Isolation and Purification of a Hydrophobic Non-Ribosomal Peptide from an Escherichia coli Fermentation Broth
Abstract
:1. Introduction
2. Materials and Methods
2.1. Production Strain and Vectors
2.2. Culture Medium
2.3. Reactor Cultivation
2.4. Cell Disruption
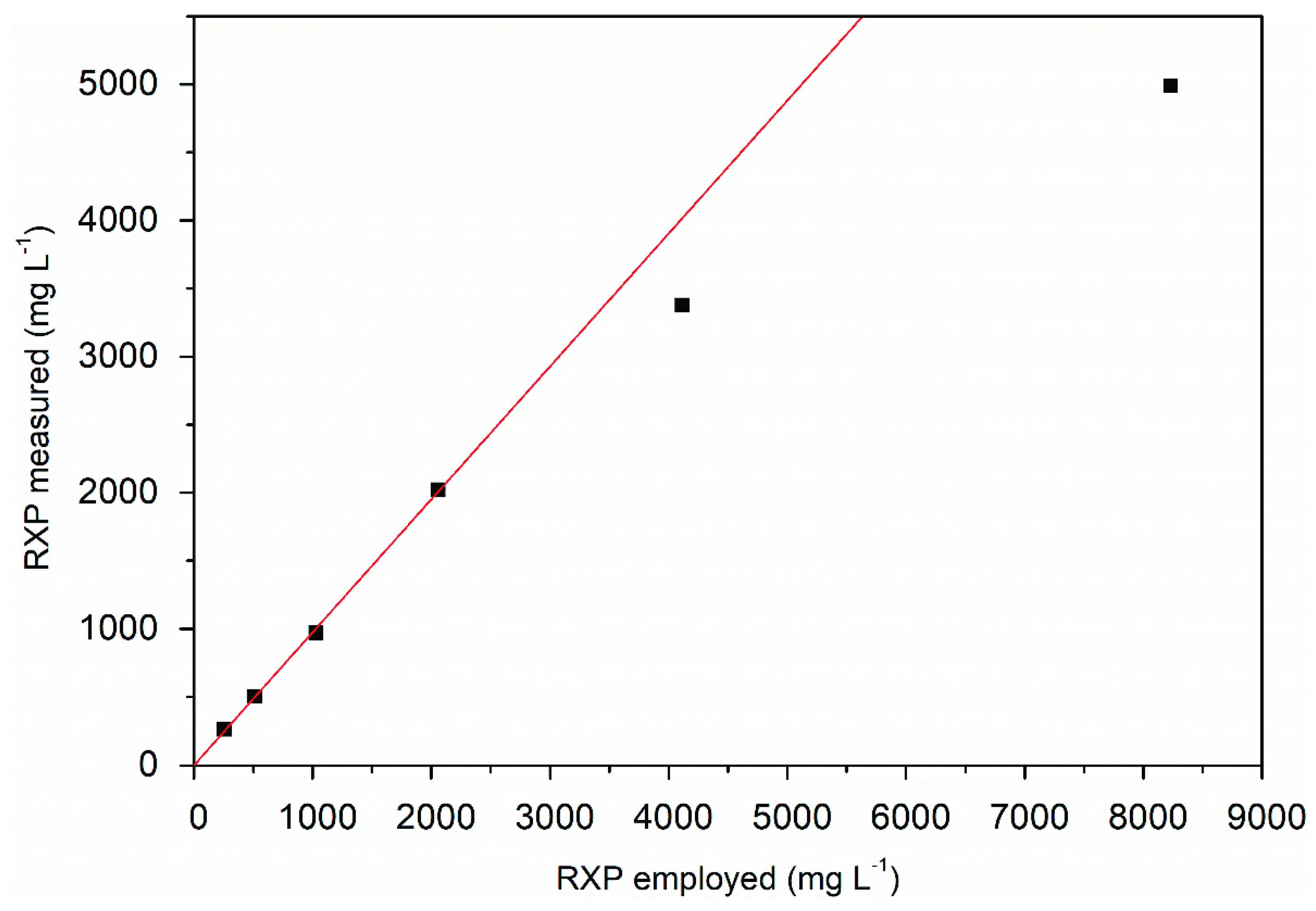
2.5. RXP Quantification
2.6. Succinate Quantification
2.7. Measurement of Cell Dry Weight
2.8. Capture Step
2.9. Experimental Designs for Intermediate Purification
2.10. Polishing
3. Results and Discussion
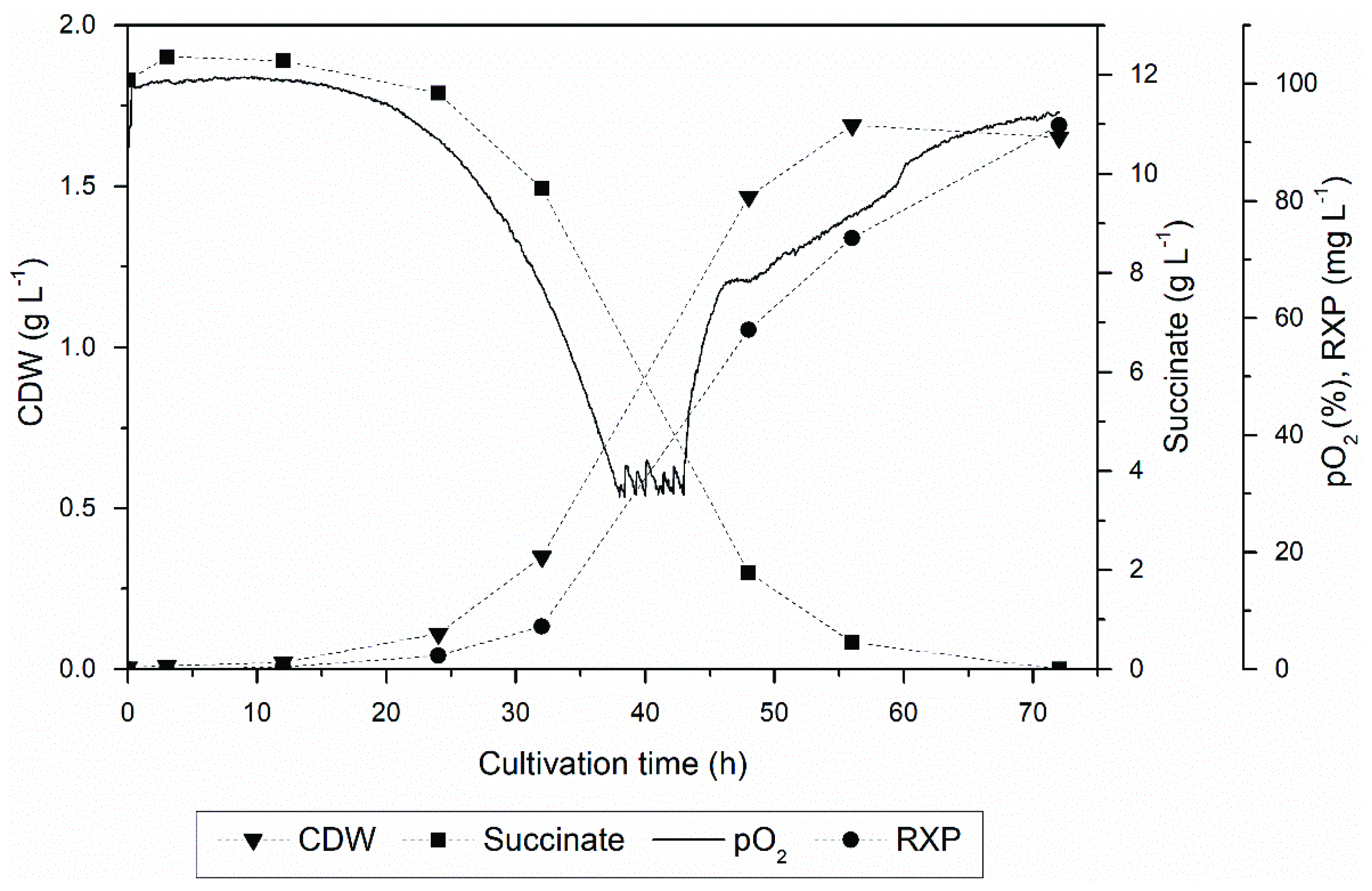
3.1. Upstream Processing
3.2. Product Localization
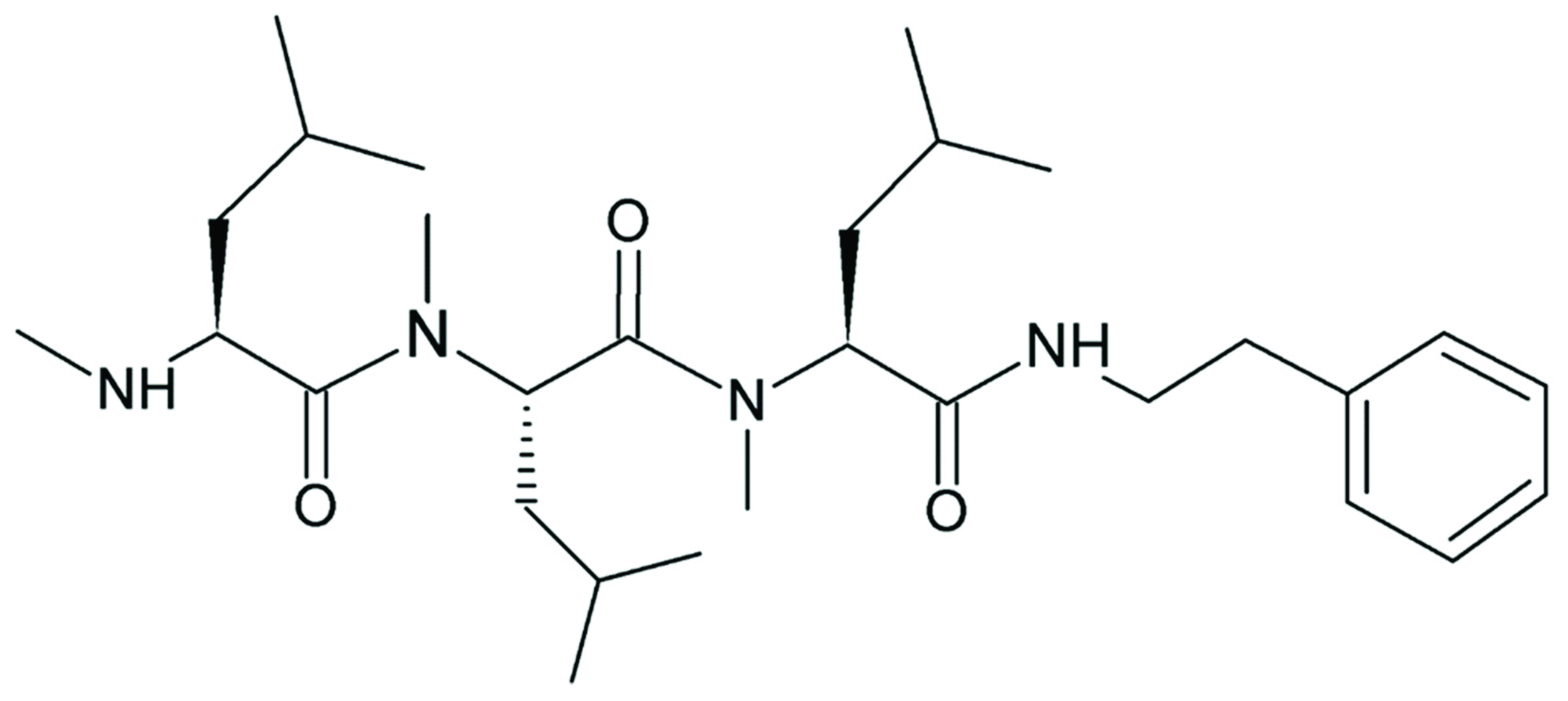
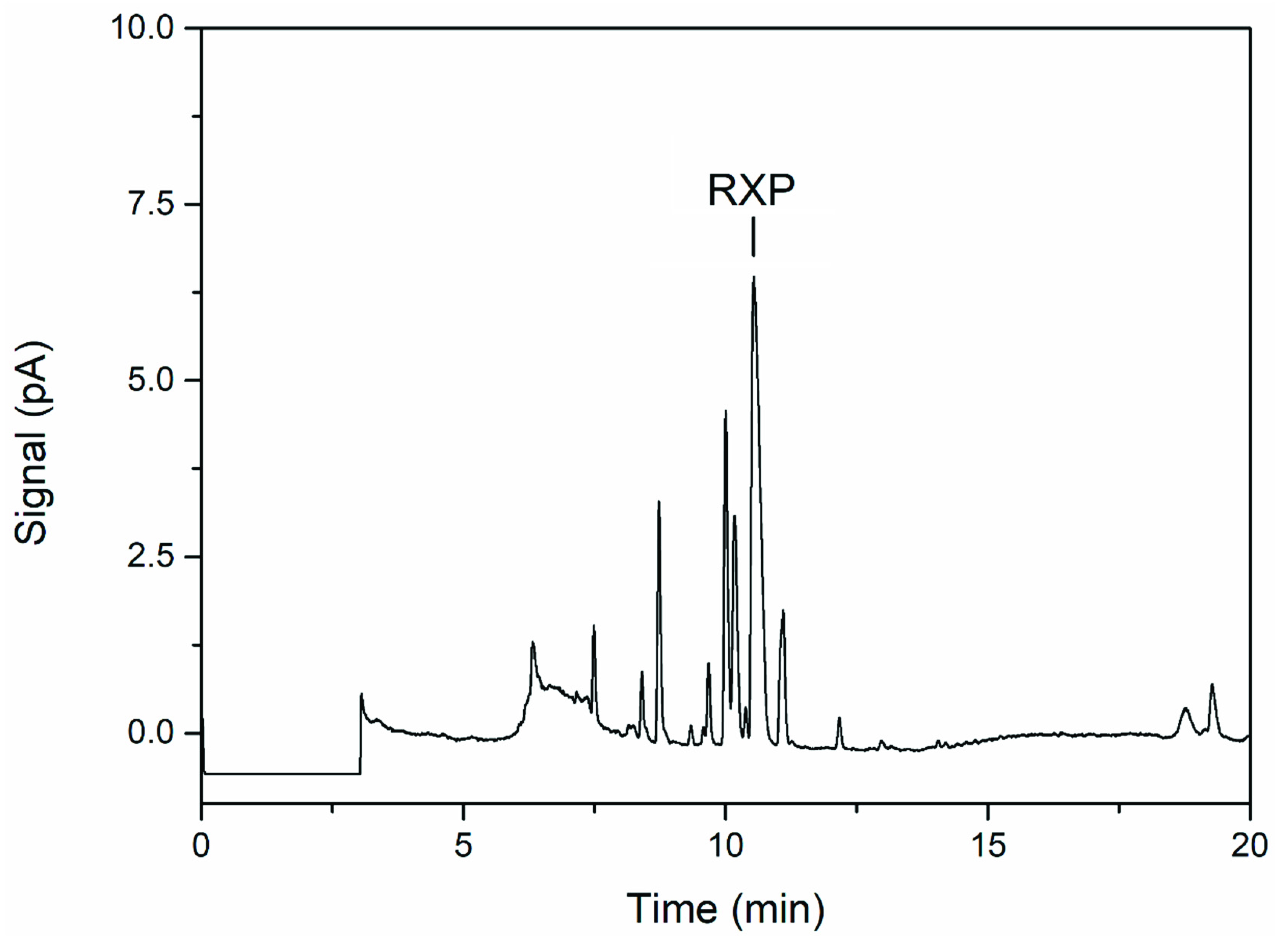
3.3. Product Analysis
3.4. Product Capture
3.5. Experimental Design for Intermediate Purification
3.6. Polishing
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Balibar, C.J.; Vaillancourt, F.H.; Walsh, C.T. Generation of D amino acid residues in assembly of arthrofactin by dual condensation/epimerization domains. Chem. Biol. 2005, 12, 1189–1200. [Google Scholar] [CrossRef] [Green Version]
- Baltz, R.H.; Miao, V.; Wrigley, S.K. Natural products to drugs: Daptomycin and related lipopeptide antibiotics. Nat. Prod. Rep. 2005, 22, 717–741. [Google Scholar] [CrossRef]
- Caboche, S.; Leclère, V.; Pupin, M.; Kucherov, G.; Jacques, P. Diversity of monomers in nonribosomal peptides: Towards the prediction of origin and biological activity. J. Bacteriol. 2010, 192, 5143–5150. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Calcott, M.J.; Ackerley, D.F. Genetic manipulation of non-ribosomal peptide synthetases to generate novel bioactive peptide products. Biotechnol. Lett. 2014, 36, 2407–2416. [Google Scholar] [CrossRef]
