Biological Activity of Cannabis sativa L. Extracts Critically Depends on Solvent Polarity and Decarboxylation
Abstract
1. Introduction
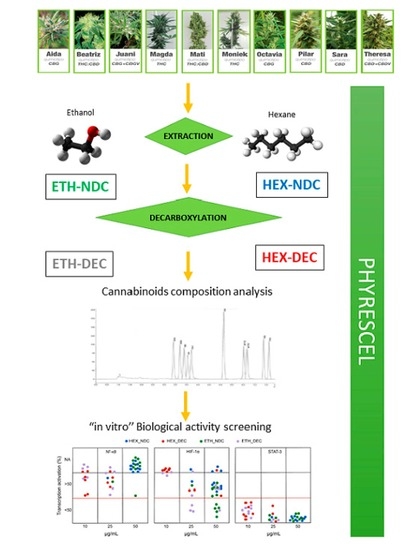
2. Materials and Methods
2.1. Plant Material
2.2. Reagents for Plant Material Extraction, Purification of Cannabinoids and Analysis of Cannabinoid Content
2.3. Extraction Procedure
2.4. Cannabinoid Purification
2.5. Analysis of Cannabinoid Content
2.6. Sample Preparation for Biological Activity
2.7. Cell Lines
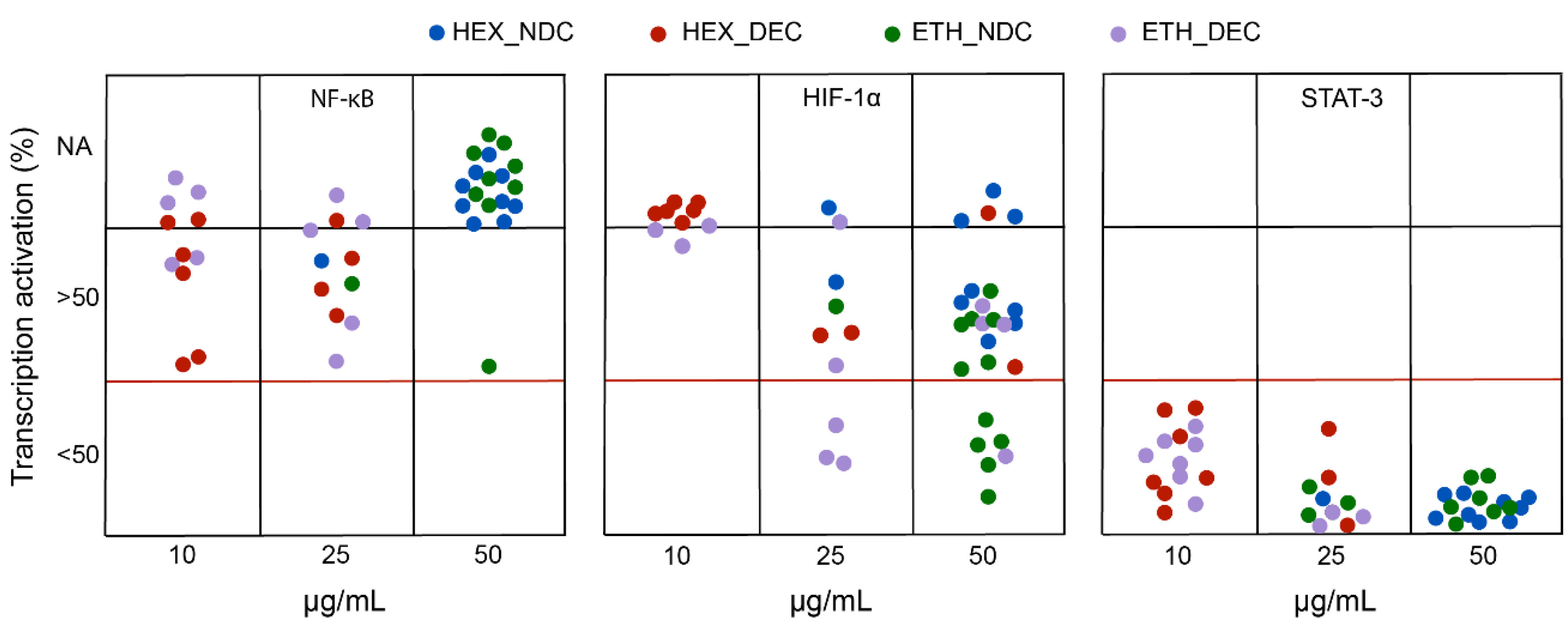
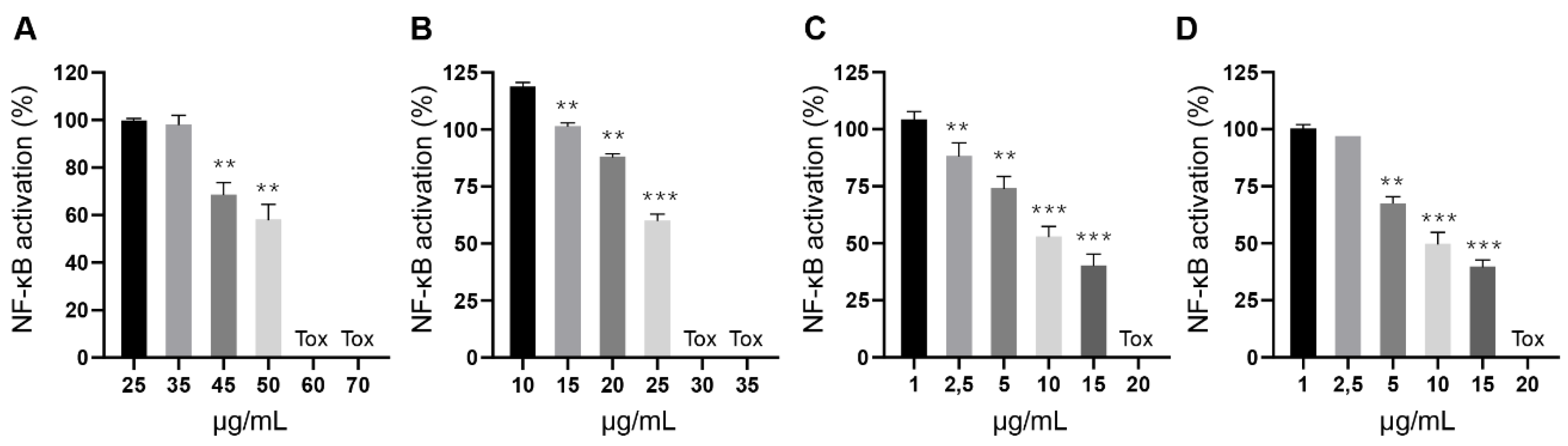
2.8. Determination of Biological Activity
2.9. Determination of Cytotoxicity
2.10. Statistical Analysis
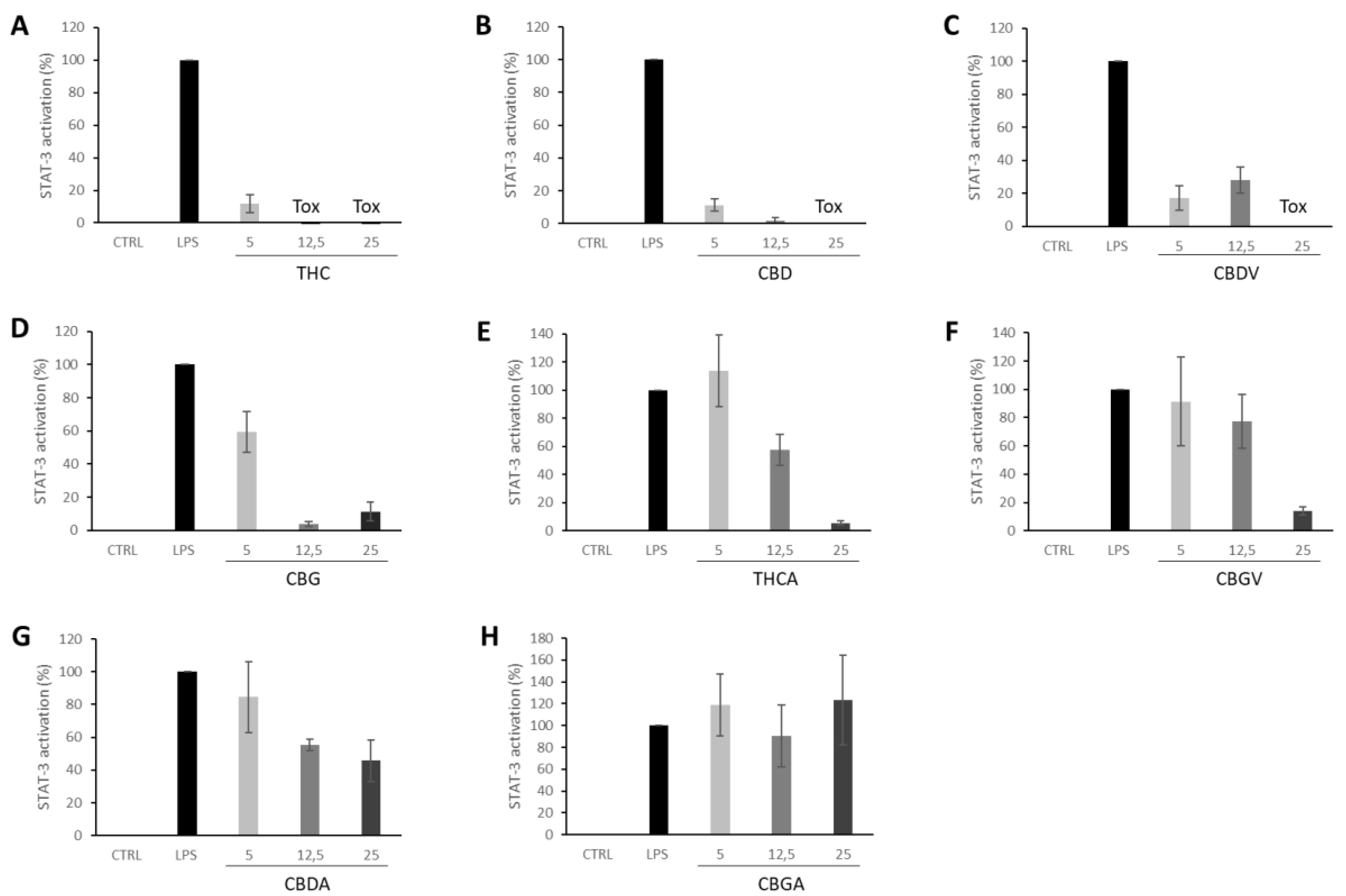
3. Results
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Hanuš, L.O.; Meyer, S.M.; Muñoz, E.; Taglialatela-Scafati, O.; Appendino, G. Phytocannabinoids: A unified critical inventory. Nat. Prod. Rep. 2016, 33, 1357–1392. [Google Scholar] [CrossRef] [PubMed]
- Russo, E.; Guy, G.W. A tale of two cannabinoids: The therapeutic rationale for combining tetrahydrocannabinol and cannabidiol. Med. Hypotheses 2006, 66, 234–246. [Google Scholar] [CrossRef] [PubMed]
- Moreno-Sanz, G. Can you pass the acid test? Critical review and novel therapeutic perspectives of δ9-tetrahydrocannabinolic acid A. Cannabis Cannabinoid Res. 2016, 1, 124–130. [Google Scholar] [PubMed]
- ElSohly, M.A.; Radwan, M.M.; Gul, W.; Chandra, S.; Galal, A. Phytochemistry of cannabis sativa L. In Progress in the Chemistry of Organic Natural Products; Springer: Berlin/Heidelberg, Germany, 2017; pp. 1–36. [Google Scholar]
- LaPrairie, R.B.; Bagher, A.M.; Kelly, M.E.M.; Denovan-Wright, E.M. Cannabidiol is a negative allosteric modulator of the cannabinoid CB1 receptor. Br. J. Pharmacol. 2015, 172, 4790–4805. [Google Scholar] [CrossRef]
