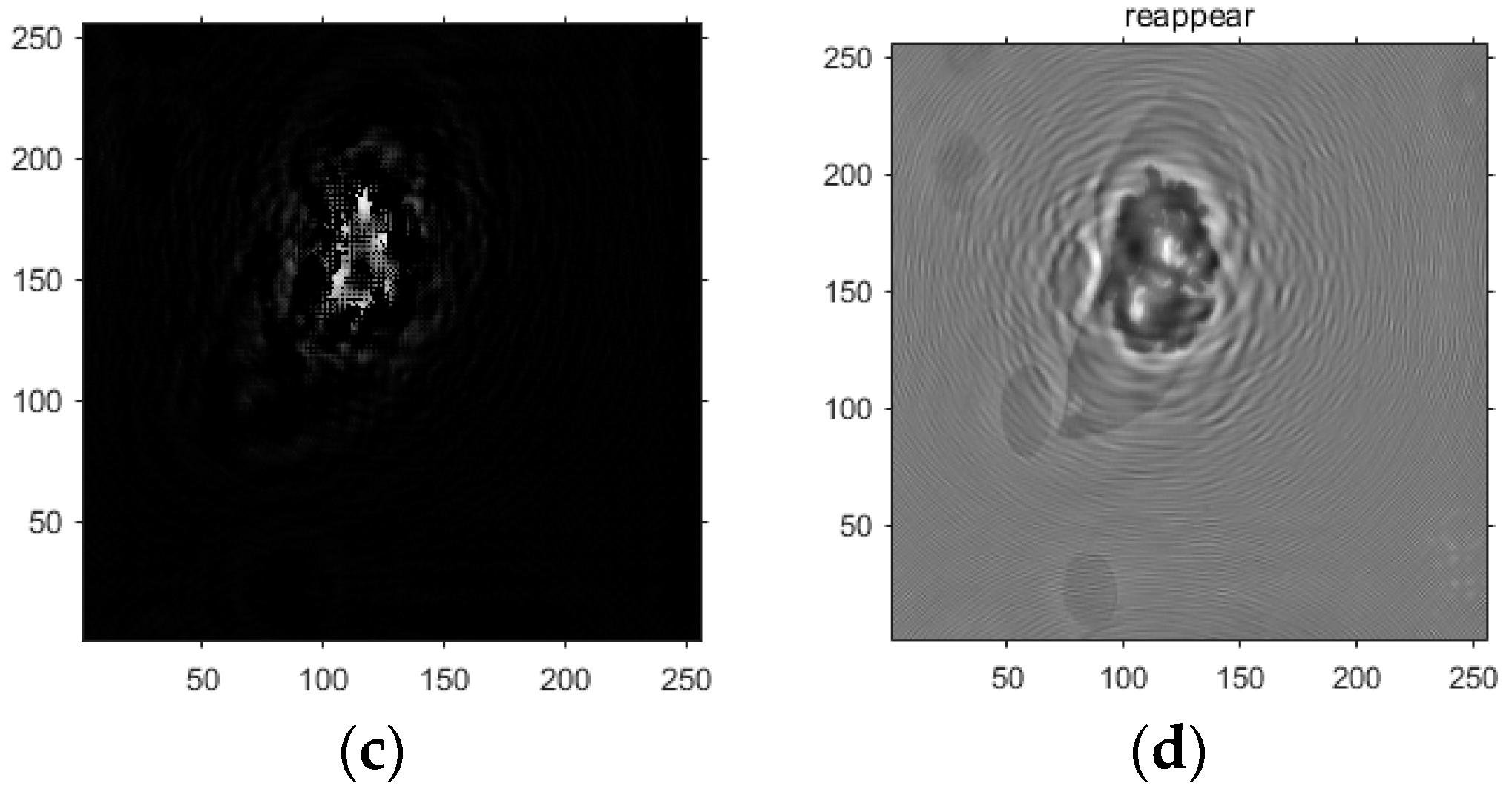
- Based on this model, the GS algorithm is combined with the improved angular spectrum method to accomplish the following steps. First, the extracted phase image is displayed. Perform normalization of the extracted phase image. Perform an inverse Fourier transform and phase extraction to display the phase from the extracted phase image. Finally, diffraction compensation is performed using the diffraction compensation function to obtain the reconstructed phase image. The results are displayed in Model 10.
