1. Introduction
Illegal wildlife trafficking is increasing at an alarming rate and is contributing to the significant decline of many threatened species, which is a major conservation concern globally.
One of these drivers is status items such as ivory used for symbolic attractiveness (e.g., ornamental objects, trophies), items used for religious purposes (e.g., ritualised practices, superstitions), nutrition (e.g., sustenance, delicacies) and perceived medicinal benefit (e.g., aphrodisiac, vitality, cure-all) [
1,
2,
3]. The domestic, national, and transnational trade of wildlife contraband is a lucrative and illicit industry that extends across ‘the tree of life’.
The illegal wildlife trade (IWT) is a high profit business with “exceptionally low risks” [
4] (p.1066). According to Wasser et al. [
4], prosecutions of those involved in the IWT are infrequent because law enforcement officers, prosecutors, and the judicial system regard wildlife crime a low priority compared to other contraband. Additionally, there is a general shortfall in resources dedicated by many governments used for follow-up investigations of wildlife crime. This warrants forensic methodologies that can cheaply and reliably aid in gathering evidence to be used in the prosecution and conviction of IWT offenders [
4,
5,
6].
The fast, accurate and reliable identification of confiscated wildlife specimens or derivatives is pivotal to wildlife crime investigations, as this is the minimum evidence needed to demonstrate that a crime was committed. Currently, methods used for identification include morphological and/or DNA analysis, each with associated limitations, including time and monetary investment, expertise required, destruction of the sample in the case of DNA and levels of specificity that can be concluded from analysis. However, the need to identify wildlife samples rapidly, accurately, and non-destructively also provides the impetus for the development of other reliable and efficient methods to protect what remains of our wildlife from further exploitation and extinction.
One recently developed method proposed by the authors as a screening method for wildlife identification is the use of volatilome profiling of wildlife contraband using headspace solid phase microextraction (HS-SPME) coupled with comprehensive two-dimensional gas chromatography–time-of-flight mass spectrometry (GC×GC-TOFMS) [
7]. Volatilomes are the collective profile of volatile organic compounds (VOCs) that are influenced by primary (i.e., genetic), secondary (i.e., diet/environment) and tertiary (i.e., unstable) factors [
8,
9]. These influences may allow for volatilomes to be both individual [
10,
11,
12,
13] and species [
7,
14] specific. The previous study conducted by this group was able to build upon this concept and demonstrate that volatilome profiling could differentiate horn specimens sourced from either black (
Diceros bicornis) or white (
Ceratotherium simum) rhinoceros through volatilome and PCA analyses. It was determined that although the black and white rhinoceros horn is made of the same material (keratin), each produced a distinct volatilome profile that differed in VOC compound classes that was used to distinguish the two species [
7]. The previous study therefore suggests the potential of volatilome analysis of wildlife samples as a screening method used in triaging of high volumes of trafficked goods through customs, mail services or trafficking hotspots.
Previous studies have analysed volatilomes through gas chromatography (GC) coupled with mass spectrometry (MS). However, these traditional methods may prevent a complete understanding of the complexity and diverse array of compounds constituting volatilomes through co-elution, requiring a more sensitive methodology. GC×GC-time of flight mass spectrometry (TOFMS) has become a preferred method for VOC analysis over the last decade [
15,
16,
17,
18]. This is due to its ability to provide a greater degree of separation than conventional GC [
19]. The addition of the TOFMS provides a faster acquisition rate and can accommodate the narrower peaks produced by the GC×GC [
20].The sensitivity and separation abilities of two-dimensional gas chromatography makes it an ideal instrument for volatilome analyses
This study aims to investigate the novel applications of GC×GC-TOFMS to volatilomes profiling in a wildlife forensic context. Though the previous study [
7] could clearly distinguish two commonly trafficked items of similar species from each other, further testing on other commonly encountered wildlife specimens has not been conducted. Additionally, the previous study was conducted only on verified reference samples with no blind testing carried out. In the current study, a presumptive volatilome identification method was developed to investigate the usefulness of this technique for ivory. Ivory is a term which encompasses any large mammalian tooth or tusk (all comprised of dentine, cementum and enamel) that can be carved or traded and thus has a commercial interest. Most commonly the term ivory is associated with traded elephant tusks (African and Asian elephant along with those from mammoth), thus this study aimed to investigate volatilome identification using known elephant ivory specimens (African elephant was used in this paper) (hereafter “ivory”), as well as examples of other mammalian teeth and commonly trafficked substitutes (i.e., bone from different species). The reliability of this method was conducted through testing a series of suspected illegal ivory seizures from the Australian border. Confirmatory testing using DNA analysis was carried out on all suspected ivory specimens to ensure the accuracy of the identifications.