- Martínez-Pinilla, E.; Varani, K.; Reyes-Resina, I.; Angelats, E.; Vincenzi, F.; Ferreiro-Vera, C.; Oyarzabal, J.; Canela, E.I.; Lanciego, J.L.; Nadal, X.; et al. Binding and signaling studies disclose a potential allosteric site for cannabidiol in cannabinoid CB2 Receptors. Front. Pharmacol. 2017, 8, 744. [Google Scholar] [CrossRef]
- Tham, M.; Yilmaz, O.; Alaverdashvili, M.; Kelly, M.E.M.; Denovan-Wright, E.M.; LaPrairie, R.B. Allosteric and orthosteric pharmacology of cannabidiol and cannabidiol-dimethylheptyl at the type 1 and type 2 cannabinoid receptors. Br. J. Pharmacol. 2019, 176, 1455–1469. [Google Scholar] [CrossRef]
- Navarro, G.; Reyes-Resina, I.; Rivas-Santisteban, R.; De Medina, V.S.; Morales, P.; Casano, S.; Ferreiro-Vera, C.; Lillo, A.; Aguinaga, D.; Jagerovic, N.; et al. Cannabidiol skews biased agonism at cannabinoid CB1 and CB2 receptors with smaller effect in CB1-CB2 heteroreceptor complexes. Biochem. Pharmacol. 2018, 157, 148–158. [Google Scholar] [CrossRef]
- Russo, E.B.; Marcu, J. Cannabis pharmacology: The usual suspects and a few promising leads. In Advances in Pharmacology; Academic Press: Cambridge, MA, USA, 2017; pp. 67–134. [Google Scholar]
- MacCallum, C.A.; Russo, E.B. Practical considerations in medical cannabis administration and dosing. Eur. J. Intern. Med. 2018, 49, 12–19. [Google Scholar] [CrossRef]
- Grinspoon, L.; Bakalar, J.B.; Zimmer, L.; Morgan, J.P.; McGregor, I.S.; Di Chiaro, G.; De Fonseca, F.R.; Navarro, M.; Carrera, M.R.A.; Koob, G.F.; et al. Marijuana addiction. Science 1997, 277, 749–752. [Google Scholar] [CrossRef]
- Russo, E.B.; McPartland, J.M. Cannabis is more than simply Δ9-tetrahydrocannabinol. Psychopharmacology 2003, 165, 431–432. [Google Scholar] [CrossRef]
- Russo, E.B. Taming THC: Potential cannabis synergy and phytocannabinoid-terpenoid entourage effects. Br. J. Pharmacol. 2011, 163, 1344–1364. [Google Scholar] [CrossRef] [PubMed]
- Santiago, M.; Sachdev, S.; Arnold, J.C.; McGregor, I.S.; Connor, M. Absence of entourage: Terpenoids commonly found in Cannabis sativa do not modulate the functional activity of Δ9-THC at human CB1and CB2 receptors. Cannabis Cannabinoid Res. 2019, 4, 165–176. [Google Scholar] [CrossRef] [PubMed]
