Characterization of the Binding Behavior of Specific Cobalt and Nickel Ion-Binding Peptides Identified by Phage Surface Display
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemicals and Buffers
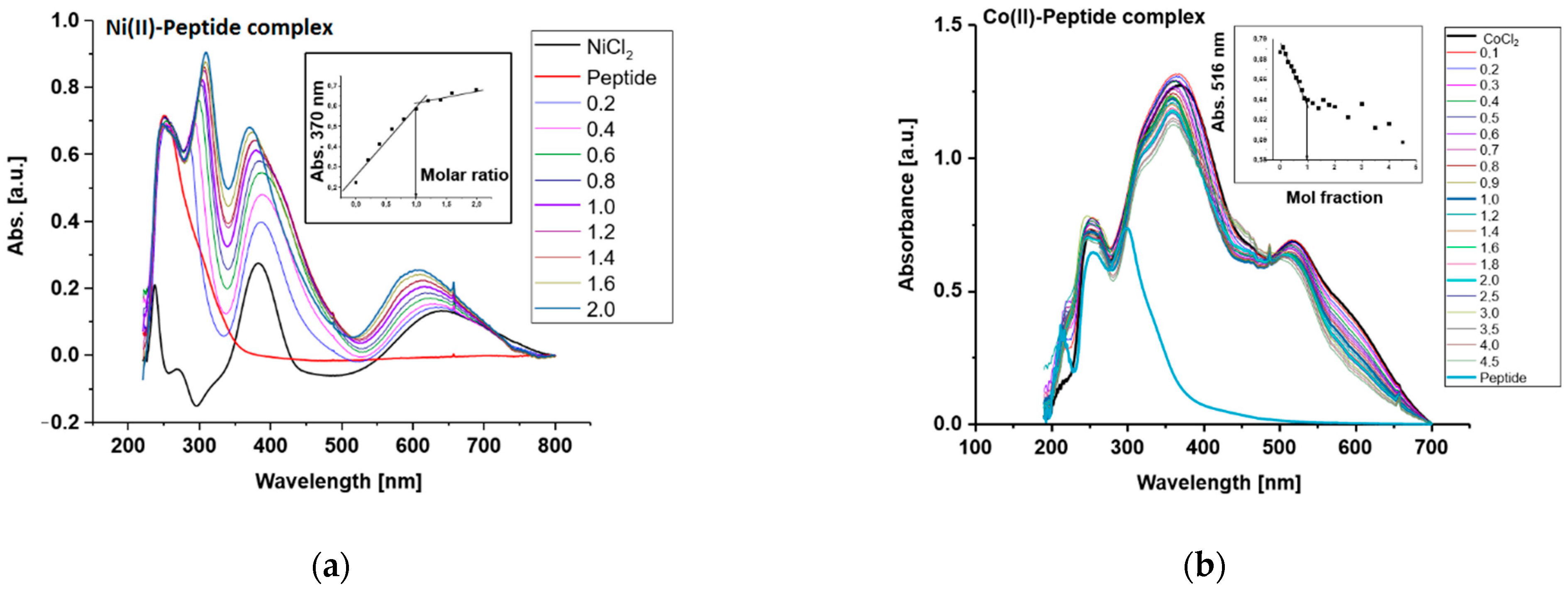
2.2. UV/VIS Spectroscopy
2.3. Isothermal Titration Calorimetry
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gislev, M.; Grohol, M. Report on Critical Raw Materials and the Circular Economy; Directorate-General for Internal Market, Industry, Entrepreneurship and SMEs, EU Commission: Brussels, Belgium, 2018. [Google Scholar]
- Graf, P. Die Werkzeuge der Bioökonomie; BMBF: Berlin, Germany, 2021. [Google Scholar]
- BMBF. Bioökonomie Als Gesellschaftlicher Wandel; BMBF: Berlin, Germany, 2021. [Google Scholar]
- Packer, M.S.; Liu, D.R. Methods for the directed evolution of proteins. Nat. Rev. Genet. 2015, 16, 379–394. [Google Scholar] [CrossRef] [PubMed]
- Cobb, R.E.; Chao, R.; Zhao, H. Directed Evolution: Past, Present and Future. AIChE J. 2013, 59, 1432–1440. [Google Scholar] [CrossRef] [PubMed]
- Bozovičar, K.; Bratkovič, T. Evolving a Peptide: Library Platforms and Diversification Strategies. Int. J. Mol. Sci. 2019, 21, 215. [Google Scholar] [CrossRef]
- Wang, Y.; Xue, P.; Cao, M.; Yu, T.; Lane, S.T.; Zhao, H. Directed Evolution: Methodologies and Applications. Chem. Rev. 2021, 121, 12384–12444. [Google Scholar] [CrossRef]
- Schmidt-Dannert, C. Directed Evolution of Single Proteins, Metabolic Pathways, and Viruses. Biochemistry 2001, 40, 13125–13136. [Google Scholar] [CrossRef] [PubMed]