4. Light Word and Chromosome Cell Computational Holography Experiments
4.1. Optical Character Computational Holography Experiment
4.2. Chromosome Cell Image Computational Holography Experiment
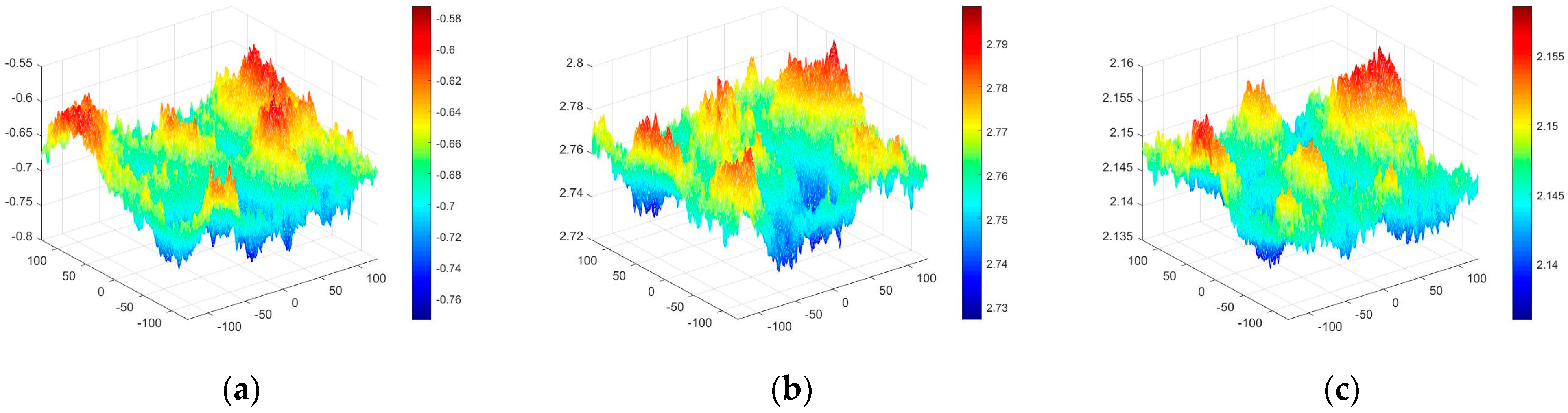
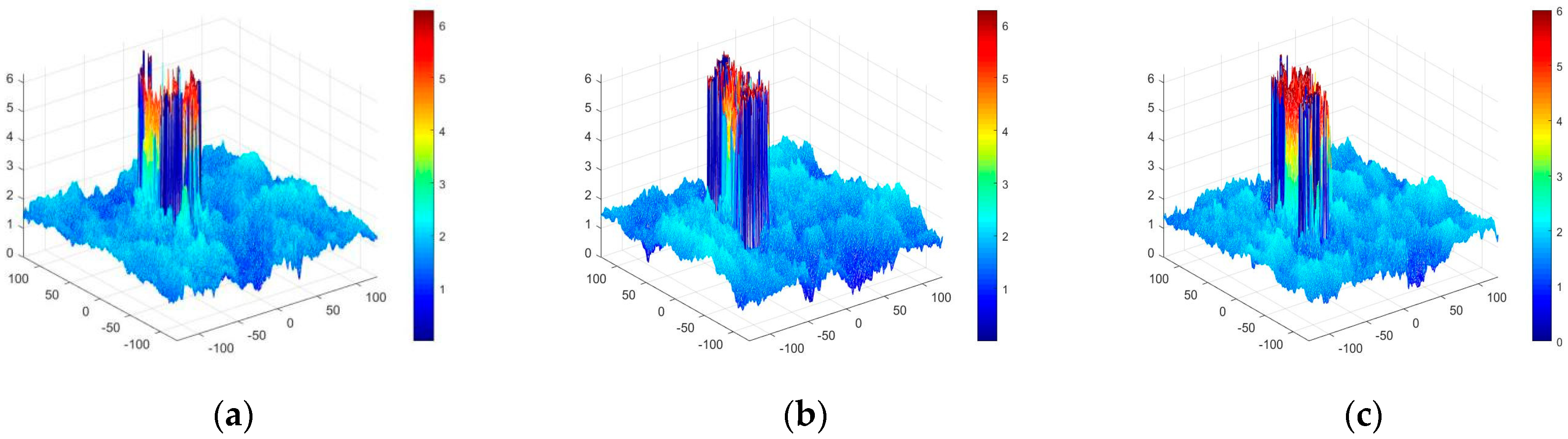
4.3. Experimental Discussion of Improved Angular Spectrum Method for Calculating Holography
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sypek, M.; Prokopowicz, C.; Gorecki, M. Image multiplying and high-frequency oscillations effects in the Fresnel region light propagation simulation. Opt. Eng. 2003, 42, 3158–3164. [Google Scholar] [CrossRef]
- Ge, B.; Lu, Q.; Zhang, Y. Particle digital in-line holography with spherical wave recording. Chin. Opt. Lett. 2003, 1, 517–519. [Google Scholar]
- Cavallini, L.; Bolognesi, G.; Di Leonardo, R. Real-time digital holographic microscopy of multiple and arbitrarily oriented planes. Opt. Lett. 2011, 36, 3491–3493. [Google Scholar] [CrossRef]
- Merola, F.; Miccio, L.; Paturzo, M.; Finizio, A.; Grilli, S.; Ferraro, P. Driving and analysis of micro-objects by digital holographic microscope in microfluidics. Opt. Lett. 2011, 36, 3079–3081. [Google Scholar] [CrossRef]
- Shin, D.; Daneshpanah, M.; Anand, A.; Javidi, B. Optofluidic system for three-dimensional sensing and identification of micro-organisms with digital holographic microscopy. Opt. Lett. 2010, 35, 4066–4068. [Google Scholar] [CrossRef]
- Kemper, B.; Von Bally, G. Digital holographic microscopy for live cell applications and technical inspection. Appl. Opt. 2008, 47, A52–A61. [Google Scholar] [CrossRef] [PubMed]
- Rappaz, B.; Marquet, P.; Cuche, E.; Emery, Y.; Depeursinge, C.; Pierre, J. Measurement of the integral refractive index and dynamic cell morphometry of living cells with digital holographic microscopy. Opt. Express 2005, 13, 9361–9373. [Google Scholar] [CrossRef] [PubMed]
- Javidi, B.; Moon, I.; Yeom, S.; Carapezza, E. Three-dimensional imaging and recognition of microorganism using single-exposure on-line (SEOL) digital holography. Opt. Express 2005, 13, 4492–4506. [Google Scholar] [CrossRef] [PubMed]