- Fitzcharles, M.A.; Eisenberg, E. Medical cannabis: A forward vision for the clinician. Eur. J. Pain 2018, 22, 485–491. [Google Scholar] [CrossRef]
- Callado, C.S.-C.; Núñez-Sánchez, N.; Casano, S.; Ferreiro-Vera, C. The potential of near infrared spectroscopy to estimate the content of cannabinoids in Cannabis sativa L.: A comparative study. Talanta 2018, 190, 147–157. [Google Scholar] [CrossRef] [PubMed]
- Roura, X.N. Methods of Purifying Cannabinoids, Compositions, and Kits Thereof. WO Patent 2016116628, 28 July 2016. [Google Scholar]
- Roura, X.N. Methods of Purifying Cannabinoids Using Liquid: Liquid Chromatography. WO Patent 2019145552A1, 1 August 2019. [Google Scholar]
- Baldino, L.; Scognamiglio, M.; Reverchon, E. Supercritical fluid technologies applied to the extraction of compounds of industrial interest from Cannabis sativa L. and to their pharmaceutical formulations: A review. J. Supercrit. Fluids 2020, 165, 104960. [Google Scholar] [CrossRef]
- Lewis-Bakker, M.M.; Yang, Y.; Vyawahare, R.; Kotra, L.P. Extractions of medical cannabis cultivars and the role of decarboxylation in optimal receptor responses. Cannabis Cannabinoid Res. 2019, 4, 183–194. [Google Scholar] [CrossRef]
- Del Prete, D.; Taglialatela-Scafati, O.; Minassi, A.; Sirignano, C.; Cruz, C.; Bellido, M.L.; Muñoz, E.; Appendino, G. Electrophilic triterpenoid enones: A comparative thiol-trapping and bioactivity study. J. Nat. Prod. 2017, 80, 2276–2283. [Google Scholar] [CrossRef]
- Minassi, A.; Rogati, F.; Cruz, C.; Prados, M.E.; Galera, N.; Jiménez, C.; Appendino, G.; Bellido, M.L.; Calzado, M.A.; Caprioglio, D.; et al. Triterpenoid hydroxamates as HIF prolyl hydrolase inhibitors. J. Nat. Prod. 2018, 81, 2235–2243. [Google Scholar] [CrossRef] [PubMed]
