Abstract
In this study, the chemical composition of different tissues of Rana temporaria chensinensis David derived from the same individual was analyzed by comparative approach. First, pre-column derivatization combined with high performance liquid chromatography (HPLC) was established to determine the content of 1-methyl hydantoin in samples, which used S1–S5 samples. The results indicated that 1-methyl hydantoin was determined in Oviductus Ranae (OR), Rana chensinensis ovum (RCO), Rana chensinensis meat (RCM), and Rana chensinensis skin (RCS), except for Rana chensinensis bone (RCB). Moreover, the content of it in RCS was the highest. In addition, the contents of six polyunsaturated fatty acids (PUFAs) in different tissues of Rana chensinensis were measured by HPLC, including eicosapentaenoic acid (EPA), α-linolenic acid (ALA), docosahexaenoic acid (DHA), arachidonic acid (ARA), linoleic acid (LA) and oleic acid (OA). The results indicated that OR, RCO, RCM, RCS, and RCB all contained the above six PUFAs. With the aid of chemometrics methods, the results of principal component analysis (PCA), hierarchical cluster analysis (HCA), and orthogonal partial least squares discriminant analysis (OPLS-DA) combined with the sequencing results of the total PUFAs content of each sample, showed that different tissues of Rana chensinensis could be divided into four categories, and the RCO sample was divided into one category because of the highest PUFAs content, which was a good source of PUFA. For comparison, OR and other tissue from the perspective of PUFAs, we also established OPLS-DA models of them. It could be found that the RCM was the most similar to the OR in the diversity and content of PUFAs. This study provided a theoretical basis for the further development and utilization of RCO, RCM, RCS, and RCB as by-products of OR.
1. Introduction
Rana temporaria chensinensis David, also known as Hashima and Xueha, is distributed in northeast China [1,2,3,4]. Oviductus Ranae (OR) is the dried product of the female Rana chensinensis fallopian tube [5,6,7,8]. OR has effects in nourishing yin, moistening lung, tonifying kidney, replenishing essence, invigorating spleen, and nourishing the stomach [9]. Due to the high price of OR, most farmers obtain OR while discarding other tissues from Rana chensinensis, such as Rana chensinensis ovum (RCO), Rana chensinensis meat (RCM), Rana chensinensis skin (RCS), and Rana chensinensis bone (RCB). However, studies have shown that the contents of fatty acids, protein and mineral elements [10] in the OR- removed Rana chensinesis are still very rich, which are attracting much attention due to their extensive physiological activities [11,12]. The content of polyunsaturated fatty acids (PUFAs) in RCO is several times higher than that of OR [11]. The existing pharmacological studies have found that RCO is rich in protein and has obvious functions of anti-fatigue and regulating immunity [12]. The antimicrobial peptide [13] and collagen [14,15,16] of Rana chensinensis can be extracted from the RCS, which has research value and development prospects. RCM has high protein, low fat and low cholesterol, which is slippery and easy to absorb. It is also rich in essential amino acids and has high nutritional value [17]. RCB is rich in natural pure organic calcium and amino acids, which is helpful for human absorption. It is therefore a good source of calcium [18]. Therefore, the OR removed Rana chensinesis can be widely used after proper processing and development. In consequence, the study of various tissues of Rana chensinensis provided a scientific theoretical basis for the comprehensive utilization of Rana chensinensis resources. In recent years, the research on OR has become increasingly comprehensive and in-depth. 1-methyl hydantoin and PUFAs, as the main pharmacodynamic components, have good pharmacological activity, which can be used as the standard of quality evaluation.
Under the standard of OR in the Chinese Pharmacopoeia (2020 edition), 1-methyl hydantoin was used as an indicator for qualitative identification [19]. As one of the main active components, 1-methyl hydantoin has good antitussive and anti-inflammatory effects [20]. However, the pre-column derivatization was used in this experiment due to the harsh chromatographic conditions (using water as mobile phase accelerates the wear on the chromatographic column) in Chinese Pharmacopoeia (2020 edition) and there were problems such as low sensitivity and poor resolution [21,22]. 1-methyl hydantoin was derivatized with benzoylchloride, having a chromogenic group to impart its chromophore in structure to form benzoyl-1-methyl hydantoin (bzmh) for HPLC detection [23]. Furthermore, it was reported that 1-methyl hydantoin was detected in RCO of Rana chensinensis [20], the existence of 1-methyl hydantoin in RCM, RCS, and RCB, which are also by-products of Rana chensinensis, had not been reported. The samples of OR, RCO, RCS, RCM, and RCB taken from the same Rana chensinensis (Figure 1) were analyzed by HPLC to determine whether there exists 1-methyl hydantoin and their content in the samples.
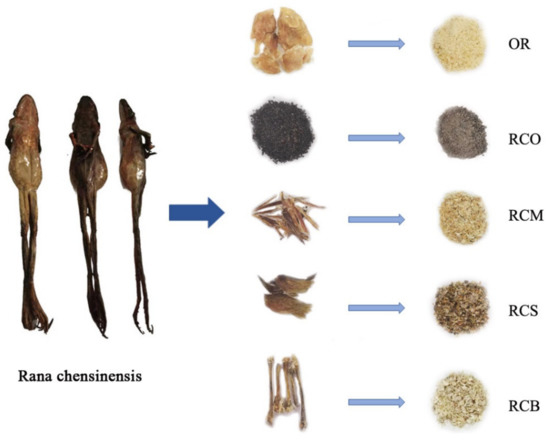
Figure 1.
The whole Rana chensinensis and samples of OR, RCO, RCS, RCM, and RCB.
In addition, OR is also rich in various unsaturated fatty acids (UFAs) [24], in which UFAs account for 40% of the total fatty acids. Polyunsaturated fatty acid (PUFA) constitutes the fat of the organism, and it is also an indispensable fatty acid for the human body, which plays an important physiological function in the organism [25]. PUFA can be classified as omega-3 series and omega-6 series according to the position and function of double keys.Omega-3 fatty acids can promote neurodevelopment [26], regulate inflammatory response, reduce the risk of cardiovascular disease [27,28], promote the health of the circulatory system and prevent cholesterol and fat from accumulating on the arterial wall, which is beneficial to metabolism [29], Besides, it is also an essential nutrient in the human diet [30,31]. In addition to reducing the risk of metabolic syndrome, omega-6 fatty acids also have the function of preventing cardiovascular and cerebrovascular disease [32]. Therefore, PUFAs are potential indicators for quantification. Previous studies have found that through research different solvent extracts (ethanol extract, petroleum ether extract and water extract) of OR, it was found that the petroleum ether extract of OR had good pharmacological activity, especially in terms of countering depression and fatigue [33]. Further composition identification revealed that the petroleum ether extract of OR was rich in PUFAs, including members of the omega-3 fatty acid family (eicosapentaenoic acid (EPA), α-linolenic acid (ALA), docosahexaenoic acid (DHA)) and the omega-6 fatty acid family (arachidonic acid (ARA), linoleic acid (LA) and oleic acid (OA)) [34]. The specific information of six PUFAs was in Supplementary Materials Table S1. To compare the difference of six PUFAs from the petroleum ether extract of OR and other tissue from the same Rana chensinensis, in this work, the OR, RCO, RCS, RCM, and RCB were taken from the same Rana chensinensis and were determined by Soxhlet extraction combined with HPLC. Hierarchical cluster analysis (HCA), principal component analysis (PCA), and stoichiometric pattern recognition methods were used to compare the content of six PUFAs in other tissue of the Rana chensinensis with that of the OR.