- Tian, J.; Xie, J.; He, Z.; Qin, D.; Wang, X. Modeling of an impedimetric biosensor with ultrasonic-assisted cell alignment for the detection of yeast. J. Sens. 2022, 2022, 4514218. [Google Scholar] [CrossRef]
- Wen, C.; Kong, Y.; Zhao, J.; Li, Y.; Yu, M.; Zeng, S.; Shi, Z.; Jiang, Q. Efficacy of the photon-initiated photoacoustic streaming combined with different solutions on Enterococcus faecalis in the root canals. Microsc. Res. Tech. 2020, 83, 647–657. [Google Scholar] [CrossRef]
- Otuboah, F.Y.; Jihong, Z.; Tianyun, Z.; Cheng, C. Design of a reduced objective Lens fluorescence dPCR Gene chip detection system with high-throughput and large field of view. Optik 2019, 179, 1071–1083. [Google Scholar] [CrossRef]
- Borovkova, M.; Trifonyuk, L.; Ushenko, V.; Dubolazov, O.; Vanchulyak, O.; Bodnar, G.; Ushenko, Y.; Olar, O.; Ushenko, O.; Sakhnovskiy, M.; et al. Mueller-matrix-based polarization imaging and quantitative assessment of optically anisotropic polycrystalline networks. PLoS ONE 2019, 14, e0214494. [Google Scholar] [CrossRef]
- Ushenko, V.; Sdobnov, A.; Syvokorovskaya, A.; Dubolazov, A.; Vanchulyak, O.; Ushenko, A.; Ushenko, Y.; Gorsky, M.; Sidor, M.; Bykov, A.; et al. 3D Mueller-matrix diffusive tomography of polycrystalline blood films for cancer diagnosis. Photonics 2018, 5, 54. [Google Scholar] [CrossRef]
- Peyvasteh, M.; Dubolazov, A.; Popov, A.; Ushenko, A.; Ushenko, Y.; Meglinski, I. Two-point Stokes vector diagnostic approach for characterization of optically anisotropic biological tissues. J. Phys. D Appl. Phys. 2020, 53, 395401. [Google Scholar] [CrossRef]
- Ushenko, V.A.; Sdobnov, A.Y.; Mishalov, W.D.; Dubolazov, A.V.; Olar, O.V.; Bachinskyi, V.T.; Ushenko, V.G.; Ushenko, Y.A.; Wanchuliak, O.Y.; Meglinski, I. Biomedical applications of Jones-matrix tomography to polycrystalline films of biological fluids. J. Innov. Opt. Health Sci. 2019, 12, 1950017. [Google Scholar] [CrossRef]
- Peyvasteh, M.; Tryfonyuk, L.; Ushenko, V.; Syvokorovskaya, A.; Dubolazov, A.; Vanchulyak, O.; Ushenko, A.; Ushenko, Y.; Gorsky, M.; Sidor, M. 3D Mueller-matrix-based azimuthal invariant tomography of polycrystalline structure within benign and malignant soft-tissue tumours. Laser Phys. Lett. 2020, 17, 115606. [Google Scholar] [CrossRef]