- Du, L.; Lou, L. PKS and NRPS release mechanisms. Nat. Prod. Rep. 2010, 27, 255–278. [Google Scholar] [CrossRef] [PubMed]
- Felnagle, E.A.; Jackson, E.E.; Chan, Y.A.; Podevels, A.M.; Berti, A.D.; McMahon, M.D.; Thomas, M.G. Nonribosomal peptide synthetases involved in the production of medically relevant natural products. Mol. Pharm. 2008, 5, 191–211. [Google Scholar] [CrossRef] [PubMed]
- Lawen, A. Biosynthesis of cyclosporins and other natural peptidyl prolyl cis/trans isomerase inhibitors. Biochim. Biophys. Acta 2015, 1850, 2111–2120. [Google Scholar] [CrossRef]
- Sieber, S.A.; Marahiel, M.A. Molecular mechanisms underlying nonribosomal peptide synthesis: Approaches to new antibiotics. Chem. Rev. 2005, 105, 715–738. [Google Scholar] [CrossRef] [PubMed]
- Grünewald, J.; Marahiel, M.A. Chemoenzymatic and template-directed synthesis of bioactive macrocyclic peptides. Microbiol. Mol. Biol. Rev. 2006, 70, 121–146. [Google Scholar] [CrossRef] [Green Version]
- Cai, X.; Nowak, S.; Wesche, F.; Bischoff, I.; Kaiser, M.; Fürst, R.; Bode, H.B. Entomopathogenic bacteria use multiple mechanisms for bioactive peptide library design. Nat. Chem. 2017, 9, 379–386. [Google Scholar] [CrossRef]