2. Materials and Methods
2.1. Sample Collection
Six African elephant ivory tusk samples confirmed by morphology and DNA analysis (methods below) were used as the known elephant ivory samples to build the model. Additionally, one vial was created in which shavings from all six known samples were deposited into one container (and therefore had a larger volume of sample) for comparative analyses.
Suspected ivory samples (seized artefacts summarized in
Table 1) were provided by the Australian Government Department of Environment and Energy (Commonwealth). All samples were labelled by the Department of Environment and Energy (DoEE) as ivory but had yet to be confirmed.
Teeth from other mammalian species including dugong (
Dugong dugon), n = 4; sperm whale (
Physeter microcephalus), n = 2; and Asian elephant (
Elephas maximus), (n = 1); as well as bones from cow (
Bos Taurus) [
21,
22], (n = 5); sheep (
Ovis aries), (n = 1); and koala (
Phascolarctos cinereus), (n = 4); were analysed to determine if separations were related to species or item composition chemical/biological structure. Details of these samples are in
Table 2.
2.2. Headspace Extraction
Dependent on size, samples were either placed into individually labelled 20 mL solid phase microextraction (SPME) vials with a screw cap containing a polytetrafluoroethylene/silicone septum (Supelco, Bellefonta, PA, USA) or into sealed stainless-steel cans (Morris Mc Mahon & Co, Wolli Creek, Australia).
Samples analysed in SPME vials were heated for 30 min in a Thermoline scientific dry block heater at 80 °C. Following, a 50/30 mm divinylbenzene/carboxen/polydimethylsiloxane (DVB/CAR/PDMS) 24 Ga Stableflex SPME fibre with manual fibre holder (Supelco, Bellefonte, PA, USA) was exposed to the headspace for 45 min. The fibre type, heating temperature, accumulation time, and sample collection time were all optimized in a previous study [
23].
For larger samples, the can size was selected to minimise the headspace in order to pre-concentrate the VOCs produced by each sample. These samples were heated in a sand bath using an Accuplate temperature-controlled hotplate (Labnet International Inc., Edison, NJ, USA) to the same specifications as above.
Prior to analyses, the SPME fibre was conditioned according to the manufacturer’s instructions (60 min at 270 °C). Blank fibre tests were performed between each specimen to monitor for carry-over.
2.3. GC×GC-TOFMS
SPME fibres were desorbed directly into the GC×GC inlet at 250 °C for 5 min. The GC×GC analyses were performed with a LECO Pegasus 4D® (LECO, Australia) consisting of an Agilent 7890A GC (Agilent Technologies, Australia) with a dual stage quad jet thermal modulator, secondary oven and Pegasus TOFMS. The column configurations consisted of a Rxi®624Sil MS (Restek Corporation, Australia) primary column (30.0 m × 0.25 mm i.d., with a 1.4 μm coating), connected to a Stabilwax® secondary column (Restek Corporation, Australia) (2 m × 0.25 mm i.d., with a 0.5 μm coating). Helium was used as the carrier gas for all analyses and held at a flow rate of 1 mL/min. All injections were performed manually and were carried out in splitless mode. The GC oven temperature was initially set to 35 °C and held for 5 min, ramping to 240 °C at a rate of 5 °C/min and held for 5 min. The secondary oven temperature offset was 10 °C. Data were acquired at a rate of 100 spectra per second and processed using ChromaTOF® (version 4.51.6.0; LECO).
2.4. GC×GC Data Processing
ChromaTOF
® (version 4.51.6.0; LECO) was used for data processing. The minimum signal-to-noise (S/N) ratio was set to 250 for the base peak and 20 for sub-peaks. The 1D and 2D peak widths were set at 25 s and 0.15 s, respectively. The 2011 National Institute of Standards and Technology (NIST) mass spectral library database was used for initial identification whereby a minimum similarity max of >800 was required. A standard test mixture containing a range of compounds covering several different compound classes was run periodically to ensure the instrument was running optimally. The statistical compare feature in ChromaTOF
® was used for data processing. Samples were classed into the following groups: Known ivory (n = 16), unknown seizures (n = 18), koala bones (n = 4), dugong teeth (n = 12), cow bone (n = 10), sperm whale teeth (n = 6), sheep bone (n = 3), and elephant teeth (n = 3). The alignment of peaks across samples was conducted where a spectral match of 600 was needed for peaks to be identified as the same compound across different chromatograms. In addition, analytes were only retained if they were detected in 50% of the samples within a class. Fisher ratio filtering was performed based on its success in previous studies [
24,
25] in order to identify class-distinguishing compounds. Compounds above the critical value (Fcrit = 2.16) were retained. The resulting chromatographic data was further processed in excel where artefacts (e.g., column bleed, fibre contaminants) were eliminated. Finally, Principal Component Analysis (PCA) was carried out using The Unscrambler X (version 10.3, CAMO Software, Oslo, Norway).