- Ma, F.Y.; Wang, X.; Bu, Y.Z.; Tian, Y.Z.; Du, Y.L.; Gong, Q.X.; Zhuang, C.Y.; Li, J.H.; Li, L. Incoherent digital holographic spectral imaging with high accuracy of image pixel registration. Chin. Phys. B 2021, 30, 044202. [Google Scholar] [CrossRef]
- Sasaki, H.; Yamamoto, K.; Wakunami, K.; Ichihashi, Y.; Oi, R.; Senoh, T. Large size three-dimensional video by electronic holography using multiple spatial light modulators. Sci. Rep. 2014, 4, 6177. [Google Scholar] [CrossRef]
- Abdelazeem, R.M.; Ghareab Abdelsalam Ibrahim, D. Discrimination between normal and cancer white blood cells using holographic projection technique. PLoS ONE 2022, 17, e0276239. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.X.; Li, Z.Y.; Tian, Y.; Wang, W.; Pang, Y.; Tam, K.Y. Two dimensional photoacoustic imaging using microfiber interferometric acoustic transducers. Opt. Commun. 2018, 419, 41–46. [Google Scholar] [CrossRef]
- Wang, X.; Jiang, Y.; Li, Z.; Wang, W.; Li, Z. Sensitivity characteristics of microfiber Fabry-Perot interferometric photoacoustic sensors. J. Light. Technol. 2019, 37, 4229–4235. [Google Scholar] [CrossRef]
- Wang, X.; Jin, L.; Li, J.; Ran, Y.; Guan, B. Microfiber interferometric acoustic transducers. Opt. Express 2014, 22, 8126–8135. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Scott, P.D. Phase retrieval and twin-image elimination for in-line Fresnel holograms. JOSA A 1987, 4, 159–165. [Google Scholar] [CrossRef]
- Cui, H.; Wang, D.; Wang, Y.; Liu, C.; Zhao, J.; Li, Y. Automatic procedure for non-coplanar aberration compensation in lensless Fourier transform digital holography. In Proceedings of the 5th International Symposium on Advanced Optical Manufacturing and Testing Technologies: Optical Test and Measurement Technology and Equipment, Dalian, China, 26–29 April 2010; SPIE: Bellingham, WA, USA, 2010; Volume 7656, pp. 213–219. [Google Scholar]
- Jin, D.; Zhou, R.; Yaqoob, Z.; Peter, T.C. Tomographic phase microscopy: Principles and applications in bioimaging. JOSA B 2017, 34, B64–B77. [Google Scholar] [CrossRef] [PubMed]
- Weng, J.; Zhong, J.; Hu, C. Digital reconstruction based on angular spectrum diffraction with the ridge of wavelet transform in holographic phase-contrast microscopy. Opt. Express 2008, 16, 21971–21981. [Google Scholar] [CrossRef] [PubMed]