- Reimer, D.; Cowles, K.N.; Proschak, A.; Nollmann, F.I.; Dowling, A.J.; Kaiser, M.; ffrench-Constant, R.; Goodrich-Blair, H.; Bode, H.B. Rhabdopeptides as insect-specific virulence factors from entomopathogenic bacteria. Chembiochem 2013, 14, 1991–1997. [Google Scholar] [CrossRef]
- Abburi, K. Adsorption of phenol and p-chlorophenol from their single and bisolute aqueous solutions on Amberlite XAD-16 resin. J. Hazard. Mater. 2003, 105, 143–156. [Google Scholar] [CrossRef] [PubMed]
- Agrawal, P.B.; Pandit, A.B. Isolation of α-glucosidase from Saccharomyces cerevisiae: Cell disruption and adsorption. Biochem. Eng. J. 2003, 15, 37–45. [Google Scholar] [CrossRef]
- Lee, J.C.; Park, H.R.; Park, D.J.; Lee, H.B.; Kim, Y.B.; Kim, C.J. Improved production of teicoplanin using adsorbent resin in fermentations. Lett. Appl. Microbiol. 2003, 37, 196–200. [Google Scholar] [CrossRef]
- Lin, S.-H.; Juang, R.-S. Adsorption of phenol and its derivatives from water using synthetic resins and low-cost natural adsorbents: A review. J. Environ. Manag. 2009, 90, 1336–1349. [Google Scholar] [CrossRef]
- Yang, S.A.; Pyle, D.L. The adsorption kinetics of cephalosporin-c on non-ionic polymeric macropore Amberlite XAD-16 resin. J. Chem. Technol. Biotechnol. 1999, 74, 216–220. [Google Scholar] [CrossRef]
- Sandhu, A.K.; Gu, L. Adsorption/desorption characteristics and separation of anthocyanins from muscadine (Vitis rotundifolia) juice pomace by use of macroporous adsorbent resins. J. Agric. Food Chem. 2013, 61, 1441–1448. [Google Scholar] [CrossRef] [PubMed]