2. Materials and Methods
Petroleum ether, methanol, cyclohexane, dichloromethane, anhydrous pyridine, benzoyl chloride, anhydrous calcium chloride and phosphoric acid were analytically pure and purchased from the Beijing Chemical Factory (Beijing, China). HPLC-grade acetonitrile and HPLC-grade methanol were purchased from Fisher (Fisher Scientific, Fair Lawn, NJ, USA). The ultrapure water was prepared from a gradient water purification system (Water Purifier, Chengdu, China). EPA (Lot No: 32510020); DHA (Lot No: 0501808) and ARA (Lot No: 2491801) were purchased from TanMo Quality Testing Technology Co., Ltd. (Beijing, China). ALA (Lot No: U-62A-J25-C), LA (Lot No: U-52A-JA3-C) and OA (Lot No: U-46A-MA6-C) were purchased from ANPEL Laboratory Technologies (Shanghai, China) Inc. (Shanghai, China). The standard of bzmh was made in laboratory with a purity of 99%. The purity of each standard compound was higher than 99%, as determined by HPLC. Six PUFAs standards were formulated to different concentrations using HPLC-grade methanol and filtrated with a 0.22 µm filter membrane prior HPLC injection.
Rana chensinensis samples were collected from the main production area of the Changbai Mountain areas (Baishan, China). Specific information is shown in Table 1.

Table 1.
Information of Rana chensinensis samples.
2.1. 1-Methyl Hydantoin Pre-Column Derivatization
2.1.1. Preparation of Standard Solution
10.00 mg bzmh standard placed in a 100 mL volumetric flask, add acetonitrile volume to scale, shake well, made each 1mL containing 0.1000 mg solution as the standard substance solution.
2.1.2. Sample Preparation
Each OR, RCO, RCS, RCM, and RCB sample from the same Rana chensinensis was pulverized into powders (passing through a 20-mesh sieve). 1.0 g powder of each sample was placed in the 150 mL plug cone bottle, after which was added 20 ml of petroleum ether and then extracted ultrasonically for 30 min. They were then filtered, and the filtrate was discarded and the filter residue collected. The filter residue was placed in a 150ml stoppered conical flask and methanol 20 mL was added and ultrasonic treatment was applied for 30 min. After this, the filtrate was collected and the sample was concentrated with methanol extract under reduced pressure.
2.1.3. Pre-Column Derivatization
8 mL dichloromethane and 2 mL anhydrous pyridine was placed in the methanol extract and stirred until fully dissolved. At room temperature, 0.1 mL benzoyl chloride was added drop by drop and the anhydrous calcium chloride drying tube was sealed. It was then stirred at room temperature for 1 h. Figure 2 shows the equation of 1-methyl hydantoin pre-column derivatization. Then the reaction solution was concentrated under reduced pressure, 10 mL cyclohexane was added and spun dry. This was repeated three times until there was no pyridine flavor. Finally, the residue was added with acetonitrile to the 1 mL volumetric flask, filtered with 0.45 mμL microporous membrane, and the OR RCO RCS RCM, and RCB derivative test solution were obtained.
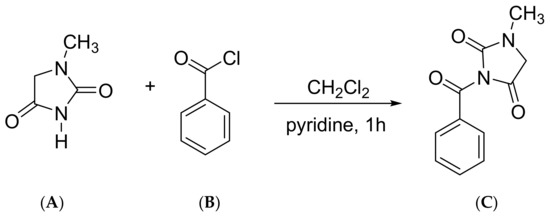
Figure 2.
1-methyl hydantoin pre-column derivatization equation. (A) 1-methyl hydantoin; (B) benzoyl chloride; (C) bzmh.
2.1.4. Blank Control and Negative Control Solution Preparation
Chromatographic acetonitrile was detected by HPLC under the aforementioned liquid phase conditions as blank control. The methanol extract of the unpre-column derivatization sample was used as the negative control.
2.1.5. HPLC Chromatographic Analysis Conditions
The HPLC analysis was performed on an Agilent Technologies 1260 Series liquid chromatograph (Agilent Technologies, Pittsburgh, PA, USA) that was equipped with a quaternary pump, autosampler, thermostatic column chamber, and diode array detector (DAD). Chromatographic separation was achieved on an Agilent HC-C18 column (250 × 4.6 mm, 5 µm). The detection wavelength was set at 254 nm, the column temperature was 20 °C, and the injection volume was 10 µL. The mobile phase consisted of HPLC-grade methanol (B) and water (D) (20:80) for the flow rate of 1.0 mL/min for 30 min. The data were recorded and processed using Agilent Chemstation (Agilent Technologies, Pittsburgh, PA, USA) software.
2.1.6. Validation of the Method
Linear relationship test: accurately weighed 12.100 mg bzmh standard, placed in a 100 mL volumetric flask, added the appropriate amount of chromatographic acetonitrile to dissolve to obtain 0.1210 mg/mL bzmh stock solution. Accurately aspirated 0.1, 0.2, 0.5, 1, 2, and 4 mL of the stock solution into a 10 mL volumetric flask and dissolved with appropriate amount of chromatographic acetonitrile to obtain the standard solutions at concentrations of 0.00121, 0.00242, 0.00605, 0.01210, 0.02420 and 0.04840 mg/mL, respectively. The standard solutions were injected into the HPLC in turn, and the chromatographic peak area was recorded according to the chromatographic conditions of “2.1.5”. The standard curve was drawn with the reference substance concentration (X) as horizontal axis and chromatographic peak area (Y) as vertical axis.
Limit of detection (LOD) and limit of quantification (LOQ) test: Stock solution containing bzmh was diluted to give a series of appropriate concentrations with acetonitrile and aliquots of the diluted solutions were injected. The LOD and LOQ of bzmh were calculated when the signal-to-noise ratios (S/N) were 3 and 10, respectively. Six parallel samples were analyzed in each test.
Precision test: 10 μL of the same sample solution was precisely absorbed and injected into the HPLC and analyzed continuously for six times to assess the precision.