- DeBenedictis, E.A.; Chory, E.J.; Gretton, D.W.; Wang, B.; Golas, S.; Esvelt, K.M. Systematic molecular evolution enables robust biomolecule discovery. Nat. Methods 2022, 19, 55–64. [Google Scholar] [CrossRef] [PubMed]
- Smith, G.P. Filamentous fusion phage: Novel expression vectors that display cloned antigens on the virion surface. Science 1985, 228, 1315–1317. [Google Scholar] [CrossRef] [PubMed]
- Smith, G.P.; Petrenko, V.A. Phage Display. Chem. Rev. 1997, 97, 391–410. [Google Scholar] [CrossRef]
- Arap, M.A. Phage display technology: Applications and innovations. Genet. Mol. Biol. 2005, 28, 1–9. [Google Scholar] [CrossRef]
- Jaroszewicz, W.; Morcinek-Orłowska, J.; Pierzynowska, K.; Gaffke, L.; Węgrzyn, G. Phage display and other peptide display technologies. FEMS Microbiol. Rev. 2022, 46, fuab052. [Google Scholar] [CrossRef]
- He, B.; Jiang, L.; Duan, Y.; Chai, G.; Fang, Y.; Kang, J.; Yu, M.; Li, N.; Tang, Z.; Yao, P.; et al. Biopanning data bank 2018: Hugging next generation phage display. Database 2018, 2018, bay032. [Google Scholar] [CrossRef] [PubMed]
- Azzazy, H.M.E.; Highsmith, W.E. Phage display technology: Clinical applications and recent innovations. Clin. Biochem. 2002, 35, 425–445. [Google Scholar] [CrossRef]
- Fack, F.; Hügle-Dörr, B.; Song, D.; Queitsch, I.; Petersen, G.; Bautz, E.K.F. Epitope mapping by phage display: Random versus gene-fragment libraries. J. Immunol. Methods 1997, 206, 43–52. [Google Scholar] [CrossRef]
- Moreira, G.M.S.G.; Fühner, V.; Hust, M. Epitope Mapping by Phage Display. In Phage Display: Methods and Protocols; Hust, M., Lim, T.S., Eds.; Springer: New York, NY, USA, 2018; pp. 497–518. [Google Scholar]