- Lepane, V. Comparison of XAD resins for the isolation of humic substances from seawater. J. Chromatogr. A 1999, 845, 329–335. [Google Scholar] [CrossRef]
- Oestreich, A.M.; Suli, M.I.; Gerlach, D.; Fan, R.; Czermak, P. Media development and process parameter optimization using statistical experimental designs for the production of nonribosomal peptides in Escherichia coli. Electron. J. Biotechnol. 2021, 52, 85–92. [Google Scholar] [CrossRef]
- Prasad, S.; Mandal, I.; Singh, S.; Paul, A.; Mandal, B.; Venkatramani, R.; Swaminathan, R. Near UV-Visible electronic absorption originating from charged amino acids in a monomeric protein. Chem. Sci. 2017, 8, 5416–5433. [Google Scholar] [CrossRef] [Green Version]
- Saraiva, M.A. Interpretation of α-synuclein UV absorption spectra in the peptide bond and the aromatic regions. J. Photochem. Photobiol. B 2020, 212, 112022. [Google Scholar] [CrossRef]
- Hazra, C.; Samanta, T.; Mahalingam, V. A resonance energy transfer approach for the selective detection of aromatic amino acids. J. Mater. Chem. C 2014, 2, 10157–10163. [Google Scholar] [CrossRef]
- Gheshlaghi, R.; Scharer, J.M.; Moo-Young, M.; Douglas, P.L. Application of statistical design for the optimization of amino acid separation by reverse-phase HPLC. Anal. Biochem. 2008, 383, 93–102. [Google Scholar] [CrossRef]
- Huang, H.S.; Wu, J.R.; Chen, M.L. Reversed-phase high-performance liquid chromatography of amphoteric beta-lactam antibiotics: Effects of columns, ion-pairing reagents and mobile phase pH on their retention times. J. Chromatogr. 1991, 564, 195–203. [Google Scholar] [CrossRef]