- Ruhaak, L.R.; Felth, J.; Karlsson, P.C.; Rafter, J.J.; Verpoorte, R.; Bohlin, L. Evaluation of the cyclooxygenase inhibiting effects of six major cannabinoids isolated from Cannabis sativa. Biol. Pharm. Bull. 2011, 34, 774–778. [Google Scholar] [CrossRef]
- Brown, I.; Cascio, M.G.; Rotondo, D.; Pertwee, R.G.; Heys, S.D.; Wahle, K.W.J. Cannabinoids and omega-3/6 endocannabinoids as cell death and anticancer modulators. Prog. Lipid Res. 2013, 52, 80–109. [Google Scholar] [CrossRef] [PubMed]
- Baram, L.; Peled, E.; Berman, P.; Yellin, B.; Besser, E.; Benami, M.; Louria-Hayon, I.; Lewitus, G.M.; Meiri, D. The heterogeneity and complexity of Cannabis extracts as antitumor agents. Oncotarget 2019, 10, 4091–4106. [Google Scholar] [CrossRef] [PubMed]
- Kozela, E.; Pietr, M.; Juknat, A.; Rimmerman, N.; Levy, R.; Vogel, Z. Cannabinoids Δ 9-tetrahydrocannabinol and cannabidiol differentially inhibit the lipopolysaccharide-activated NF-κB and Interferon-β/STAT proinflammatory pathways in BV-2 microglial cells. J. Biol. Chem. 2010, 285, 1616–1626. [Google Scholar] [CrossRef] [PubMed]
- Jüttler, E.; Potrovita, I.; Tarabin, V.; Prinz, S.; Dong-Si, T.; Fink, G.; Schwaninger, M. The cannabinoid dexanabinol is an inhibitor of the nuclear factor-kappa B (NF-κB). Neuropharmacology 2004, 47, 580–592. [Google Scholar] [CrossRef] [PubMed]
- Blanco, E.M.; Appendino, G.; Fiebich, B.L.; Grassi, G. A Composition Containing Non-Psychotropic Cannabinoids for the Treatment of Inflammatory Diseases. WO Patent 2009043836A1, 9 April 2007. [Google Scholar]
- Sánchez-Duffhues, G.; Calzado, M.A.; de Vinuesa, A.G.; Appendino, G.; Fiebich, B.L.; Loock, U.; Lefarth-Risse, A.; Krohn, K.; Muñoz, E. Denbinobin inhibits nuclear factor-kappaB and induces apoptosis via reactive oxygen species generation in human leukemic cells. Biochem. Pharmacol. 2009, 77, 1401–1409. [Google Scholar] [CrossRef]
- Kaur, B.; Khwaja, F.W.; Severson, E.A.; Matheny, S.L.; Brat, D.J.; van Meir, E.G. Hypoxia and the hypoxia-inducible-factor pathway in glioma growth and angiogenesis. NeuroOncology 2005, 7, 134–153. [Google Scholar] [CrossRef]