- Nemudraya, A.A.; Richter, V.A.; Kuligina, E.V. Phage Peptide Libraries As a Source of Targeted Ligands. Acta Naturae 2016, 8, 48–57. [Google Scholar] [CrossRef]
- Qi, H.; Ma, M.; Lai, D.; Tao, S.-C. Phage display: An ideal platform for coupling protein to nucleic acid. Acta Biochim. Biophys. Sin. 2021, 53, 389–399. [Google Scholar] [CrossRef] [PubMed]
- Nagano, K.; Tsutsumi, Y. Phage Display Technology as a Powerful Platform for Antibody Drug Discovery. Viruses 2021, 13, 178. [Google Scholar] [CrossRef]
- Sidhu, S.S.; Fairbrother, W.J.; Deshayes, K. Exploring protein-protein interactions with phage display. Chembiochem 2003, 4, 14–25. [Google Scholar] [CrossRef] [PubMed]
- Sundell, G.N.; Ivarsson, Y. Interaction analysis through proteomic phage display. Biomed. Res. Int. 2014, 2014, 176172. [Google Scholar] [CrossRef]
- Huang, W.; Soeung, V.; Boragine, D.M.; Palzkill, T. Mapping Protein–Protein Interaction Interface Peptides with Jun-Fos Assisted Phage Display and Deep Sequencing. ACS Synth. Biol. 2020, 9, 1882–1896. [Google Scholar] [CrossRef]
- Rakonjac, J.; Bennett, N.J.; Spagnuolo, J.; Gagic, D.; Russel, M. Filamentous bacteriophage: Biology, phage display and nanotechnology applications. Curr. Issues Mol. Biol. 2011, 13, 51–76. [Google Scholar]
- Seker, U.O.S.; Demir, H.V. Material Binding Peptides for Nanotechnology. Molecules 2011, 16, 1426–1451. [Google Scholar] [CrossRef] [PubMed]
- Paczesny, J.; Bielec, K. Application of Bacteriophages in Nanotechnology. Nanomaterials 2020, 10, 1944. [Google Scholar] [CrossRef] [PubMed]