- Kaliszan, R.; Wiczling, P.; Markuszewski, M.J. pH gradient reversed-phase HPLC. Anal. Chem. 2004, 76, 749–760. [Google Scholar] [CrossRef]
- Roses, M.; Canals, I.; Allemann, H.; Siigur, K.; Bosch, E. Retention of Ionizable Compounds on HPLC. 2. Effect of pH, Ionic Strength, and Mobile Phase Composition on the Retention of Weak Acids. Anal. Chem. 1996, 68, 4094–4100. [Google Scholar] [CrossRef] [PubMed]
- Egeberg, P.K.; Alberts, J.J. Determination of hydrophobicity of NOM by RP-HPLC, and the effect of pH and ionic strength. Water Res. 2002, 4997–5004. [Google Scholar] [CrossRef]
- Afsah-Hejri, L.; Jinap, S.; Arzandeh, S.; Mirhosseini, H. Optimization of HPLC conditions for quantitative analysis of aflatoxins in contaminated peanut. Food Control 2011, 22, 381–388. [Google Scholar] [CrossRef]
- Issaq, H.J.; Fox, S.D.; Lindsey, K.; McConnell, J.H.; Weiss, D.E. Effect of Temperature on HPLC Separations Using C 1, C 4, C 8, and C 18 Alkyl Chain Bonded Silica Columns. J. Liq. Chromatogr. 1987, 10, 49–70. [Google Scholar] [CrossRef]
- Martín, J.; Méndez, R.; Negro, A. Effect of Temperature on HPLC Separations of Penicillins. J. Liq. Chromatogr. 1988, 11, 1707–1716. [Google Scholar] [CrossRef]
- Pappa-Louisi, A.; Nikitas, P.; Zisi, C.; Papachristos, K. Combined effect of temperature and organic modifier concentration on the retention under single mode gradient conditions in reversed-phase HPLC. J. Sep. Sci. 2008, 31, 2953–2961. [Google Scholar] [CrossRef]