- Langehanenberg, P.; Bally, G.; Kemper, B. Application of partially coherent light in live cell imaging with digital holographic microscopy. J. Mod. Opt. 2010, 57, 709–717. [Google Scholar] [CrossRef]
- Shaked, N.T.; Micó, V.; Trusiak, M.; Kuś, A.; Mirsky, S.K. Off-axis digital holographic multiplexing for rapid wavefront acquisition and processing. Adv. Opt. Photonics 2020, 12, 556–611. [Google Scholar] [CrossRef]
- Picazo-Bueno, J.Á.; Barroso, Á.; Ketelhut, S.; Schnekenburger, J.; Micó, V.; Kemper, B. Single capture bright field and off-axis digital holographic microscopy. Opt. Lett. 2023, 48, 876–879. [Google Scholar] [CrossRef]
- Zhang, W.; Li, B.; Zhang, X.; Shi, C. Off-axis digital holography based on the Sagnac interferometer. Laser Phys. Lett. 2021, 18, 035202. [Google Scholar] [CrossRef]
- Chen, H.C.; Cheng, C.J. Holographic Optical Tweezers: Techniques and Biomedical Applications. Appl. Sci. 2022, 12, 10244. [Google Scholar] [CrossRef]
- Wang, X.; Yang, Y.; Wu, T.; Zhu, H.; Yu, J.; Tian, J.; Li, H. Energy minimization segmentation model based on MRI images. Front. Neurosci. 2023, 17, 1175451. [Google Scholar] [CrossRef] [PubMed]
- Schnars, U.; Jüptner, W.P.O. Digital recording and numerical reconstruction of holograms. Meas. Sci. Technol. 2002, 13, R85. [Google Scholar] [CrossRef]
- Goodman, J.W.; Sutton, P. Introduction to Fourier optics. Quantum Semiclassical Opt.-J. Eur. Opt. Soc. Part B 1996, 8, 1095. [Google Scholar] [CrossRef]
- Sahu, P.; Mazumder, N. Advances in adaptive optics–based two-photon fluorescence microscopy for brain imaging. Lasers Med. Sci. 2020, 35, 317–328. [Google Scholar] [CrossRef]
- Balasubramani, V.; Kuś, A.; Tu, H.Y.; Cheng, C.J.; Baczewska, M.; Krauze, W.; Kujawińska, M. Holographic tomography: Techniques and biomedical applications. Appl. Opt. 2021, 60, B65–B80. [Google Scholar] [CrossRef]
- May, M.A.; Barré, N.; Kummer, K.K.; Kress, M.; Ritsch-Marte, M.; Jesacher, A. Fast holographic scattering compensation for deep tissue biological imaging. Nat. Commun. 2021, 12, 4340. [Google Scholar] [CrossRef]