2.5. Compound Classification
Resulting VOCs from each specimen were manually classified into chemical classes (i.e., Alcohols, Aldehydes, Aromatics, Carboxylic acids, Esters, Ethers, Halogenated, Hydrocarbons, Ketones, N-containing, P-containing, Sulfides, Terpenes). The responses of each compound were averaged per class, across all triplicates and compared across all samples.
2.6. Discriminant Analyses
Two discriminant analyses were conducted on resulting data. The first categorized and sorted all ivory samples as “0” and all other tooth samples (e.g., dugong, whale, Asian elephant) as “1”. The independent variables (i.e., compound classes) were regressed to the dependent variable (i.e., category number). Individual predictions were conducted using the following equation, where X represents individual wildlife specimens and b
0 represents the intercept and b
x represents regression coefficients. Average discriminant score:
Cutoff classification scores were determined using Equation 2, where N
0 were the number of ivory samples, đ
0 is the average discriminant score for ivory, N
1 is the number of other tooth samples and đ
1 is the average discriminant score for other tooth samples.
Classifications were conducted through determining if samples met the cutoff criteria for each group. Misclassifications and performance checks were conducted through counting the number of misclassified and correctly classified samples and determining the percentage of correctly identified samples. The second discriminant analysis again categorized and sorted all ivory samples as “0” and all samples that were bone (e.g., cow, koala, sheep) as “1” and followed the same procedures as outlined above.
2.7. Morphology and DNA Analysis
To confirm the identification of the ivory samples used in this study, known and unknown items were assessed for morphological features distinguishing elephant ivory (schreger lines) and other forms of ivory [
26] and DNA sequenced.
DNA extraction was conducted in the Australian Museum’s Australian Centre for Wildlife Genomics (ACWG) designated ultra clean low-template DNA laboratory. This facility has positive air pressure and HEPA filtered air handling system, isolating it from other ACWG laboratories, minimizing potential contamination. A 1 mm drill bit was used to drill a small sample from two sections of each item (and A and B subsample). DNA was extracted following the protocol outlined in [
27]. A partial section of the cytochrome oxidase b region of the mitochondrial DNA was amplified using the primers L14841 & H15149 [
28] and those in [
29]. PCRs were carried out in 25 μL reactions using 100 ng of genomic DNA, 1 × Reaction Buffer (Bioline MyTaq Red Reagent Buffer; Bioline, Australia), 2 pmol of each primer and 0.5U Bioline MyTaq Red DNA polymerase. PCR products were purified using ExoSAP-IT (Thermofisher Scientific, Scoresby, Victoria, Australia). Sanger sequencing was carried out on an AB3730xl Sequencer at the Australian Genome Research Facility, Sydney. Sequences were quality checked and edited with reference to chromatograms using Sequencher v 5.2.4 (Gene Codes Corporation, AnnArbor, MI, USA). Species identification of edited sequences was carried out via blast search of the NCBI database, as well as via phylogenetic tree analyses of nearest neighbor species based on the NCBI blast results.
3. Results
3.1. Volatilome Analysis
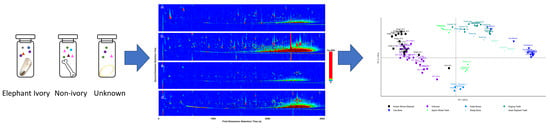
The volatilomes of various wildlife specimens were analysed to determine the applicability of volatilome profiling as a non-destructive, forensic screening method to resolve the identity of confiscated wildlife contraband. To do so, a small volatilome library was constructed using known ivory specimens, the other teeth and bone samples, and unknown, suspected ivory specimens. The initial separate characterisations of the volatilome profile of each sample was completed using GC×GC-TOFMS and a range of compound groups, including known odour-producing compounds, were reported. The total ion chromatograms (TIC) (
Figure 1a–d) display the data obtained from the analysis. Clear visual distinctions were observed from the generated chromatograms of the bone samples, represented here by koala bone (
Figure 1b) and the DoEE seizures suspected ivory sample (
Figure 1c,d). The known ivory samples had an average of 320 identified compounds (
Figure 1a). The number of identifiable compounds in the koala bone samples was considerably higher, totaling 450 VOCs. Approximately 360 compounds were identified in the unknown samples.