Stability test: the same sample solution was placed at room temperature for 0, 2, 4, 6, 8, 12, 24 h respectively and injected into HPLC for analysis to assess the stability of the sample solution.
Repeatability test: Six samples of the Rana chensinensis sample from the same origin were prepared according to the method of “2.1.2”, and analyzed to assess the repeatability.
Accuracy test: The accuracy was expressed as the recovery of standard compounds added to the pre-analyzed samples. The reference substance bzmh was added to the sample solution with ratio 1:1 and the sample recovery experiment was performed to determine the accuracy. Six samples for each tissue of Rana chensinensis were analyzed.
2.2. Six Kind of PUFAs
2.2.1. Sample Preparation
Each OR, RCO, RCS, RCM, and RCB sample from the same Rana chensinensis was pulverized into powders (passing through a 20-mesh sieve). 0.8 g powder of each sample was wrapped with filter paper and placed in Soxhlet extractors. Added 70 mL petroleum ether to a 100 mL round bottom flask. The Soxhlet extractor was placed in a constant temperature water bath at 90 °C for 6 h [35]. After the extraction was completed, the petroleum ether was recovered by rotary evaporator under the condition of 45 °C. Finally, the extract was dissolved with methanol and fixed volume into the 10 mL volumetric flask. Before HPLC detection, the solution was filtered through a filter of 0.22 μm. Moreover, 500 μL of the solution from each sample was pooled to prepare a QC sample to assure the stability and reproducibility of the HPLC analysis [36]. Through the whole worklist, 5 QC injections were implemented randomly. The injected volumes of the QC and testing samples were 10 µL for each operation.
2.2.2. HPLC Chromatographic Analysis Conditions
The HPLC analysis was performed on an Agilent Technologies 1260 Series liquid chromatograph (Agilent Technologies, Pittsburgh, PA, USA), which was equipped with a quaternary pump, autosampler, thermostatic column chamber, and ultraviolet (UV) detector. Chromatographic separation was achieved on an Agilent TC-C18 column (250 × 4.6 mm, 5 µm). The mobile phase consisted of HPLC-grade acetonitrile (A) and 1% aqueous phosphoric acid (B) while using a gradient program of 86–93% A in 0–10 min, 93% A in 10–20 min and 93–100% A in 20–30 min. A gradient flow rate of 1.0–0.5 mL/min in 0–14 min and 0.5 mL/min in 14–30 min was used. The detection wavelength was set at 203 nm, the column temperature was 30 °C, and the injection volume was 10 µL. The standards and samples were determined under the same HPLC conditions. The data were recorded and processed using Agilent Chemstation (Agilent Technologies, Pittsburgh, PA, USA) software.
2.2.3. Validation of the Method
We refer to and quote the previous methodological experiments done by our laboratory to the quality evaluation of Oviductus Ranae based on PUFAs using HPLC fingerprint techniques combined with chemometric methods [24].
2.3. Date Analysis
In this study, all samples were analyzed by HPLC in triplicate. The relative standard deviation (RSD) of three repetitions of the same sample was always less than 5%. Before chemometric analysis, we normalized the content of six PUFAs of all samples to meet the requirements of analysis.
2.3.1. Hierarchical Cluster Analysis (HCA)
HCA is a multivariable analysis method that simplifies complex data through data modeling. The analysis results are plot as tree families, which can clearly display the classification results of the samples and facilitate intuitive analysis of the similarities and differences between the samples [37]. In this study, the data of PUFAs in different tissues of Rana chensinensis were analyzed by HCA. The program was processed on Metaboanalyst (http://www.metaboanalyst.ca/, accessed on 27 July 2020) online analysis. The cluster analysis was carried out by using the inter-group connection clustering method and the Squared Euclidean Distance interval.
2.3.2. Principal Component Analysis (PCA)
As a statistical method of dimension reduction, PCAs try to combine many original indexes with certain correlation into a group of new comprehensive indexes which are irrelevant to each other to replace that original indexes [38]. PCA helps to visualize the overall clustering trend between individual samples, and the dispersion of points indicates similarities or differences between sample data [39], which simplifies the complex problem [40]. In this work, PCA was performed while using SIMCA (version 14.1, MKS Umetrics, Malmö, Sweden), and the score plot was interpreted by the relationship between PC1 and PC2 for visual analysis of the data matrix to analyze the principal components of PUFAs in different tissues of the Rana chensinensis.
2.3.3. Orthogonal Partial Least Squares Discriminant Analysis (OPLS-DA)
In the past decade, unsupervised methods such as HCA and PCA have been frequently applied to the classification of natural products [41]. Accordingly, the supervised method uses class members to guide the grouping to the specified direction, so that it is more convenient to find the mark responsible for the specified effect. OPLS-DA is a supervised statistical method of discriminant analysis and a regression modeling method of the multi-dependent variable to the multi-independent variable [42]. Its greatest feature is that the data variation that is irrelevant with the classification variable Y in the independent variable X can be removed [43] so that the classification information is mainly concentrated in a principal component, and thus the model becomes simple and easy to explain [41]. An OPLS-DA model of different parts of Rana chensinensis was established by using PUFAs data after orthogonal signal correction, which can improve the reliability of the model. In this work, PCA was performed while using SIMCA (version 14.1, MKS Umetrics, Malmö, Sweden). On the basis of PUFAs data, orthogonal signal correction (OSC) combined with the partial least squares (PLS) method was used to establish the analysis and discrimination model of different parts of Rana chensinensis, which was compared with Oviductus Ranae and classified and analyzed.
3. Results and Discussion
3.1. Results of 1-Methyl Hydantoin Pre-Column Derivatization
3.1.1. Methodology Validation
The standard curves of the bzmh standard were used for the quantitative analysis of bzmh in all samples. The regression equation of bzmh was Y = 42718X + 2.8163 (R2 = 1), where X and Y respectively were the concentration and peak area, showed good linearity in the test range (0.001~0.04 mg/mL).
In the precision experimental results, the relative standard deviation (RSD) of retention time and peak area were 0.14% and 0.31%, respectively. The RSD of retention time and peak area of repetitive results were 2.98% and 4.79%, respectively. The stability test results showed that the RSD of the retention time and peak area of the sample within 24 h were 0.97% and 1.18%, respectively. The LOD and LOQ for bzmh were 0.08 ng/mL and 0.26 ng/mL, respectively. The recovery was found to be in the range of 98.00–104.96%. Table 2 shows the methodology in detail. The results showed that the pre-column derivation combined with HPLC could meet the requirement for the analysis of bzmh content in different tissue of Rana chensinensis.

Table 2.
Methodological validation of pre-column derivatization of 1-methyl hydantoin combined with HPLC.