- Dar, A.U.; Ahanger, A.B.; Rasool, M.; Singh, M.; Assad, A.; Macha, M.A.; Aalam, S.W.; Ahanger, A.N. Applications of Artificial Intelligence and Digital Holography in Biomedical Microscopy. Authorea 2023. [Google Scholar] [CrossRef]
- Jiang, Y.; Li, H.; Pang, Y.; Ling, J.; Wang, H.; Yang, Y.; Li, X.; Tian, Y.; Wang, X. Cell image reconstruction using digital holography with an improved GS algorithm. Front. Physiol. 2022, 13, 1040777. [Google Scholar] [CrossRef]
- Vishniakou, I.; Seelig, J.D. Differentiable Adaptive Optics and Light Shaping in Two-Photon Microscopy. In Proceedings of the Optics and the Brain, Vancouver, BC, Canada, 24–27 April 2023; Optica Publishing Group: Bellingham, WA, USA, 2023; p. BW3B.5. [Google Scholar]
- Vishniakou, I.; Seelig, J.D. Differentiable optimization of the Debye-Wolf integral for light shaping and adaptive optics in two-photon microscopy. Opt. Express 2023, 31, 9526–9542. [Google Scholar] [CrossRef]
- Rubart, M. Two-photon microscopy of cells and tissue. Circ. Res. 2004, 95, 1154–1166. [Google Scholar] [CrossRef]
- Yoon, S.; Cheon, S.Y.; Park, S.; Lee, D.; Lee, Y.; Han, S.; Kim, M.; Koo, H. Recent advances in optical imaging through deep tissue: Imaging probes and techniques. Biomater. Res. 2022, 26, 1–27. [Google Scholar] [CrossRef] [PubMed]
- Qin, Z.; He, S.; Yang, C.; Yung, J.S.Y.; Chen, C.; Leung, C.K.S.; Liu, K.; Qu, J.Y. Adaptive optics two-photon microscopy enables near-diffraction-limited and functional retinal imaging in vivo. Light Sci. Appl. 2020, 9, 79. [Google Scholar] [CrossRef] [PubMed]
- Di, J.; Song, Y.; Xi, T.; Zhang, J.; Li, Y.; Ma, C.; Wang, K.; Zhao, J. Dual-wavelength common-path digital holographic microscopy for quantitative phase imaging of biological cells. Opt. Eng. 2017, 56, 111712. [Google Scholar] [CrossRef]
- Liao, R.; Han, D.; Zeng, Y. Laser Speckle Micro-angiography. In Proceedings of the Digital Holography and Three-Dimensional Imaging, Shanghai, China, 24–28 May 2015; Optica Publishing Group: Bellingham, WA, USA, 2015; p. DW2A.21. [Google Scholar] [CrossRef]
- Cheng, H.; Luo, Q.; Zeng, S.; Chen, S.; Cen, J.; Gong, H. Modified laser speckle imaging method with improved spatial resolution. J. Biomed. Opt. 2003, 8, 559–564. [Google Scholar] [CrossRef] [PubMed]