- Kossatz, E.; Maldonado, R.; Robledo, P. CB2 cannabinoid receptors modulate HIF-1α and TIM-3 expression in a hypoxia-ischemia mouse model. Eur. Neuropsychopharmacol. 2016, 26, 1972–1988. [Google Scholar] [CrossRef]
- Solinas, M.; Massi, P.; Cinquina, V.; Valenti, M.; Bolognini, D.; Gariboldi, M.; Monti, E.; Rubino, T.; Parolaro, D. Cannabidiol, a non-psychoactive cannabinoid compound, inhibits proliferation and invasion in U87-MG and T98G glioma cells through a multitarget effect. PLoS ONE 2013, 8, e76918. [Google Scholar] [CrossRef] [PubMed]
- Deng, X.; Peng, Y.; Zhao, J.; Lei, X.; Zheng, X.; Xie, Z.-Z.; Tang, G. Anticancer activity of natural flavonoids: Inhibition of HIF-1α signaling pathway. Curr. Org. Chem. 2019, 23, 2945–2959. [Google Scholar] [CrossRef]
- Yu, H.; Lee, H.; Herrmann, A.; Buettner, R.; Jove, R. Revisiting STAT3 signalling in cancer: New and unexpected biological functions. Nat. Rev. Cancer 2014, 14, 736–746. [Google Scholar] [CrossRef]
- Zhou, H.; Zhang, Z.; Wei, H.; Wang, F.; Guo, F.; Gao, Z.; Marsicano, G.; Wang, Q.; Xiong, L. Activation of STAT3 is involved in neuroprotection by electroacupuncture pretreatment via cannabinoid CB1 receptors in rats. Brain Res. 2013, 1529, 154–164. [Google Scholar] [CrossRef]
- Ciaglia, E.; Torelli, G.; Pisanti, S.; Picardi, P.; D’Alessandro, A.; Laezza, C.; Malfitano, A.M.; Fiore, D.; Zottola, A.C.P.; Proto, M.C.; et al. Cannabinoid receptor CB1 regulates STAT3 activity and its expression dictates the responsiveness to SR141716 treatment in human glioma patients’ cells. Oncotarget 2015, 6, 15464–15481. [Google Scholar] [CrossRef] [PubMed]
- Montecucco, F.; Lenglet, S.; Braunersreuther, V.; Burger, F.; Pelli, G.; Bertolotto, M.B.; Mach, F.; Steffens, S. CB (2) cannabinoid receptor activation is cardioprotective in a mouse model of ischemia/reperfusion. J. Mol. Cell. Cardiol. 2009, 46, 612–620. [Google Scholar] [CrossRef] [PubMed]