After the initial visual comparison of the chromatograms, PCA analysis was carried out (
Figure 2) to visualise any grouping or separations in the dataset. The pre-identified known ivory samples were clustered tightly together, exhibiting similar volatilomes. The different bone samples also clustered together forming separate groups, distinctly different from the ivory samples in the PCA plot. The unknown suspected ivory samples predominately overlapped with the pre-identified known ivory samples. One of the unknown samples, IWT 276A clustered closer towards the koala bone cluster, however, was still close to the pre-identified known ivory mixture. The volatiles causing this separation of this particular unknown from the other samples were found to be mostly aromatic compounds, a ketone and hydrocarbons, more specifically 2-(2,5-dimethylphenyl)-1,4-dimethylbenzene, 3-methylbipheny, 4,6,8-trimethylazulene, benzophenone; cyclotetradecane; 2,6,11-trimethyldodecane, 1,6,7-trimethylnaphthalene, 2-isopropylnaphthalene, nonylcyclohexane, nonadecane, phenol, amilfenol (emitted by plants), 4-tert-butylphenol (a flavour ingredient), undecanal and a reduced presence of cyclotridecane.
PCA analysis demonstrated that the majority of the sample groups could be distinguished from each other. The classifications were based on the four first principal components which accounted for a total of 55% of the variation in the data set. The Asian elephant tooth sample was found to form a cluster separately from the known African elephant derived ivory samples. None of the unknown seizures were found to cluster with the Asian elephant tooth, suggesting these samples were likely African elephant.
Other teeth samples, also comprising dentine, cementum and enamel like ivory were compared to ensure the screening method was not prone to false positives. None of the known elephant ivory samples clustered with any of these false positive groups of the same biological makeup. This indicates that volatilomes can be influenced by factors outside of primary biological makeup. This observation strengthens the potential of the volatilome technique in classifying closely related biological species from one another. Other mammalian teeth, including the sperm whale teeth were selected as items thought to provide a false positive in result. In comparison to the elephant ivory samples, in which a majority of the samples from the same species clustered together with little individual variation, the PCA analyses clearly distinguished the two tooth samples from each other (
Figure 2).
In addition to clear separation due to chemical and biological makeup, PCA analyses could also reasonably differentiate most species from one another, although the African elephants ivory was only identified to genus (
Loxodonta sp.) and it is possible that the two species, forest elephant (
Loxodonta cyclotis) and savanna elephant (
Loxodonta africana), are both present in the samples tested. Additionally the one cow bone sample and the sheep leg, had some overlap with the dugong teeth cluster when looking at PC-1 and PC-2. This clustering was driven by the presence of azulene, 1,4-dichlorobenzene, benzothiophene, methylnaphthalene, and 2,4,6-trichlorophenol. Further principal components were also investigated, and when exploring PC-4, the dugong teeth were clearly separated from the cow bone and sheep leg samples (
Figure 2b). Further research will need to be conducted in order to create a more thorough library for stringent identification, however, preliminarily this data shows potential that volatilome analyses could be informative enough for species identification.
3.2. Compound Classification
In order to further evaluate similarities and differences between the species, the volatiles were grouped according to their compound class (
Figure 3). A visual comparison of the group characteristics showed that the sheep bone, the two sperm whale teeth and the dugong teeth had very different group profiles from any of the unknown seized samples or the known ivory samples. The cow bone samples were found to be reasonably similar to the unknown samples in their class composition, however, the cow bones were found to contain a large number of ester compounds that were not detected in any of the unknowns.
The eight unknown seized samples from the DoEE consisted of the same compound groups (
Figure 3). In addition, these appeared to be in similar ratios, with the exception of carboxylic acid volatiles. In IWT 276 and 233, the component percentage of this compound class was higher than in the other unknown samples. The ivory mixture had a higher component percentage of aldehydes than the average pre-identified known ivory and unknown seized samples. The pre-identified known ivory and the Asian elephant tooth were visually comparable to the unknown seized samples, though the Asian elephant tooth had a greater abundance of hydrocarbon volatiles and a smaller abundance of esters than any of the unknowns (
Figure 3). The unknown seized samples could therefore also potentially be eliminated as being of Asian elephant origin, strengthening the view that these unknown samples are most likely of African origin based on this volatilome classification.
From the initial analysis of the class volatilomes composition, the unknown samples appear to be most closely related to the pre-identified known ivory. However, further statistical analysis was required to confirm this.