- Hyman, P.; Denyes, J. Bacteriophages in Nanotechnology: History and Future. In Bacteriophages: Biology, Technology, Therapy; Harper, D.R., Abedon, S.T., Burrowes, B.H., McConville, M.L., Eds.; Springer International Publishing: Cham, Switzerland, 2021; pp. 657–687. [Google Scholar]
- Vreuls, C.; Zocchi, G.; Genin, A.; Archambeau, C.; Martial, J.; Van de Weerdt, C. Inorganic-binding peptides as tools for surface quality control. J. Inorg. Biochem. 2010, 104, 1013–1021. [Google Scholar] [CrossRef] [PubMed]
- Kriplani, U.; Kay, B.K. Selecting peptides for use in nanoscale materials using phage-displayed combinatorial peptide libraries. Curr. Opin. Biotechnol. 2005, 16, 470–475. [Google Scholar] [CrossRef] [PubMed]
- Sawada, T. Filamentous virus-based soft materials based on controlled assembly through liquid crystalline formation. Polym. J. 2017, 49, 639–647. [Google Scholar] [CrossRef]
- Machera, S.J.; Niedziółka-Jönsson, J.; Szot-Karpińska, K. Phage-Based Sensors in Medicine: A Review. Chemosensors 2020, 8, 61. [Google Scholar] [CrossRef]
- Peltomaa, R.; Benito-Peña, E.; Barderas, R.; Moreno-Bondi, M.C. Phage Display in the Quest for New Selective Recognition Elements for Biosensors. ACS Omega 2019, 4, 11569–11580. [Google Scholar] [CrossRef]
- Wu, J.; Park, J.P.; Dooley, K.; Cropek, D.M.; West, A.C.; Banta, S. Rapid Development of New Protein Biosensors Utilizing Peptides Obtained via Phage Display. PLoS ONE 2011, 6, e24948. [Google Scholar] [CrossRef]
- Tan, Y.; Tian, T.; Liu, W.; Zhu, Z.; Yang, C. Advance in phage display technology for bioanalysis. Biotechnol. J. 2016, 11, 732–745. [Google Scholar] [CrossRef]