- Jafari, C.Z.; Crouzet, C.; Phan, T.; Wilson, R.H.; Shin, T.J.; Choi, B. Intrinsic, widefield optical imaging of hemodynamics in rodent models of Alzheimer’s disease and neurological injury. Neurophotonics 2023, 10, 020601. [Google Scholar]
- Abud, M.M. Laser Doppler Using Holographic. Int. J. Emerg. Trends Technol. Comput. Sci. 2014, 3, 97–101. [Google Scholar]
- Qureshi, M.M.; Allam, N.; Im, J.; Kwon, H.S.; Chung, E.; Vitkin, I.A. Advances in Laser Speckle Imaging: From qualitative to quantitative hemodynamic assessment. J. Biophotonics 2023, e202300126. [Google Scholar] [CrossRef]
- Fong, R.Y.; Yuan, F.G. Phase estimation via riesz transform in laser speckle interferometry for large-area damage imaging. NDT E Int. 2022, 132, 102711. [Google Scholar] [CrossRef]
- Vaz, P.G.; Humeau-Heurtier, A.; Figueiras, E.; Correia, C.; Cardoso, J. Laser speckle imaging to monitor microvascular blood flow: A review. IEEE Rev. Biomed. Eng. 2016, 9, 106–120. [Google Scholar] [CrossRef]




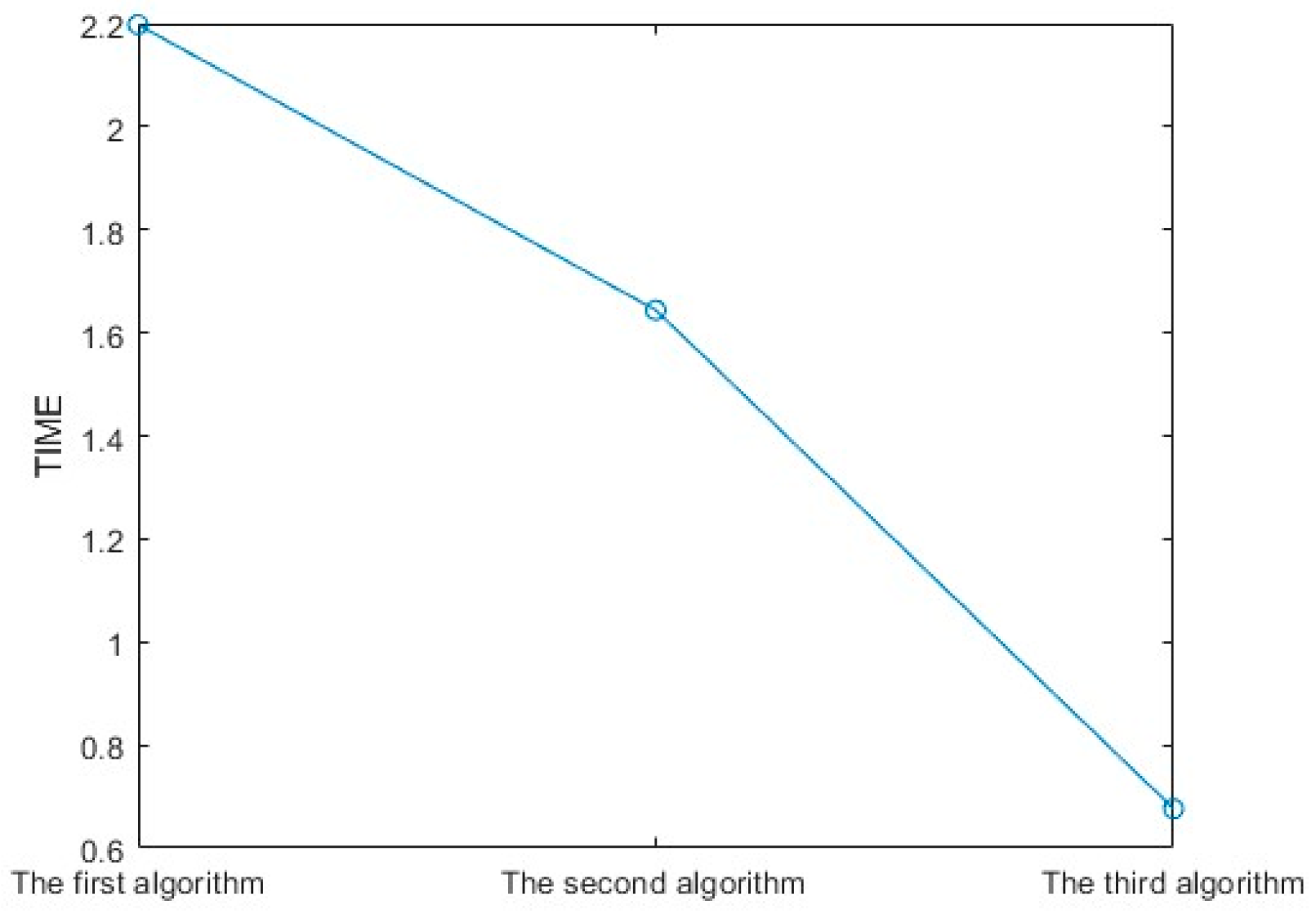
| RMSE of the Angular Spectrum Method | RMSE of the Angular Spectrum Method Combined with the GS Algorithm | RMSE of the Improved Angular Spectrum Method |
|---|---|---|
| 0.4610 | 0.2836 | 0.1361 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, T.; Yang, Y.; Wang, H.; Chen, H.; Zhu, H.; Yu, J.; Wang, X. Investigation of an Improved Angular Spectrum Method Based on Holography. Photonics 2024, 11, 16. https://doi.org/10.3390/photonics11010016
Wu T, Yang Y, Wang H, Chen H, Zhu H, Yu J, Wang X. Investigation of an Improved Angular Spectrum Method Based on Holography. Photonics. 2024; 11(1):16. https://doi.org/10.3390/photonics11010016
Chicago/Turabian StyleWu, Ting, Yuling Yang, Hao Wang, Hao Chen, Hao Zhu, Jisheng Yu, and Xiuxin Wang. 2024. "Investigation of an Improved Angular Spectrum Method Based on Holography" Photonics 11, no. 1: 16. https://doi.org/10.3390/photonics11010016
APA StyleWu, T., Yang, Y., Wang, H., Chen, H., Zhu, H., Yu, J., & Wang, X. (2024). Investigation of an Improved Angular Spectrum Method Based on Holography. Photonics, 11(1), 16. https://doi.org/10.3390/photonics11010016