3.3. Discriminant Analyses
Statistical analysis comparing the compound classes of the different species was conducted in order to determine if the unknown DoEE seizures could be reliably distinguished as elephant ivory, other mammalian teeth, or bone substitute. Two classification models were tested. In the first model, samples were parsed and classed as either elephant ivory (n = 35) or mammalian teeth (n = 21) specimens (i.e., all teeth). These classifications had a 71.4% success rate of discrimination, with the following samples being misclassified: sperm whale tooth (M45150), ivory mix, IWT 276A, C and one replicate of IWT 276B, IWT 298, one replicate of 348C, and two dugong teeth replicates.
A second classification was then tested whereby samples were parsed and classed either as bone or elephant ivory. In this model, classifications were either elephant ivory (n = 35) or bone specimens (cow, koala, sheep (n = 17)). This analysis had a 75% success rate, with two cow bone replicates, the sheep bone replicates, and IWT 276C being misclassified.
3.4. Morphology and DNA Results
Seven out of the eight unknown ivory samples were identified as being African Elephant (
Loxodonta sp). The tests used do not provide sufficient resolution to distinguish between African Savanah Elephants (
L. africana) or African Forest Elephant (
L. cyclotis) but is likely informative to genus level (
Loxodonta sp.). Out of these seven, four (IWT 348A, IWT 348B, IWT 348C and IWT 233) also had morphological features consistent with elephant ivory in the form of schreger lines [
30]. One of the necklace pendants (IWT 276B) was unable to be identified as elephant ivory based on DNA.
4. Discussion
A volatilome method used for the identification of illegal wildlife samples was developed. Initial visual comparison of the chromatographs between the unknown seizures and pre-identified known ivory (African elephant) did not show any distinct similar patterns. Similarly, the TIC of the unknown seizures was not visually comparable to any of the mammalian teeth samples or the bone samples analysed. It was hypothesized that the increased number of total compounds in the unknown samples compared to the pre-identified known ivory may be due to background noise (i.e., varnishes, glues, metal pendants) as the pre-identified known ivory was from raw blocks and the unknown ivory was in the form of processed jewelry. This background can vary from sample to sample and produce its own volatile organic compounds. Additionally, as one of the pieces failed both DNA extractions and was inconclusive based on its volatilome, it may well be that the piece is not elephant ivory. A series of previously identified odour compounds were found in all of the different sample groups, including butanal, pentanal, heptanal, hexanal, nonanal, ethanol, acetophenone, benzaldehyde, benzene, heptanal, hexanal, nonanal, ethanol, 1-butanol, ethylbenzene, 2-nonanone, 1-decene, and toluene [
31,
32,
33,
34,
35,
36,
37]. The presence of several previously reported and unreported odour compounds show the potential for volatilome analysis as a successful tool to analyse wildlife specimens.
The PCA analysis further highlighted the usefulness of the volatilomes technique and showed a separation between the sample groups. Seven out of the eight unknown samples were classified as ivory based on the developed model. The pre-identified known African elephant ivory samples were tightly clustering and distinctly different from the samples from other mammalian teeth. This separation from samples of the same material illustrated that this model is not likely prone to false positives. None of the unknown seizures were found to cluster with the Asian elephant tooth, suggesting the samples were more closely related with African elephant. However, caution must be taken with interpreting these preliminary results as only one replicate of Asian elephant was analysed and further analysis of additional Asian elephant teeth and tusk is needed to improve the method. The sperm whale teeth that were analysed as another mammalian species known to have traded teeth, were found to be separated into two groups based on the two teeth samples analysed. Notably, the two sperm whale teeth that were analysed were from the same species (Physeter macrocephalus) but had very different volatilome profiles. It was hypothesized that this difference in volatilomes profile was due to one of the teeth samples having been treated with varnishes prior to ending up in the museum collection and that this separation was a result of additional compounds. As illegally traded wildlife specimens can be significantly modified (i.e., ground powder, preserved, etc.), there is potential for several treatment methods to be encountered. Different treatment methods and the effects of glues, varnishes, and metals should be characterized and their effect on the volatilomes monitored for both known and potential false positive giving samples and fakes to ensure reliable results. Another potential reason for this difference could be due to the animals coming from different regions of the world and living in different environments and potentially having a different diet. This further highlights the need to increase the volatilome database with diverse samples.
The discriminant analysis based on total compound class across biological materials had between 71%–75% success rates. In this model, all pendants were misclassified, potentially because metal clasps were not removed prior to analyses. Additionally, the sheep bone had glue on all of the joints, which may have complicated results. For future discriminant analyses, a larger sample size representing all forms of treated specimens, as well as more ornate jewelry items should be tested.