Figure 3 shows the HPLC chromatograms of 1-methyl hydantoin pre-column derivatization. As can be seen from the chromatograms, the solution of the sample was well separated, and the retention time of the chromatographic peak was consistent with that of the bzmh standard, and there was no chromatographic peak of this component in the solution of the negative control and blank control.
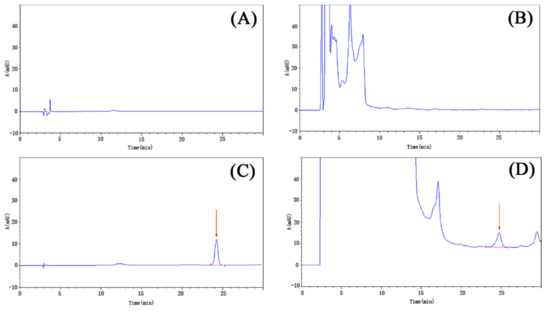
Figure 3.
The high performance liquid chromatography (HPLC) chromatograms of 1-methyl hydantoin pre-column derivatization. (A) blank control; (B) negative control; (C) benzoyl-1-methyl hydantoin (bzmh) standard; (D) sample.
3.1.2. Results of Bzmh Content in Each Sample
Quantitative analysis of bzmh in OR, RCO, RCS, RCM, and RCB was carried out by standard curve Y = 42718X + 2.8163. The results are shown in Table 3.

Table 3.
The contents of bzmh in OR, RCO, RCM, RCS, and RCB (X ± SD).
The results showed that the signal peak of bzmh could be detected in OR, RCO, RCM and RCS in the same Rana chensinensis (Figure 4), which further indicated that there was 1-methyl hydantoin in theirs and individual difference existed, and no characteristic peak of bzmh was detected in the HPLC chromatogram of RCB. Table 3 and Figure 5 show the content of bzmh in RCS was the highest in the same Rana chensinensis, and the content in RCM and RCO was also abundant.
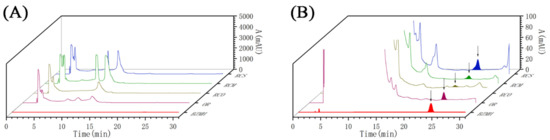
Figure 4.
The HPLC chromatogram of bzmh standard, OR, RCO, RCM, RCS, and RCB. (A) A: 0–5000 mAU; (B) A: 0–100 mAU.
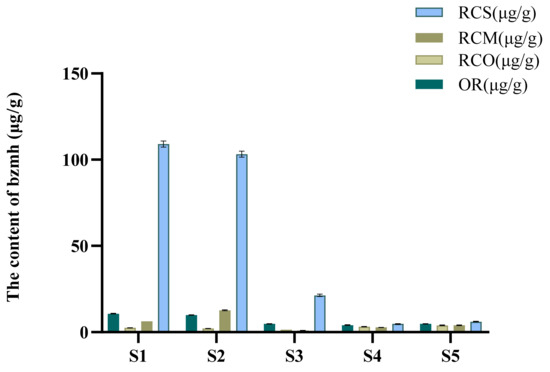
Figure 5.
Contents of bzmh in OR, RCO, RCM, and RCS from five Rana chensinensis samples.
3.2. Determination Results of PUFAs Content
3.2.1. Methodology Verification
The standard curves of six kinds PUFAs standards were used for the quantitative analysis of six PUFAs in Rana chensinensis samples. The regression equations of six components were calculated in the form of standard curve (Y = aX + b), where X and Y respectively were the concentration and peak area. Table 4 shows the results of the regression equations. All of the standard curves showed good linearity in the test range (R2 > 0.9982). The standard curves of six PUFAs were in Supplementary Materials Figure S1.

Table 4.
Regression Equations of six PUFAs standards.
The precision, stability and repeatability of the method meet the requirements of analysis [24]. The methodological verification of HPLC analytical methods of PUFAs were in Supplementary Materials Table S2.
3.2.2. Results of Contents of Six PUFAs in Each Sample
Six PUFAs identified in OR were quantitatively analyzed to explore the difference of PUFAs among OR, RCO, RCM, RCS, and RCB. Figure 6 shows the HPLC of mixed standard of six PUFAs, and OR, RCO, RCM, RCS, and RCB. The results showed that in addition to OR, six PUFAs could also be detected in RCO, RCM, RCS, RCB, and six PUFAs could be separated in 30 min. Table 5 shows the determination results of six PUFAs in all samples. Mirror comparison of HPLC chromatograms of mixed standards of six PUFAs to OR, RCO, RCM, and RCS were in Supplementary Materials Figure S2.
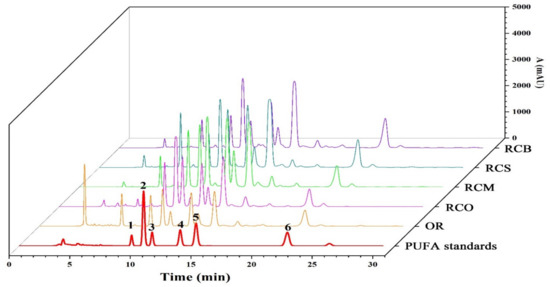
Figure 6.
The HPLC chromatogram of mixed standard of six PUFAs, OR, RCO, RCM, RCS, and RCB. (1. ALA; 2. ALA; 3. DHA; 4. ARA; 5. LA; 6. OA).

Table 5.
The contents of PUFAs in Different Tissue of Rana chensinensis samples. (X ± SD).
In order to compare the contents of six PUFAs in OR and other four parts tissue of the same Rana chensinensis, we compared the contents of six PUFAs in OR with the average contents of six PUFAs in RCO, RCM, RCS, and RCB. That means we treated other tissue as a whole and compared them with OR. The results are shown in Figure 7. It could be clearly seen that in the five Rana chensinensis samples except S13 the average contents of six PUFAs in RCO, RCM, RCS, and RCB were higher than those in OR. It was further proved that other tissue of OR removed Rana chensinensis also contain rich PUFAs. Detail data were presented in Supplementary Materials Table S3.
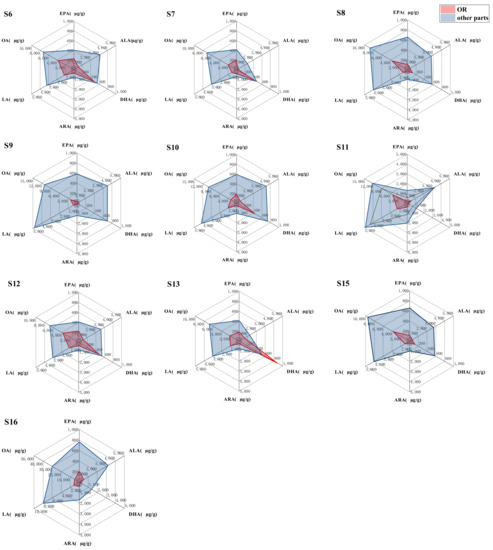
Figure 7.