- Stančić, A.; Jandl, K.; Hasenöhrl, C.; Reichmann, F.; Marsche, G.; Schuligoi, R.; Heinemann, A.; Storr, M.A.; Schicho, R. The GPR55 antagonist CID16020046 protects against intestinal inflammation. Neurogastroenterol. Motil. 2015, 27, 1432–1445. [Google Scholar] [CrossRef] [PubMed]
- Chang, X.; Bian, Y.; He, Q.; Yao, J.; Zhu, J.; Wu, J.; Wang, K.; Duan, T. Suppression of STAT3 signaling by Δ9-Tetrahydrocannabinol (THC) induces trophoblast dysfunction. Cell. Physiol. Biochem. 2017, 42, 537–550. [Google Scholar] [CrossRef] [PubMed]

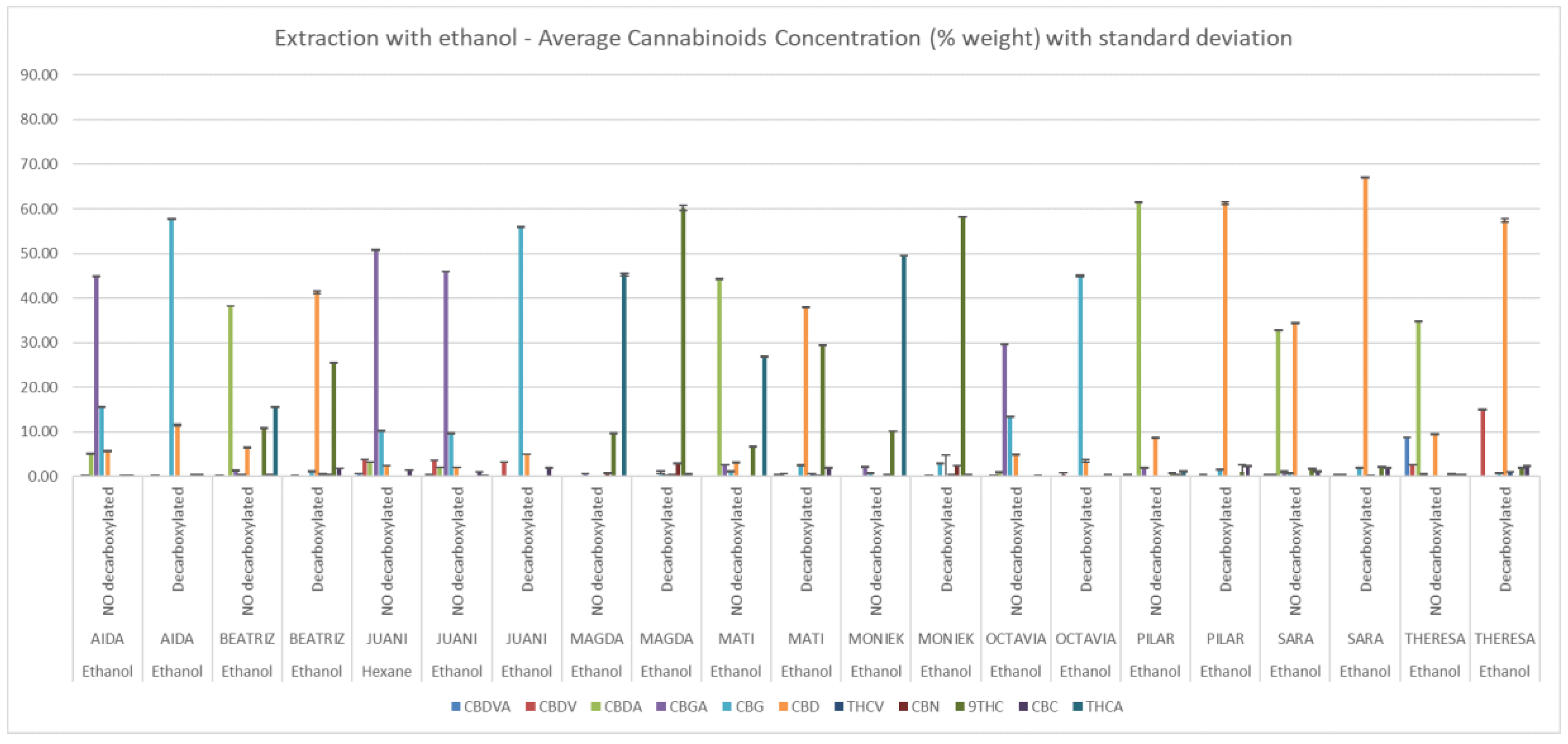

| VARIETY | Main Cannabinoid | Phenotype | Biomass Yield (g/m2) | Δ9-THC (%) | Cannabinoids Total (%) | Terpenes Total (%) | Terpene Profile | Yield Ethanol Extraction (%) | Yield Hexane Extraction (%) |
|---|---|---|---|---|---|---|---|---|---|
| MONIEK | Δ9-THC | I | 1.000 (flower only) | 15.5–22.0 | 19.0–26.0 | 0.75–0.90 | Limonene:Linalool:β-Caryophyllene:Guaiol ~1 > Myrcene > α-Terpineol:Fenchol:β Pinene ~1 | 28 | 28.4 |
| MAGDA | Δ9-THC | I | 500 (flower only) | 11.0–14.0 | 13.0–16.5 | 0.50–0.60 | Myrcene:β-Caryophyllene ~1 > α-Pinene > β-Pinene > Limonene:Linalool ~1 | 20 | 20 |
| MATI | Δ9-THC+CBD | II | 1.000 (flower only) | 5.00–6.00 | 10.5–14.5 | ND | ND | 20 | 19.6 |
| BEATRIZ | Δ9-THC+CBD | II | 500 (flower only) | 4.50–6.50 | 11.0–15.0 | ND | ND | 18.8 | 17.8 |
| SARA | CBD | III | 515 (leaf and flower) | 0.40–0.60 | 7.0–10.5 | 0.30–0.45 | Myrcene > Phytol > β-Caryophyollene | 20.1 | 18.7 |
| PILAR | CBD | III | 605 (leaf and flower) | 0.30–0.65 | 4.5–8.5 | 0.25–0.50 | α-Pinene > Myrcene:β-Caryophyllene ~1 > Guaiol:β-Pinene:Phytol ~1 | 12 | 11.7 |
| THERESA | CBD+CBDV | III | 780 (leaf and flower) | 0.30 –0.70 | 6.0–11.0 | 0.25–0.80 | Myrcene > β-Caryophyllene > Phytol > Limonene:Linalool ~1 | 8.8 | 8 |
| AIDA | CBG | IV | 555 (leaf and flower) | 0.05 –0.11 | 3.5–8.5 | 0.30–0.40 | Myrcene > Guaiol:Phytol ~1 > β-Caryophyllene: α-Pinene ~1 | 9.5 | 7.6 |