The confirmatory DNA analysis conducted on the seized samples determined that seven out of the eight unknown ivory samples were African Elephant (Loxodonta sp.). One sample was unable to be conclusively identified as the DNA analysis for species ID failed. This sample was a very tiny amulet and it is possible that there was not sufficient material in the drill shaving for the DNA extraction to be successful.
In order to further develop the volatilomes model and ensure results are accurate, reliable and comparable to DNA, the gold standard in wildlife forensics, more samples (including more vouchered specimens) need to be tested using the method developed herein. Additional samples from African Forest elephants (L. cyclotis), African Savanna elephants (L. africana), Asian elephants (E. maximus) samples, as well as examples of other species which also have traded ivory including mammoth (Mammuthus sp), walrus (Odobenus. rosmarus), orca (Orcinus orca), narwhal (Monodon monoceros), hippopotamus (Hippopotamus. amphibious), pygmy hippopotamus (Hexaprotodon liberiensis), and warthogs (Phacochoerus africanus) or other pig species.
In the current study there was only one potential fake sample (i.e., not identified as ivory) amongst the seized samples from DoEE. The model created used a series of bone samples as the fake material as this has been used in the past as substitutes [
21,
22]. For the method to be further improved, additional bone samples, including other species, should be investigated. In addition, other ivory substitutes such as plastics, organic resin, casein resin, and the new synthetic ivory [
38] will be tested.
5. Conclusions
In the present study, a GC×GC-TOFMS screening method was developed for the classification of confiscated suspected ivory samples using volatilomes profiling. For all unknown seized specimens, DNA analyses was conducted for confirmatory analysis. The results from the volatilome screening method suggested that seven of the eight suspected seizures were determined to be ivory from African Elephant (Loxodonta sp). One of the unknown seized samples (IWT 276A) was slightly separated from the known ivory cluster using the PCA approach. This sample was later confirmed as being ivory through DNA confirmatory analysis. DNA analysis confirmed that six out of the seven samples preliminarily identified as ivory through volatilome screening (excluding IWT 276A as mentioned above) were correctly classified. IWT 276B was not confirmed as being ivory though the DNA analysis as the sample taken for analysis was likely too small. All other unknown samples classed as ivory through volatilome profiling were confirmed as African elephant ivory through DNA analysis. This research also suggests that volatilomes of wildlife products may be species specific. Using the volatilomes compound group characteristics, both PCA and discriminant analysis demonstrated that the dentine and bone samples could be reasonably separated from elephant ivory, yielding an over 70% correct classification model. In order to run discriminant analyses for species identity, more samples will have to be analysed. In the future, other fake materials such as plastic and synthetic ivory will be added to the volatilome model so that fake samples can be more easily detected without the need for additional DNA analysis. Overall the screening method correctly classed seven out of the eight seized samples (87.5% accuracy) showing its potential as a non-destructive screening method for the detection of illegal ivory products.
Developing a non-destructive screening method for illegal wildlife products is widely beneficial in a forensic context in the fight to combat illegal wildlife trade. A fast and reliable method would allow for the triaging of large sample volumes at entry and exit trafficking hotspots such as mail centres, borders and airports.
GC×GC-TOFMS has the potential to provide robust analysis for classifying the species identity of confiscated wildlife items via the volatilome profile due to its specificity and separation ability. It has been clearly demonstrated that these separations are needed, as both individual compounds and compound classes significantly contribute to the ability to distinguish wildlife parts from each other. Further, GC×GC-TOFMS volatilome studies of wildlife specimens will form the basis for portable, industrial based initiatives, such as applications using the electronic nose (e-nose) devices in the future. These devices have the potential to be considerably cheaper than current analyses and can be used at the frontline as a screening tool. This study supports the application of GC×GC-TOFMS as a new analytical tool for wildlife forensic investigations.
Author Contributions
Conceptualization, M.U., R.N.J. and S.L.F.; Investigation and performing experiments, M.U., A.B., C.B. and G.J.F.; Data curation, M.U. and G.J.F.; Writing—original draft preparation, M.U.; Writing—review and editing, M.U., A.B., G.J.F., R.N.J. and S.L.F. All authors have read and agree to the published version of the manuscript.
Funding
This project was supported by the Wildlife Crime Tech Challenge (WCTC) and funded by the United States Agency for International Development (USAID).