Comparison of the contents of six PUFAs in OR and their average contents in RCO, RCM, RCS, and RCB in the same Rana chensinensis.
3.3. Hierarchical Cluster Analysis (HCA)
The results of cluster analysis showed that there were significant differences in the contents of six PUFAs in OR, RCO, RCM, RCS, and RCB. From the clustering results (Figure 8A), it was observed that the content differences of six kinds of PUFAs in OR, RCO, RCM, RCS and RCB were significant. From the clustering heat map, it could be seen that OR, RCO, RCM, RCS and RCB could be divided into four categories according to the differences of the contents of the six kinds of PUFAs. The first category mainly included part of OR and most of the RCM samples; almost all of the RCO and a small amount of RCM samples were classified into one category; most of the RCB and RCS samples were classified into one category, and the last category included OR, RCB and a small amount of RCS samples. Based on the total content sequence of the statistical charts (Figure 8B) of the total contents of six kinds PUFAs in the samples, it could be seen that the PUFA contents in most of the OR and some of the RCM and RCB were lower in front, while the RCO samples were higher in the back, and the rest of the RCB and RCS samples were between the two, which were generally consistent with the results of clustering analysis. Based on the content of six kinds of PUFAs in the samples, HCA successfully classified the OR, RCO, RCM, RCS, and RCB of all samples, which was related to the total amount of PUFAs in the samples. Therefore, compared with OR, other tissues of Rana chensinensis had more abundant PUFAs, which had development value as a by-product of this aspect.
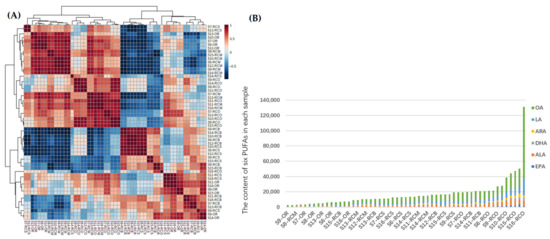
Figure 8.
(A) Clustering heatmap of six PUFAs in each sample. (B) Total content sequence of six PUFAs in each sample.
3.4. Principal Component Analysis (PCA)
The PCA of the sample was carried out by using SIMCA software. As shown in Figure 9, the quality control (QC) samples in PCA are centrally distributed and tightly clustered, indicating that the system has satisfactory stability. According to the content of six PUFAs in the sample based on the characteristic value > 1, the variance contribution rates of the first two principal components (PC1 and PC2) were 79.0% (R2X [1]) and 12.7% (R2X [2]), respectively. The cumulative variance contribution rate was 91.7%. It showed that the two principal components contain the most information of all variables and fully reflect the original data. The score plot was drawn when visualizing the results using two principal components (PC1 and PC2) (Figure 9). Corresponding to the results of HCA analysis, the scores of six PUFAs in five Rana chensinensis, such as OR, RCO, RCM, RCS, and RCB, could be divided into four regions in Figure 9 except that a few sample points were too discrete to gather in the corresponding area. Area a contained a portion of OR and most of RCM. Area b contained almost all RCO and a small amount of RCM, and area c contained most of the RCB and RCS samples. Area d included OR, RCB, and a small sample of RCS. Except for a few samples scattered in regions, the classification results of PCA model are consistent with those of HCA analysis. The samples of OR, RCO, RCM, RCS, and RCB were successfully classified. The combination of HAC and PCA is an effective tool to evaluate the content of PUFAs in different tissue of Rana chensinensis [44]. The similarities and differences in the contents of six PUFAs between OR and other tissue were shown by classification.
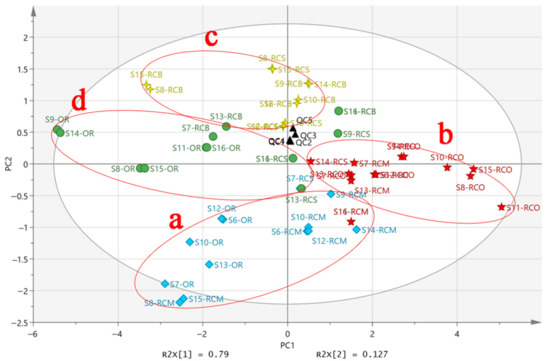
Figure 9.
The PCA score plot of six PUFAs in different tissue of Rana chensinensis.
3.5. Orthogonal Partial Least Squares Discriminant Analysis (OPLS-DA)
As a supervised pattern recognition model, OPLS-DA can maximize the differences between different categories and help filter the tags responsible for category separation [42,45]. The OPLS-DA model of different tissue of Rana chensinensis based on the PUFAs data after orthogonal signal correction can improve the reliability of the prediction model. In the OPLS-DA model, the samples of different tissue of the Rana chensinensis were pre-classified into four categories according to the results of PCA, and the score is shown in Figure 10A. The OPLS-DA model had satisfactory descriptiveness. The OPLS-DA score plot of the different tissue of Rana chensinensis was consistent with the results of HCA and PCA, and the score chart showed that the four predetermined categories were closely distributed.
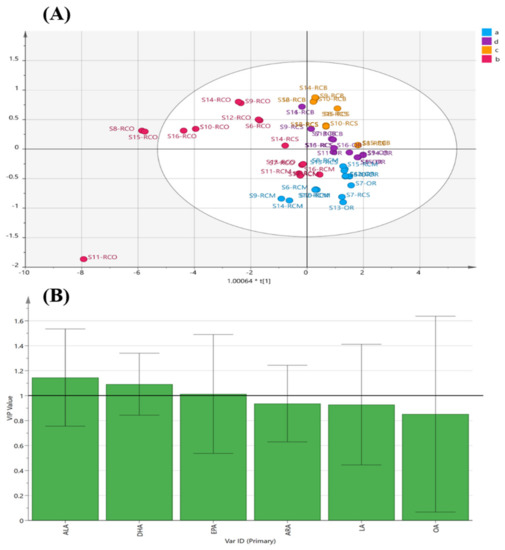
Figure 10.
Supervised pattern recognition model. (A) The OPLS-DA score plot of the different tissue of Rana chensinensis. (B) The variable importance for the project (VIP) plot of OPLS-DA.
The variable importance for the projection (VIP) diagram (Figure 10B) was used to determine the importance of each variable to the classification, which summarized the importance of the variable to interpret X and associating it with Y. Chemical variables with a VIP value greater than 1 are considered to play a significant role in classification [46,47]. In this OPLS-DA model, the VIP values of three variables (ALA, DHA, and EPA) were greater than 1, which further indicated that the content of PUFAs played an important role in distinguishing the different tissue of Rana chensinensis and evaluating their quality.