- Nian, R.; Kim, D.S.; Nguyen, T.; Tan, L.; Kim, C.-W.; Yoo, I.-K.; Choe, W.-S. Chromatographic biopanning for the selection of peptides with high specificity to Pb2+ from phage displayed peptide library. J. Chromatogr. A 2010, 1217, 5940–5949. [Google Scholar] [CrossRef]
- Schönberger, N.; Braun, R.; Matys, S.; Lederer, F.L.; Lehmann, F.; Flemming, K.; Pollmann, K. Chromatopanning for the identification of gallium binding peptides. J. Chromatogr. A 2019, 1600, 158–166. [Google Scholar] [CrossRef]
- Braun, R.; Bachmann, S.; Schönberger, N.; Matys, S.; Lederer, F.; Pollmann, K. Peptides as biosorbents—Promising tools for resource recovery. Res. Microbiol. 2018, 169, 649–658. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Dong, W.; Liu, Y.; Zhang, H.; Wang, G. Enhanced Biosorption of Nickel Ions on Immobilized Surface-Engineered Yeast Using Nickel-Binding Peptides. Front. Microbiol. 2019, 10, 1254. [Google Scholar] [CrossRef] [PubMed]
- Matys, S.; Schönberger, N.; Lederer, F.L.; Pollmann, K. Characterization of specifically metal-binding phage clones for selective recovery of cobalt and nickel. J. Environ. Chem. Eng. 2020, 8, 103606. [Google Scholar] [CrossRef]
- Ghai, R.; Falconer, R.J.; Collins, B.M. Applications of isothermal titration calorimetry in pure and applied research—Survey of the literature from 2010. J. Mol. Recognit. 2011, 25, 32–52. [Google Scholar] [CrossRef] [PubMed]
- Brynn Hibbert, D.; Thordarson, P. The death of the Job plot, transparency, open science and online tools, uncertainty estimation methods and other developments in supramolecular chemistry data analysis. ChemComm 2016, 52, 12792–12805. [Google Scholar] [CrossRef]