Acknowledgments
The authors would like to acknowledge Dr Sandy Ingleby, collection manager, Mammals Section from the Australian Museum for allowing access to the different wildlife species and the Australian Department of Environment and Energy for the seized samples.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Hansen, A.L.S.; Li, A.; Joly, D.; Mekaru, S. Brownstein Digital surveillance: A novel approach to monitoring the illegal wildlife trade. PLoS ONE 2012, 7, e51156. [Google Scholar]
- Patel, N.G.; Rorres, C.; Joly, D.O.; Brownstein, J.S.; Boston, R.; Levy, M.Z.; Smith, G.J. Quantitative methods of identifying the key nodes in the illegal wildlife trade network. Proc. Natl. Acad. Sci. USA 2015, 112, 7948–7953. [Google Scholar] [CrossRef]
- Rojas, M.; González, I.; Pavón, M.Á.; Pegels, N.; Hernández, P.E.; García, T.; Martín, R.J. Development of a real-time PCR assay to control the illegal trade of meat from protected capercaillie species (Tetrao urogallus). Forensic Sci. Int. 2011, 210, 133–138. [Google Scholar] [CrossRef] [PubMed]
- Wasser, S.K.; William, J.C.; Drori, O.; Kisamo, E.S.; Mailand, C.; Mutayoba, B.; Stephens, M. Combating the illegal trade in African elephant ivory with DNA forensics. J. Conserv. Biol. 2008, 22, 1065–1071. [Google Scholar] [CrossRef] [PubMed]
- Wasser, S.K.; Mailand, C.; Booth, R.; Mutayoba, B.; Kisamo, E.; Clark, B.; Stephens, M.J. Using DNA to track the origin of the largest ivory seizure since the 1989 trade ban. Proc. Natl. Acad. Sci. USA 2007, 104, 4228–4233. [Google Scholar] [CrossRef] [PubMed]
- Huffman, J.E.; Wallace, J.R. Wildlife Forensics: Methods and Applications; John Wiley & Sons: New Jersey, NJ, USA, 2012; Volume 6. [Google Scholar]
- Ueland, M.; Ewart, K.; Troobnikoff, A.N.; Frankham, G.; Johnson, R.N.; Forbes, S.L. A rapid chemical odour profiling method for the identification of rhinoceros horns. Forensic Sci. Int. 2016, 266, e99–e102. [Google Scholar] [CrossRef] [PubMed]
- Curran, A.M.; Rabin, S.I.; Prada, P.A.; Furton, K.G. Comparison of the volatile organic compounds present in human odor using SPME-GC/MS. J. Chem. Ecol. 2005, 31, 1607–1619. [Google Scholar] [CrossRef]
- Curran, A.M.; Prada, P.A.; Furton, K.G. The differentiation of the volatile organic signatures of individuals through SPME-GC/MS of characteristic human scent compounds. Forensic Sci. Int. 2010, 55, 50–57. [Google Scholar] [CrossRef]
- Deshpande, K.; Furton, K.G.; De Etta, K.J. The equine volatilome: Volatile organic compounds as discriminatory markers. J. Equine Vet. Sci. 2018, 62, 47–53. [Google Scholar] [CrossRef]
- Curran, A.M.; Rabin, S.I.; Furton, K.G. Analysis of the uniqueness and persistence of human scent. Forensic Sci. Commun. 2005, 7. Gale Document Number: GALE|A139298569. [Google Scholar]
- Martin, S.J.; Shemilt, S.; Drijfhout, F.J.N. Effect of time on colony odour stability in the ant Formica exsecta. Naturwissenschaften 2012, 99, 327–331. [Google Scholar] [CrossRef] [PubMed]
- Schoon, G.A.; De Bruin, J. The ability of dogs to recognize and cross-match human odours. Forensic Sci. Int. 1994, 69, 111–118. [Google Scholar] [CrossRef]
- Bos, L.D.; Sterk, P.J.; Schultz, M.J. Volatile metabolites of pathogens: A systematic review. PLOS Pathog. 2013, 9, e1003311. [Google Scholar] [CrossRef]
- Dekeirsschieter, J.; Stefanuto, P.-H.; Brasseur, C.; Haubruge, E.; Focant, J.-F. Enhanced characterization of the smell of death by comprehensive two-dimensional gas chromatography-time-of-flight mass spectrometry (GCxGC-TOFMS). PLoS ONE 2012, 7, e39005. [Google Scholar] [CrossRef]
- Stadler, S.; Stefanuto, P.-H.; Brokl, M.; Forbes, S.L.; Focant, J.-F. Characterization of volatile organic compounds from human analogue decomposition using thermal desorption coupled to comprehensive two-dimensional gas chromatography–time-of-flight mass spectrometry. Anal. Chem. 2012, 85, 998–1005. [Google Scholar] [CrossRef] [PubMed]