In order to further compare the similarities and differences between OR and other parts of Rana chensinensis, we established models for the relationship between the contents of six PUFAs in OR and other parts, respectively. According to the OPLS-DA score plot (Figure 11A), the OR samples were located at the end of the t[1] direction, and the RCO samples were located at the t[2] direction, which is completely separated from the OR. The RCM was located near the center of the model plane and partially overlapped with the OR overlapped in the t[1] direction (Figure 11B). The RCS samples were located near the center of the model plane, and significantly separated from the OR (Figure 11C). The RCB samples were located at the end in the t[2] direction, and did not intersect with the OR sample points (Figure 11D). The results showed that the content of six PUFAs in OR and RCM was similar, and the difference was small compared with other samples.
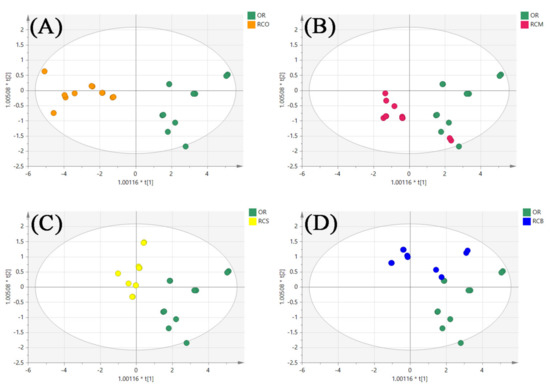
Figure 11.
The OPLS-DA score plot of OR and other parts of Rana chensinensis. (A) OR and RCO. (B) OR and RCM. (C) OR and RCS. (D) OR and RCB.
4. Conclusions
This study was divided into two parts. First, the contents of 1-methyl hydantoin in different tissue (OR, RCO, RCM, RCS, and RCB) of the same Rana chensinensis (S1–S5) were determined by pre-column derivatization combined with HPLC. The results showed that OR, RCO, RCM, and RCS all contained 1-methyl hydantoin, except for RCB. Moreover, the content of 1-methyl hydantoin in RCS was the highest. From the perspective of obtaining 1-methyl hydantoin, RCS has the most development value. Second, the contents of six PUFAs in different parts (OR, RCO, RCM, RCS, and RCB) of the same Rana chensinensis (S6–S16) were determined by HPLC and was used to further compare the contents of six PUFAs in OR and other tissue of Rana chensinensis. Two unsupervised pattern recognition models (PCA and HCA) and an unsupervised pattern recognition model (OPLS-DA) were established to analyze and classify the contents of six PUFAs in all samples. We combined the ranking results of the total amount of six kinds of PUFAs in all samples; it could be concluded that from the perspective of obtaining PUFAs, RCO had the most development value, and RCM was the most similar to expensive OR.
1-methyl hydantoin in OR had good antitussive and anti-inflammatory effects. The PUFAs in OR make it have high nutritional value. The results of two parts of this study indicated that RCO, RCM, and RCS, which were the by-products of OR, also contained the above components and were abundant, while the RCB contained rich PUFAs. This investigation could change the present situation of Rana chensinensis residue as waste after obtaining OR, and provide a theoretical basis for the further development of RCO, RCS, RCM, and RCB, which improves the utilization rate of the by-products of OR, and makes greater use of Rana chensinensis resources to promote the development of the Rana chensinensis industry.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/separations8100164/s1, Figure S1: The standard curves of eicosapentaenoic acid (EPA), α-linolenic acid (ALA), docosahexaenoic acid (DHA), arachidonic acid (ARA), linoleic acid (LA), oleic acid (OA) and benzoyl-1-methyl hydantoin (bzmh), Figure S2: Comparison of HPLC chromatograms of mixed standards of six PUFAs to OR, RCO, RCM, and RCS. (OR, Oviductus Ranae; RCO, Rana chensinensis ovum; RCM, Rana chensinensis meat; RCS, Rana chensinensis skin; RCB, Rana chensinensis bone.), Table S1: The six kinds of polyunsaturated fatty acids (PUFAs), Table S2: The methodological verification of HPLC analytical methods of PUFAs, Table S3: The contents of six PUFAs in OR and their average contents in RCO, RCM, RCS, and RCB in the same Rana chensinensis.
Author Contributions
Conceptualization, Y.W. and J.Z.; methodology, J.Z. and Z.W.; software, J.Z. and S.W.; validation, J.Z., S.W. and Y.W.; formal analysis, J.Z., Z.W., C.Z., D.X. and Y.Y.; investigation, J.Z., Z.W., C.Z. and N.L.; resources, Y.W. and S.W.; data curation, J.Z., Z.W. and S.W.; writing—original draft preparation, J.Z., Z.W., S.W., C.Z., N.L., D.X. and Y.Y.; writing—review and editing, J.Z., Z.W., S.W., C.Z. and D.X.; visualization, Z.W. and N.L.; supervision, Y.W.; project administration, Y.W.; funding acquisition, Y.W. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by Science and Technology Development Planning Project of Jilin Province, and the grant numbers was 20200404037YY.
Institutional Review Board Statement
The study was conducted according to the guidelines of the Declaration of Helsinki, and approved by Institutional Animal Care and Use Committee of Jilin University School of Pharmaceutical Science (the protocol code is 20200042 and the date of approval is 16 September 2020).
Informed Consent Statement
Not applicable.