| OCTAVIA | CBG | IV | 1.000 (leaf and flower) | 0.02 –0.15 | 5.0–11.0 | 0.15–0.20 | Phytol > Myrcene > β-caryophyllene: Ocimene ~1 | 6.8 | 4.9 |
| JUANI | CBG+CBGV | IV | 880 (leaf and flower) | 0.03–0.05 | 3.0–6.0 | 0.15–0.20 | Phytol > Myrcene > β-caryophyllene:guaiol ~1 > α-Pinene: Ocimene ~1 | 8.2 | 5.7 |
| Group | Variety | % of Transcriptional Activation | CBDV | CBD | CBDA | THCV | Δ-9-THC | THCA | CBG | CBGA | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 10 μg·mL−1 | 25 μg·mL−1 | 50 μg·mL−1 | ||||||||||
| ETH_NDC | MAGDA | 107.9 ± 4.0 | 90.1 ± 17.0 | 53.5 ± 1.98 | 0 | 0 | 0.67 | 0 | 9.71 | 45.34 | 0.1 | 0.67 |
| ETH_DEC | BEATRIZ | 131.01 | 54.21 | 0.19 | 41.36 | 0 | 0.42 | 25.5 | 0 | 1.34 | 0 | |
| HEX_DEC | PILAR | 56.8 | - | - | 0.4 | 68.0 | 0 | 0 | 1.25 | 0 | 1.88 | 0 |
| SARA | 57.9 ± 12.3 | - | - | 0.99 | 83.79 | 0 | 0.8 | 3.0 | 0 | 2.48 | 0 | |
| 5 μg·mL−1 | 12.5 μg·mL−1 | 25 μg·mL−1 | ||||||||||
| ISOLATED STANDARDS | CBD | 102.7 ± 3.6 | - | - | ||||||||
| THC | 111.3 ±12.4 | - | - | |||||||||
| CBDA | 105.3 ± 7.5 | 102 ± 13.8 | 109.1 ± 3.1 | |||||||||
| THCA | 107.9 ± 4.7 | 109.3 ± 11.7 | 112.4 ± 8.9 | |||||||||
| Group | Variety | % of Transcriptional Activation | CBDV | CBD | CBDA | THCV | Δ-9-THC | THCA | CBG | CBGA | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 10 μg·mL−1 | 25 μg·mL−1 | 50 μg·mL−1 | ||||||||||
| ETH_NDC | MONIEK | 89.1 ± 6.3 | 58.7 ± 10.5 | 30.5 ± 1.8 | 0 | 0 | 0 | 0 | 10.45 | 49.52 | 0.72 | 2.11 |
| MAGDA | 85.8 ± 9.1 | 42.2 ± 1.9 | 12.6 ± 3.1 | 0 | 0 | 0 | 0 | 9.71 | 45.34 | 0.10 | 0.67 | |
| MATI | 104.8 ± 4.8 | 70.2 ± 3.6 | 38.5 ± 17.3 | 0 | 3.16 | 44.26 | 0 | 6.76 | 26.95 | 1.14 | 2.70 | |
| PILAR | 100.6 ± 18 | 67.6 ± 8.6 | 30.3 ± 26.5 | 0 | 8.71 | 61.60 | 0 | 0.79 | 1.26 | 0.14 | 2.12 | |
| THERESA | 93.3 ± 27.1 | 61.1 ± 34 | 26.1 ± 16.2 | 2.59 | 9.48 | 34.90 | 0.08 | 0.65 | 0.42 | 0.08 | 0.53 | |
| ETH_DEC | AÍDA | 103.2 ± 9.8 | 82.2 ± 7.7 | 29.3 ± 2.7 | 0.35 | 11.54 | 0 | 0 | 0.35 | 0 | 57.86 | 0 |