- Schellman, J.A. Temperature, stability, and the hydrophobic interaction. Biophys. J. 1997, 73, 2960–2964. [Google Scholar] [CrossRef] [Green Version]
- Southall, N.T.; Dill, K.A.; Haymet, A.D.J. A View of the Hydrophobic Effect. J. Phys. Chem. B 2002, 106, 521–533. [Google Scholar] [CrossRef]
- Bischof, J.C.; He, X. Thermal stability of proteins. Ann. N. Y. Acad. Sci. 2005, 1066, 12–33. [Google Scholar] [CrossRef]
- Wijayanti, H.B.; Bansal, N.; Deeth, H.C. Stability of Whey Proteins during Thermal Processing: A Review. Compr. Rev. Food Sci. Food Saf. 2014, 13, 1235–1251. [Google Scholar] [CrossRef]
- Peng, L.; Jayapalan, S.; Chankvetadze, B.; Farkas, T. Reversed-phase chiral HPLC and LC/MS analysis with tris(chloromethylphenylcarbamate) derivatives of cellulose and amylose as chiral stationary phases. J. Chromatogr. A 2010, 1217, 6942–6955. [Google Scholar] [CrossRef] [PubMed]
- Carr, P.W.; Wang, X.; Stoll, D.R. Effect of pressure, particle size, and time on optimizing performance in liquid chromatography. Anal. Chem. 2009, 81, 5342–5353. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qin, Q.; Zhang, S.; Zhang, W.-G.; Zhang, Z.-B.; Xiong, Y.-J.; Guo, Z.-Y.; Fan, J.; Run-Zheng, S.; Finlow, D.; Yin, Y. The impact of silica gel pore and particle sizes on HPLC column efficiency and resolution for an immobilized, cyclodextrin-based, chiral stationary phase. J. Sep. Sci. 2010, 33, 2582–2589. [Google Scholar] [CrossRef]