Data Availability Statement
The authors confirm that the data supporting the findings of this study are available within the article and its Supplementary Materials.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Wang, S.; Xu, Y.; Wang, Y.; Yang, H.; Lv, Z.; Jin, X.; Wang, Y. Simultaneous Determination of Six Active Components in Oviductus Ranae via Quantitative Analysis of Multicomponents by Single Marker. J. Anal. Methods Chem. 2017, 2017, 9194847. [Google Scholar] [CrossRef] [PubMed]
- Gan, Y.S.; Xiao, Y.; Wang, S.H.; Guo, H.Y.; Liu, M.; Wang, Z.H.; Wang, Y.S. Protein-Based Fingerprint Analysis for the Identification of Ranae Oviductus Using RP-HPLC. Molecules 2019, 24, 1687. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Jia, X.-Y.; Ma, Y.-D.; Ma, J.Z. Genetic diversity and differentiation of the Dybowski’s frog (Rana dybowskii) in Northeast China. J. For. Res. 2010, 21, 239–245. [Google Scholar] [CrossRef]
- Wang, S.A.; Gan, Y.S.; Mao, X.X.; Kan, H.; Li, N.; Zhang, C.L.; Wang, Z.H.; Wang, Y.S. Antioxidant Activity Evaluation of Oviductus Ranae Protein Hydrolyzed by Different Proteases. Molecules 2021, 26, 1625. [Google Scholar]
- Xie, C.; Zhang, L.-J.; Zhang, W.-Y.; Yang, X.; Fan, L.; Li, X. Immunomodulatory effect of Oviductus Ranae on the mice. Chin. J. Gerontol. 2010, 30, 3132–3133. [Google Scholar]
- Xiao, Y.; Ni, S.L.; Wang, S.H.; Gan, Y.S.; Zhou, Y.; Guo, H.Y.; Liu, M.; Wang, Z.H.; Wang, Y.S. Environmental influences on quality features of Oviductus Ranae in the Changbai Mountains. RSC Adv. 2019, 9, 36050–36057. [Google Scholar] [CrossRef]
- Xu, Y.; Wang, S.H.; Luo, Y.; Wang, Y.S.; Qu, X.B. Evaluation of the Merits of the New Method of Oviductus Ranae by HPLC-DAD. J. Liq. Chromatogr. Relat. Technol. 2015, 38, 1218–1222. [Google Scholar] [CrossRef]
- Wang, S.; Gan, Y.; Kan, H.; Mao, X.; Wang, Y. Exploitation of HPLC Analytical Method for Simultaneous Determination of Six Principal Unsaturated Fatty Acids in Oviductus Ranae Based on Quantitative Analysis of Multi-Components by Single-Marker (QAMS). Molecules 2021, 26, 479. [Google Scholar]
- Wang, D.H.; Wu, W.; Tian, J.M.; Wang, Z.H.; Wang, D.T.; Xiang, K.; Zhu, G.Y.; Han, T. Effect of Oviductus Ranae and Oviductus Ranae eggs on bone metabolism and osteoporosis. Chin. J. Integr. Med. 2013, 19, 532–538. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wang, Y.F.; Li, M.Z.; Liu, S.Y.; Yu, J.L.; Yan, Z.W.; Zhou, H.L. Traditional Uses, Bioactive Constituents, Biological Functions, and Safety Properties of Oviductus Ranae as Functional Foods in China. Oxidative Med. Cell. Longev. 2019, 2019, 1–24. [Google Scholar] [CrossRef] [PubMed]
- Gan, Y.; Xu, D.; Zhang, J.; Wang, Z.; Wang, S.; Guo, H.; Zhang, K.; Li, Y.; Wang, Y. Rana chensinensis Ovum Oil Based on CO2 Supercritical Fluid Extraction: Response Surface Methodology Optimization and Unsaturated Fatty Acid Ingredient Analysis. Molecules 2020, 25, 4170. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Zhao, H.; Zhang, Y. Research Progress in Active Ingredient Extraction Methods and Pharmacological Effects of Skin and Eggs Derived from Rana chensinensis. J. Jilin Inst. Chem. Technol. 2019, 36, 15–20. [Google Scholar]
- Wang, Z.; Zhao, Y.; Su, T.; Zhang, J.; Wang, F. Characterization and antioxidant activity in vitro and in vivo of polysaccharide purified from Rana chensinensis skin. Carbohydr. Polym. 2015, 126, 17–22. [Google Scholar] [CrossRef] [PubMed]
- Ling-Ling, L.; Jin, X.S.; Shi, S.Y. Development, Extraction of Rana chensinensis Skin Collagen by Papain and its Antioxidant Activity. Food Res. Dev. 2013, 34, 22–24+94. [Google Scholar]
- Miao, Z.; Zhou, Y.; Min, Z. Measurement of Collagen Protein Contents in the Skin of Rana dybowskii and Extraction Methods. J. Northeast For. Univ. 2008, 36, 81–83. [Google Scholar]
- Wang, Z.Y.; Zhao, Y.Y.; Su, T.T. Extraction and antioxidant activity of polysaccharides from Rana chensinensis skin. Carbohyd. Polym. 2015, 115, 25–31. [Google Scholar] [CrossRef]
- Zhao, G.H.; Liang, Y.; Wang, Y.; Shang, D. Analysis and Evaluation of the Nutritional Components of Frog Flesh of Rana chensinensis. Acta Nutr. Sin. 2007, 29, 623–624. [Google Scholar]
- Jing, Z.; Hu, T.; Yin, Y.; Qin, F.; Wei, C. Optimization of High Voltage Pulse Electric Field-Assisted Extraction Technology of Calcium from Rana Bone by Response Surface Methodology. Food Ind. 2018, 39, 52–56. [Google Scholar]
- Commission, N.P. Pharmacopoeia of the People’s Republic of China (2020); China Medical Science Press: Beijing, China, 2020. [Google Scholar]
- Bao, H.W.; Yang, X.U.; Wang, Y.S.; Wang, S.H. 1-Methyl Hydantoin Content Comparison between Oviductus Ranae and Wood Frogs Spawn through HPLC. Spec. Wild Econ. Anim. Plant Res. 2019, 41, 89–92. [Google Scholar]
- Xiong, N.; Li, Q.; Dong, Y.; Wei, S.; Hu, Z.C.; Xue, Y.P.; Zheng, Y.G. Development of a Simple and Sensitive Pre-column Derivatization HPLC Method for the Quantitative Analysis of Miglitol Intermediates. Chromatographia 2021, 5512, 347–358. [Google Scholar] [CrossRef]
- Burakham, R.; Grudpan, K. Flow Injection and Sequential Injection On-line Pre-column Derivatization for Liquid Chromatography. J. Chromatogr. Sci. 2009, 47, 631–635. [Google Scholar] [CrossRef][Green Version]
- Gatti, R. Simultaneous Determination of Taurine, N-Acetylcysteine, Glycine and Methionine in Commercial Formulations by High-Performance Liquid Chromatography. Chromatographia 2019, 82, 1833–1837. [Google Scholar] [CrossRef]
- Guo, H.; Gan, Y.; Liu, M.; Wang, S.; Ni, S.; Zhou, Y.; Xiao, Y.; Wang, Z.; Wang, Y. Quality Evaluation of Oviductus Ranae Based on PUFAs Using HPLC Fingerprint Techniques Combined with Chemometric Methods. Foods 2019, 8, 322. [Google Scholar] [CrossRef]