| JUANI | 80.2 ± 7.7 | 40.1 ± 14.3 | - | 3.27 | 5.10 | 0 | 0 | 0 | 0 | 56.14 | 0 | |
| MATI | 99.6 ± 35.9 | 29.2 ± 14.7 | - | 0.13 | 37.91 | 0 | 0.62 | 29.53 | 0 | 2.71 | 0 | |
| THERESA | 101.4 ± 22.7 | 28.7 ± 9.9 | - | 15.05 | 57.47 | 0 | 0.98 | 1.97 | 0 | 0.79 | 0 | |
| 5 μg·mL−1 | 12.5 μg·mL−1 | 25 μg·mL−1 | ||||||||||
| ISOLATED STANDARDS | CBD | 55.6 ± 19.5 | 0.59 ± 1.83 | - | ||||||||
| CBDV | 66.1 ± 28.3 | 26 ± 31.7 | - | |||||||||
| CBDA | 80.8 ± 21.9 | 86.6 ± 11.1 | 81.3 ± 10.9 | |||||||||
| CBG | 89.2 ± 10.5 | 59.7 ± 22 | 41.5 ± 1.2 | |||||||||
| CBGA | 75.8 ± 23.8 | 86.4 ± 41.6 | 61.4 ± 18.1 | |||||||||
| THCA | 106.4 ± 9.1 | 103.1 ± 3.9 | 85.2 ± 25.7 | |||||||||
| THC | 107.9 ± 4.7 | 109.3 ± 11.7 | 112.4 ± 8.9 | |||||||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Moreno-Sanz, G.; Ferreiro Vera, C.; Sánchez-Carnerero, C.; Nadal Roura, X.; Sánchez de Medina Baena, V. Biological Activity of Cannabis sativa L. Extracts Critically Depends on Solvent Polarity and Decarboxylation. Separations 2020, 7, 56. https://doi.org/10.3390/separations7040056
Moreno-Sanz G, Ferreiro Vera C, Sánchez-Carnerero C, Nadal Roura X, Sánchez de Medina Baena V. Biological Activity of Cannabis sativa L. Extracts Critically Depends on Solvent Polarity and Decarboxylation. Separations. 2020; 7(4):56. https://doi.org/10.3390/separations7040056
Chicago/Turabian StyleMoreno-Sanz, Guillermo, Carlos Ferreiro Vera, Carolina Sánchez-Carnerero, Xavier Nadal Roura, and Verónica Sánchez de Medina Baena. 2020. "Biological Activity of Cannabis sativa L. Extracts Critically Depends on Solvent Polarity and Decarboxylation" Separations 7, no. 4: 56. https://doi.org/10.3390/separations7040056
APA StyleMoreno-Sanz, G., Ferreiro Vera, C., Sánchez-Carnerero, C., Nadal Roura, X., & Sánchez de Medina Baena, V. (2020). Biological Activity of Cannabis sativa L. Extracts Critically Depends on Solvent Polarity and Decarboxylation. Separations, 7(4), 56. https://doi.org/10.3390/separations7040056