| Run | X1: Formic Acid (%) | X2: Gradient Slope (% min−1) | X3: Temperature (°C) | Resolution |
|---|---|---|---|---|
| 1 | 0.6 | 2.5 | 40 | 1.56 |
| 2 | 0.2 | 1.5 | 45 | 1.21 |
| 3 | 1 | 2.5 | 30 | 1.67 |
| 4 | 1 | 4.5 | 35 | 1.19 |
| 5 | 0.6 | 4.5 | 50 | 1.07 |
| 6 | 0.2 | 2.5 | 35 | 1.02 |
| 7 | 0.2 | 4.5 | 30 | 0.95 |
| 8 | 0.4 | 0.5 | 50 | 1.95 |
| 9 | 0,6 | 2.5 | 40 | 1.57 |
| 10 | 0.2 | 0.5 | 35 | 1.56 |
| 11 | 1 | 2.5 | 50 | 1.55 |
| 12 | 0.6 | 4.5 | 50 | 1.00 |
| 13 | 0.4 | 0.5 | 50 | 1.81 |
| 14 | 1 | 2.5 | 50 | 1.59 |
| 15 | 1 | 0.5 | 40 | 2.03 |
| 16 | 0.8 | 1.5 | 45 | 1.76 |
| 17 | 0.6 | 0.5 | 30 | - |
| 18 | 0.8 | 1.5 | 35 | 1.75 |
| 19 | 0.6 | 2.5 | 40 | 1.54 |
| 20 | 0.6 | 2.5 | 40 | 1.62 |
| 21 | 0.6 | 3.5 | 30 | 1.26 |
| 22 | 0.2 | 3.5 | 50 | 0.99 |
| 23 | 1 | 4.5 | 35 | 1.35 |
| RXP Concentration (mg L−1) | Volume (mL) | RXP Mass (mg) | |
|---|---|---|---|
| Supernatant | 92.90 ± 0.52 | 800 | 74.32 ± 0.42 |
| Flow through | 4.34 ± 0.19 | 800 | 3.47 ± 0.15 |
| 20% ethanol wash fraction | 5.50 ± 0.18 | 105.5 | 0.58 ± 0.02 |
| 100% ethanol elution | 1369.18 ± 23.79 | 48.6 | 68.31 ± 1.16 |
| Source | Sum of Squares | df | Mean Square | F-Value | p-Value |
|---|---|---|---|---|---|
| Model | 2.0933 | 5 | 0.4187 | 52.8973 | <0.0001 |
| X1 | 0.6121 | 1 | 0.6121 | 77.3328 | <0.0001 |
| X2 | 1.3689 | 1 | 1.3689 | 172.9577 | <0.0001 |
| X3 | 0.0006 | 1 | 0.0006 | 0.0752 | 0.7874 |
| X2×3 | 0.0435 | 1 | 0.0435 | 5.5014 | 0.0322 |
| X12 | 0.1052 | 1 | 0.1052 | 13.2963 | 0.0022 |