- Olson, E.J.; Bühlmann, P. Getting More out of a Job Plot: Determination of Reactant to Product Stoichiometry in Cases of Displacement Reactions and n:n Complex Formation. J. Org. Chem. 2011, 76, 8406–8412. [Google Scholar] [CrossRef]
- Renny, J.S.; Tomasevich, L.L.; Tallmadge, E.H.; Collum, D.B. Method of continuous variations: Applications of job plots to the study of molecular associations in organometallic chemistry. Angew. Chem. Int. Ed. 2013, 52, 11998–12013. [Google Scholar] [CrossRef]
- Dragan, A.I.; Read, C.M.; Crane-Robinson, C. Enthalpy–entropy compensation: The role of solvation. Eur. Biophys. J. 2017, 46, 301–308. [Google Scholar] [CrossRef]
- Starikov, E.B.; Nordén, B. Enthalpy−Entropy Compensation: A Phantom or Something Useful? J. Phys. Chem. B 2007, 111, 14431–14435. [Google Scholar] [CrossRef]
- Ryde, U. A fundamental view of enthalpy–entropy compensation. MedChemComm 2014, 5, 1324–1336. [Google Scholar] [CrossRef]
- Bou-Abdallah, F.; Giffune, T.R. The thermodynamics of protein interactions with essential first row transition metals. Biochim. Biophys. Acta 2016, 1860, 879–891. [Google Scholar] [CrossRef]
- Johnson, R.A.; Manley, O.M.; Spuches, A.M.; Grossoehme, N.E. Dissecting ITC data of metal ions binding to ligands and proteins. Biochim. Biophys. Acta 2016, 1860, 892–901. [Google Scholar] [CrossRef]
- Grossoehme, N.E.; Spuches, A.M.; Wilcox, D.E. Application of isothermal titration calorimetry in bioinorganic chemistry. J. Biol. Inorg. Chem. 2010, 15, 1183–1191. [Google Scholar] [CrossRef] [PubMed]