- Racey, M.; MacFarlane, A.J.; Carlson, S.; Stark, K.D.; Plourde, M.; Field, C.J.; Yates, A.A.; Wells, G.A.; Grantham, A.; Bazinet, R.P.; et al. Dietary Reference Intakes Based on Chronic Disease Endpoints: Outcomes from a case study workshop for omega 3’s EPA and DHA. Appl. Physiol. Nutr. Metab. 2021, 46, 530–539. [Google Scholar] [CrossRef] [PubMed]
- Kulzow, N.; Witte, A.V.; Kerti, L.; Grittner, U.; Schuchardt, J.P.; Hahn, A.; Floel, A. Impact of Omega-3 Fatty Acid Supplementation on Memory Functions in Healthy Older Adults. J. Alzheimer’s Dis. 2016, 51, 713–725. [Google Scholar] [CrossRef] [PubMed]
- Endo, J.; Arita, M. Cardioprotective mechanism of omega-3 polyunsaturated fatty acids. J. Cardiol. 2016, 67, 22–27. [Google Scholar] [CrossRef]
- Rodriguez-Alcala, L.M.; Calvo, M.V.; Fontecha, J.; Alonso, L. Alterations in the Fatty Acid Composition in Infant Formulas and 3-PUFA Enriched UHT Milk during Storage. Foods 2019, 8, 163. [Google Scholar] [CrossRef]
- Gogus, U.; Smith, C. n-3 Omega fatty acids: A review of current knowledge. Int. J. Food Sci. Technol. 2010, 45, 417–436. [Google Scholar] [CrossRef]
- Liu, B.; Yan, W. Quantitative Polyunsaturated Fatty Acid Analysis of Chia Seed Oil by High-Performance Liquid Chromatography. J. Chromatogr. Sci. 2021, 59, 120–127. [Google Scholar] [CrossRef]
- Shahidi, F.; Ambigaipalan, P. Omega-3 Polyunsaturated Fatty Acids and Their Health Benefits. Annu. Rev. Food Sci. Technol. 2018, 9, 345–381. [Google Scholar] [CrossRef]
- Wang, D.D. Dietary n-6 polyunsaturated fatty acids and cardiovascular disease: Epidemiologic evidence. Prostaglandis Leukot. Essent. Fat. Acids 2018, 135, 5–9. [Google Scholar] [CrossRef]
- You, J.S.; Shi, J.L.; Zhang, S.F.; Duan, H.H.; Shi, S.N.; Guo, J.Y. Antidepressant Effects of Petroleum Ether Extracts from Ranae Oviductus. Chin. J. Exp. Tradit. Med. Formulae 2013, 19, 271–274. [Google Scholar]
- Wang, S. Study on the Quality Evalutation and the Active Components Derivatives of Oviductus ranae. Ph.D. Thesis, Jilin University, Changchun, China, 2017. [Google Scholar]
- Yu, G.; Qin, J.; Li, J.; Wang, W.; Bi, Y.; Wang, Z.; Xiao, W. Determination of α-linolenic acid, linoleic acid and oleic acid in Oviductus ranae by HPLC. Chin. J. Exp. Tradit. Med. 2013, 19, 82–85. [Google Scholar]
- Liu, Y.H.; Wang, Z.Y.; Wang, C.X.; Si, H.R.; Yu, H.; Li, L.; Fu, S.Z.; Tan, L.Y.; Li, P.Y.; Liu, J.P.; et al. Comprehensive phytochemical analysis and sedative-hypnotic activity of two Acanthopanax species leaves. Food Funct. 2021, 12, 2292–2311. [Google Scholar] [CrossRef] [PubMed]
- Fraige, K.; Pereira, E.R.; Carrilho, E. Fingerprinting of anthocyanins from grapes produced in Brazil using HPLC-DAD-MS and exploratory analysis by principal component analysis. Food Chem. 2014, 145, 395–403. [Google Scholar] [CrossRef] [PubMed]
- Granato, D.; Santos, J.S.; Escher, G.B.; Ferreira, B.L.; Maggio, R.M. Use of principal component analysis (PCA) and hierarchical cluster analysis (HCA) for multivariate association between bioactive compounds and functional properties in foods: A critical perspective. Trends Food Sci. Technol. 2018, 72, 83–90. [Google Scholar] [CrossRef]
- Lee, J.W.; Choi, B.R.; Kim, Y.C.; Choi, D.J.; Lee, Y.S.; Kim, G.S.; Baek, N.I.; Kim, S.Y.; Lee, D.Y. Comprehensive Profiling and Quantification of Ginsenosides in the Root, Stem, Leaf, and Berry of Panax ginseng by UPLC-QTOF/MS. Molecules 2017, 22, 2147. [Google Scholar] [CrossRef]
- Ragusa, A.; Centonze, C.; Grasso, M.E.; Latronico, M.F.; Mastrangelo, P.F.; Sparascio, F.; Fanizzi, F.P.; Maffia, M. A Comparative Study of Phenols in Apulian Italian Wines. Foods 2017, 6, 24. [Google Scholar] [CrossRef]
- Zhang, H.M.; Li, S.L.; Zhang, H.; Wang, Y.; Zhao, Z.L.; Chen, S.L.; Xu, H.X. Holistic quality evaluation of commercial white and red ginseng using a UPLC-QTOF-MS/MS-based metabolomics approach. J. Pharm. Biomed. Anal. 2012, 62, 258–273. [Google Scholar] [CrossRef]
- Huang, B.M.; Chen, T.B.; Xiao, S.Y.; Zha, Q.L.; Luo, P.; Wang, Y.P.; Cui, X.M.; Liu, L.; Zhou, H. A new approach for authentication of four ginseng herbs and their related products based on the simultaneous quantification of 19 ginseng saponins by UHPLC-TOF/MS coupled with OPLS-DA. RSC Adv. 2017, 7, 46839–46851. [Google Scholar] [CrossRef]
- Rohman, A.; Wijayanti, T.; Windarsih, A.; Riyanto, S. The Authentication of Java Turmeric (Curcuma xanthorrhiza) Using Thin Layer Chromatography and(1)H-NMR Based-Metabolite Fingerprinting Coupled with Multivariate Analysis. Molecules 2020, 25, 3928. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.F.; Liang, W.Y.; Chen, W.J.; Li, S.; Cui, Y.P.; Qi, Q.; Zhang, L.Z. Screening and Analysis of the Marker Components in Ganoderma lucidum by HPLC and HPLC-MSn with the Aid of Chemometrics. Molecules 2017, 22, 584. [Google Scholar] [CrossRef] [PubMed]
- Bylesjo, M.; Rantalainen, M.; Cloarec, O.; Nicholson, J.K.; Holmes, E.; Trygg, J. OPLS discriminant analysis: Combining the strengths of PLS-DA and SIMCA classification. J Chemom. 2006, 20, 341–351. [Google Scholar] [CrossRef]
- Lin, H.Q.; Zhu, H.L.; Tan, J.; Wang, H.; Wang, Z.Y.; Li, P.Y.; Zhao, C.F.; Liu, J.P. Comparative Analysis of Chemical Constituents of Moringa oleifera Leaves from China and India by Ultra-Performance Liquid Chromatography Coupled with Quadrupole-Time-Of-Flight Mass Spectrometry. Molecules 2019, 24, 942. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Zhang, C.X.; Shao, C.F.; Li, C.W.; Liu, S.H.; Peng, X.P.; Xu, Y.Q. Chemical Fingerprint Analysis for the Quality Evaluation of Deepure Instant Pu-erh Tea by HPLC Combined with Chemometrics. Food Anal. Methods 2016, 9, 3298–3309. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).