| Residual | 0.1266 | 16 | 0.0079 | ||
| Lack of fit | 0.0973 | 9 | 0.0108 | 2.5809 | 0.1123 |
| Pure error | 0.0293 | 7 | 0.0042 | ||
| Total | 2.2199 | 21 |
| Factor | Coefficient Estimate | df | Error |
|---|---|---|---|
| Intercept | 1.5303 | 1 | 0.0303 |
| X1 | 0.2289 | 1 | 0.0260 |
| X2 | −0.3867 | 1 | 0.0294 |
| X3 | −0.0077 | 1 | 0.0282 |
| X2X3 | −0.0893 | 1 | 0.0381 |
| X12 | −0.1533 | 1 | 0.0421 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Oestreich, A.M.; Reinhardt, S.; Gerlach, D.; Fan, R.; Czermak, P. Isolation and Purification of a Hydrophobic Non-Ribosomal Peptide from an Escherichia coli Fermentation Broth. Separations 2021, 8, 241. https://doi.org/10.3390/separations8120241
Oestreich AM, Reinhardt S, Gerlach D, Fan R, Czermak P. Isolation and Purification of a Hydrophobic Non-Ribosomal Peptide from an Escherichia coli Fermentation Broth. Separations. 2021; 8(12):241. https://doi.org/10.3390/separations8120241
Chicago/Turabian StyleOestreich, Arne Michael, Sebastian Reinhardt, Doreen Gerlach, Rong Fan, and Peter Czermak. 2021. "Isolation and Purification of a Hydrophobic Non-Ribosomal Peptide from an Escherichia coli Fermentation Broth" Separations 8, no. 12: 241. https://doi.org/10.3390/separations8120241
APA StyleOestreich, A. M., Reinhardt, S., Gerlach, D., Fan, R., & Czermak, P. (2021). Isolation and Purification of a Hydrophobic Non-Ribosomal Peptide from an Escherichia coli Fermentation Broth. Separations, 8(12), 241. https://doi.org/10.3390/separations8120241