- Wyrzykowski, D.; Pilarski, B.; Jacewicz, D.; Chmurzyński, L. Investigation of metal–buffer interactions using isothermal titration calorimetry. J. Therm. Anal. Calorim. 2013, 111, 1829–1836. [Google Scholar] [CrossRef]
- Nastyshyn, S.; Pop-Georgievski, O.; Stetsyshyn, Y.; Budkowski, A.; Raczkowska, J.; Hruby, M.; Lobaz, V. Protein corona of SiO2 nanoparticles with grafted thermoresponsive copolymers: Calorimetric insights on factors affecting entropy vs. enthalpy-driven associations. Appl. Surf. Sci. 2022, 601, 154201. [Google Scholar] [CrossRef]
- Prozeller, D.; Morsbach, S.; Landfester, K. Isothermal titration calorimetry as a complementary method for investigating nanoparticle-protein interactions. Nanoscale 2019, 11, 19265–19273. [Google Scholar] [CrossRef]
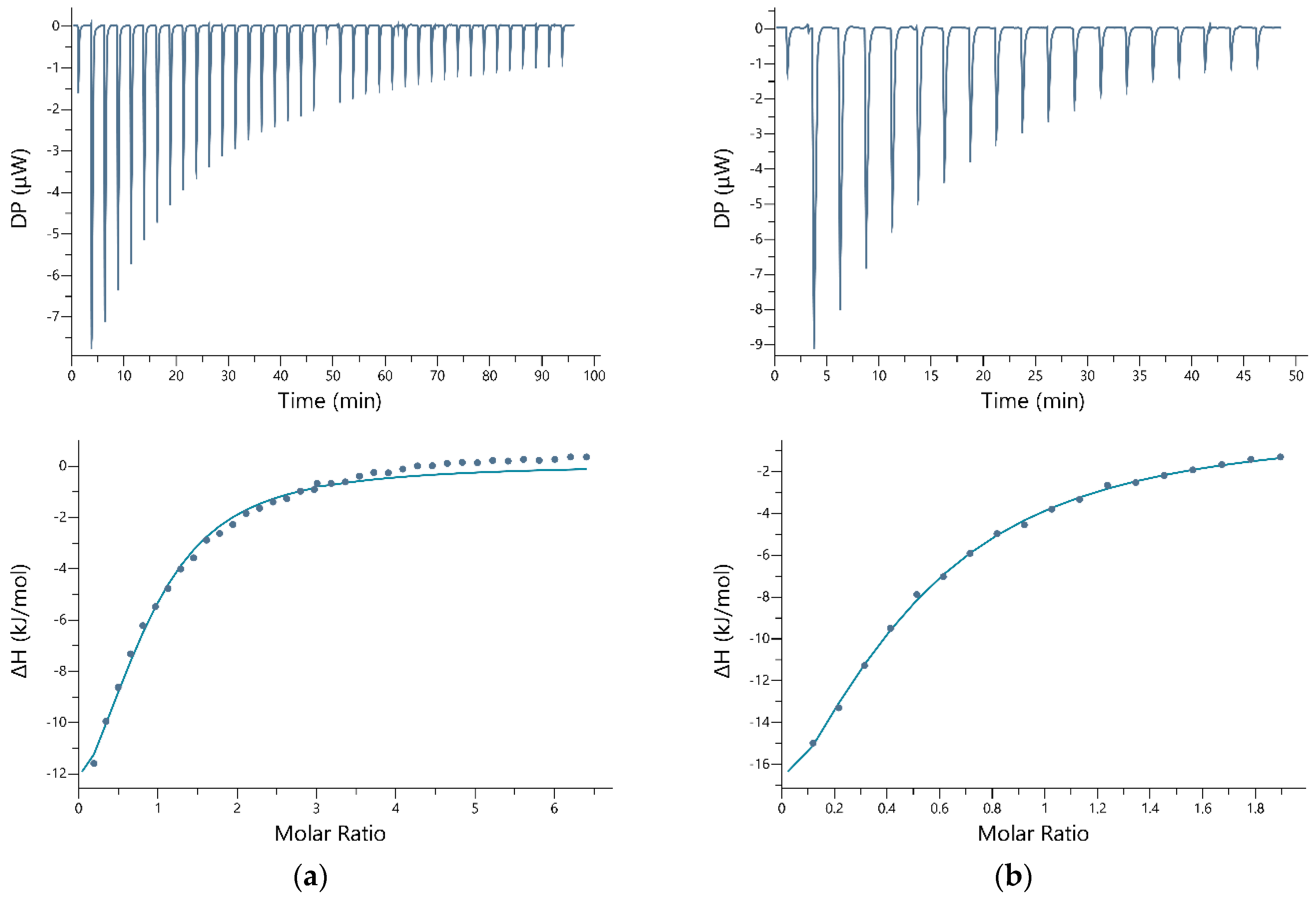
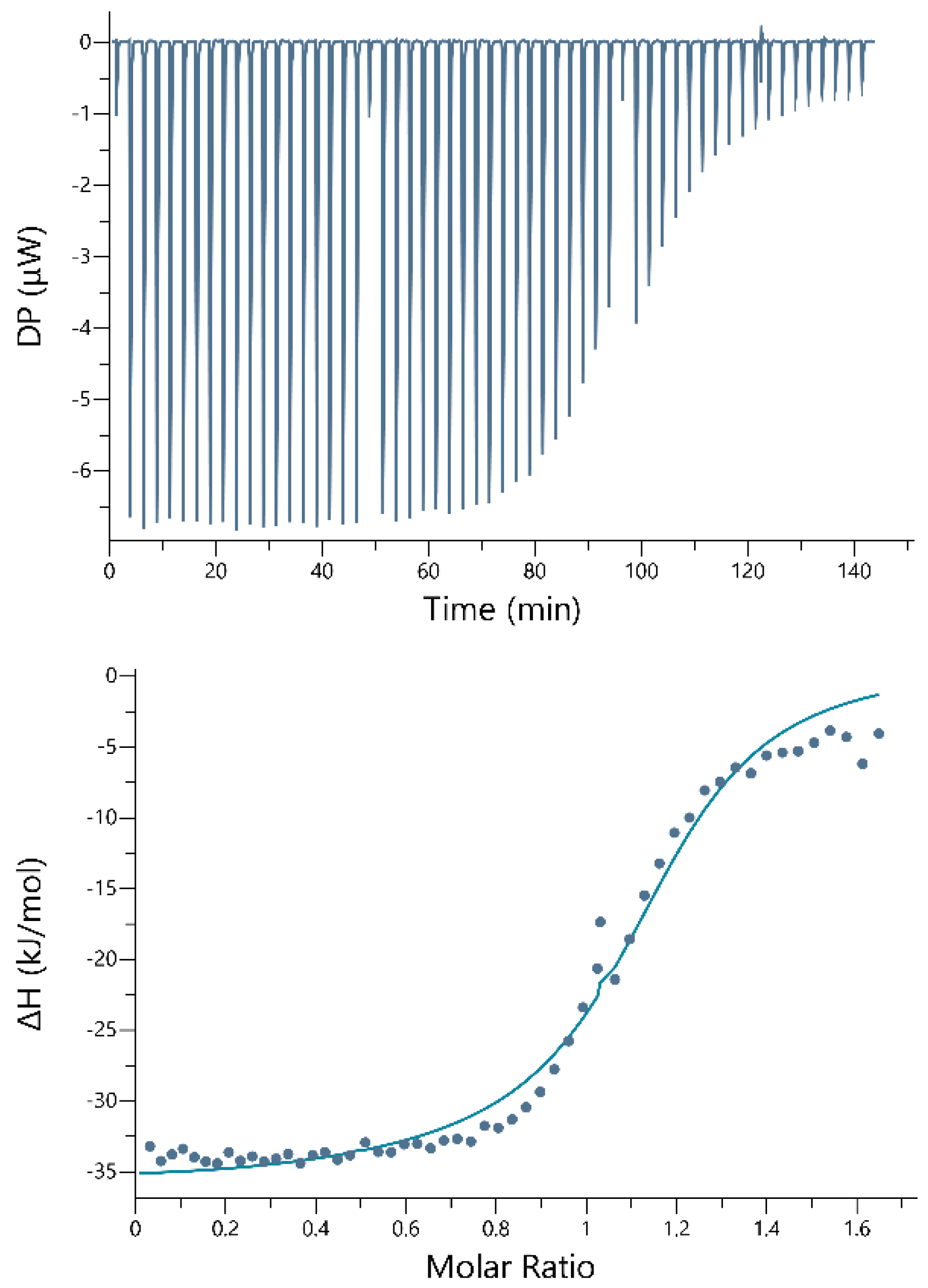
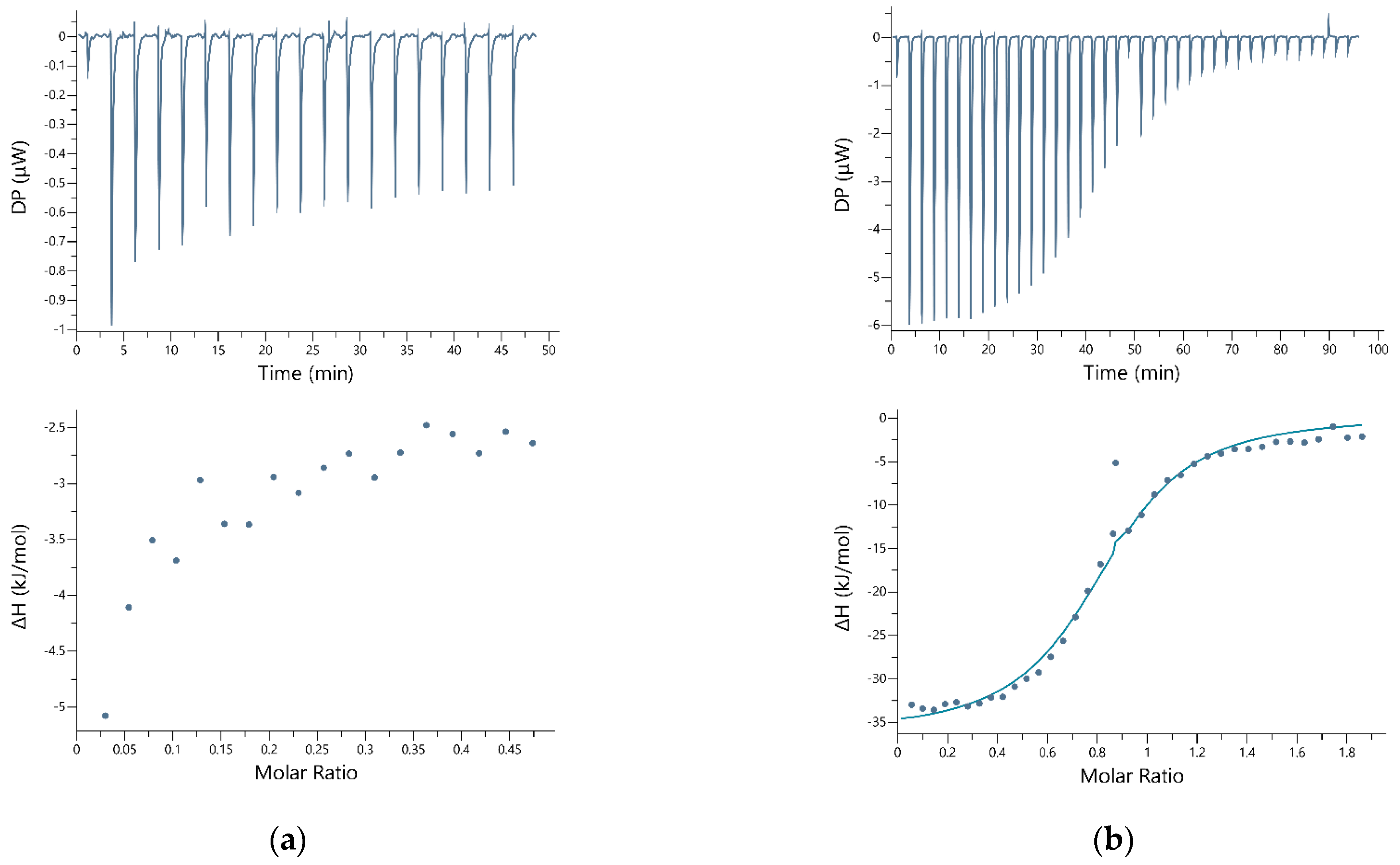
| Peptide | Conc. (g/L) | Metal Ion | Conc. (g/L) | Buffer (g/L) | N | KD (M) | ΔH (kJ/mol) | ΔG (kJ/mol) | −TΔS (kJ/mol) |
|---|---|---|---|---|---|---|---|---|---|
| CNAKHHPRCGGG | 0.284 | Ni2+ | 0.2113 | 6.057 Tris-HCl 8.766 NaCl | 0.822 ± 5.9 × 10−2 | 1.04 × 10−4 ± 1.38 × 10−5 | −18.7 ± 1.70 | −22.8 | −4.03 |
| 0.495 | Ni2+ | 0.2348 | 10.463 MOPS 8.766 NaCl | 0.397 ± 2.4 × 10−2 | 1.87 × 10−4 ± 2.27 × 10−5 | −36.4 ± 4.14 | −21.3 | 15.1 | |
| 0.495 | Ni2+ | 0.2348 | 2.383 HEPES 1.753 NaCl | 0.564 ± 3.1 × 10−2 | 2.88 × 10−4 ± 1.76 × 10−5 | −26.3 ± 1.86 | −20.2 | 6.08 | |
| 0.495 | Ni2+ | 0.2348 | 1.211 Tris-HCl 1.753 NaCl | 0 | 0 | 0 | 0 | 0 | |
| CNAKHHPRCGGG | 0.284 | Co2+ | 0.0589 | 6.057 Tris-HCl 8.766 NaCl | 0 | 0 | 0 | 0 | 0 |
| CTQMLGQLCGGG | 0.552 | Co2+ | 0.0589 | 1.211 Tris-HCl 1.753 NaCl | 1.04 ± 7.8 × 10−3 | 5.09 × 10−6 ± 6.03 × 10−7 | −35.6 ± 0.342 | −30.2 | 5.35 |
| 0.502 | Co2+ | 0.236 | 6.057 Tris-HCl 8.766 NaCl | 0 | 0 | 0 | 0 | 0 | |
| 0.502 | Co2+ | 0.236 | 10.463 MOPS 8.766 NaCl | 0 | 0 | 0 | 0 | 0 | |
| 0.502 | Co2+ | 0.236 | 2.383 HEPES 1.753 NaCl | 0 | 0 | 0 | 0 | 0 | |
| CTQMLGQLCGGG | 0.2760 | Ni2+ | 0.0587 | 1.211 Tris-HCl 1.753 NaCl | 0.837 ± 7.8 × 10−3 | 7.69 × 10−6 ± 6.59 × 10−7 | −35.6 ± 0.434 | −29.2 | 6.40 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Matys, S.; Morawietz, L.-M.; Lederer, F.; Pollmann, K. Characterization of the Binding Behavior of Specific Cobalt and Nickel Ion-Binding Peptides Identified by Phage Surface Display. Separations 2022, 9, 354. https://doi.org/10.3390/separations9110354
Matys S, Morawietz L-M, Lederer F, Pollmann K. Characterization of the Binding Behavior of Specific Cobalt and Nickel Ion-Binding Peptides Identified by Phage Surface Display. Separations. 2022; 9(11):354. https://doi.org/10.3390/separations9110354
Chicago/Turabian StyleMatys, Sabine, Lisa-Marie Morawietz, Franziska Lederer, and Katrin Pollmann. 2022. "Characterization of the Binding Behavior of Specific Cobalt and Nickel Ion-Binding Peptides Identified by Phage Surface Display" Separations 9, no. 11: 354. https://doi.org/10.3390/separations9110354
APA StyleMatys, S., Morawietz, L.-M., Lederer, F., & Pollmann, K. (2022). Characterization of the Binding Behavior of Specific Cobalt and Nickel Ion-Binding Peptides Identified by Phage Surface Display. Separations, 9(11), 354. https://doi.org/10.3390/separations9110354