- Brasseur, C.; Dekeirsschieter, J.; Schotsmans, E.M.; de Koning, S.; Wilson, A.S.; Haubruge, E.; Focant, J.-F. Comprehensive two-dimensional gas chromatography–time-of-flight mass spectrometry for the forensic study of cadaveric volatile organic compounds released in soil by buried decaying pig carcasses. J. Chromatogr. A 2012, 1255, 163–170. [Google Scholar] [CrossRef] [PubMed]
- Perrault, K.A.; Rai, T.; Stuart, B.H.; Forbes, S.L.J.A.M. Seasonal comparison of carrion volatiles in decomposition soil using comprehensive two-dimensional gas chromatography–time of flight mass spectrometry. Anal. Methods 2015, 7, 690–698. [Google Scholar] [CrossRef]
- Perrault, K.A.; Nizio, K.D.; Forbes, S.L. A comparison of one-dimensional and comprehensive two-dimensional gas chromatography for decomposition odour profiling using inter-year replicate field trials. Chromatographia 2015, 78, 1057–1070. [Google Scholar] [CrossRef]
- Cotter, R.J. Time-of-flight mass spectrometry for the structural analysis of biological molecules. Anal. Chem. 1992, 64, 1027A–1039A. [Google Scholar] [CrossRef]
- Santiapillai, C.; Silva, A.; Karyawasam, C.; Esufali, S.; Jayaniththi, S.; Basnayake, M.; Unantenne, V.; Wijeyamohan, S. Trade in Asian elephant ivory in Sri Lanka. Oryx 1999, 33, 176–180. [Google Scholar] [CrossRef]
- Sims, M.E.; Baker, B.W.; Hoesch, R.M. Tusk or bone? An example of ivory substitute in the wildlife trade. Ethnobiol. Lett. 2011, 2, 40–44. [Google Scholar] [CrossRef]
- Bartos, C. University of Technology Sydney, Sydney, Australia, Unpublished word. 2015.
- Pierce, K.M.; Hoggard, J.C.; Hope, J.L.; Rainey, P.M.; Hoofnagle, A.N.; Jack, R.M.; Wright, B.W.; Synovec, R.E. Fisher ratio method applied to third-order separation data to identify significant chemical components of metabolite extracts. Anal. Chem. 2006, 78, 5068–5075. [Google Scholar] [CrossRef] [PubMed]
- Brokl, M.; Bishop, L.; Wright, C.G.; Liu, C.; McAdam, K.; Focant, J.-F. Multivariate analysis of mainstream tobacco smoke particulate phase by headspace solid-phase micro extraction coupled with comprehensive two-dimensional gas chromatography–time-of-flight mass spectrometry. J Chromatogr. A 2014, 1370, 216–229. [Google Scholar] [CrossRef] [PubMed]
- Espinoza, E.O.; Mann, M.-J. Identification guide for ivory and ivory substitutes. Available online: https://www.bcin.ca/bcin/detail.app?id=108360&wbdisable=true (accessed on 3 December 2019).
- Mailand, C.; Wasser, S.K.J.N.P. Isolation of DNA from small amounts of elephant ivory. Nat. Protoc. 2007, 2, 2228. [Google Scholar] [CrossRef] [PubMed]
- Kocher, T.D.; Thomas, W.K.; Meyer, A.; Edwards, S.V.; Pääbo, S.; Villablanca, F.X.; Wilson, A.C. Dynamics of mitochondrial DNA evolution in animals: Amplification and sequencing with conserved primers. Proc. Natl. Acad. Sci. USA 1989, 86, 6196–6200. [Google Scholar] [CrossRef] [PubMed]
- Hauf, J.; Baur, A.; Chalwatzis, N.; Zimmermann, F.; Joger, U.; Lazarev, P. Selective amplification of a mammoth mitochondrial cytochrome b fragment using an elephant-specific primer. Curr. Genet. 1995, 27, 486–487. [Google Scholar] [CrossRef]
- Espinoza, E.O.N.; Mann, M.-J. The history and significance of the Schreger pattern in proboscidean ivory characterization. J. Am. Inst. Conserv. 1993, 32, 241–248. [Google Scholar] [CrossRef]
- Vass, A.A.; Smith, R.R.; Thompson, C.V.; Burnett, M.N.; Wolf, D.A.; Synstelien, J.A.; Dulgerian, N.; Eckenrode, B.A. Decompositional odor analysis database. J. Forensic Sci. 2004, 49, 1–10. [Google Scholar] [CrossRef]
- Vass, A.A.; Smith, R.R.; Thompson, C.V.; Burnett, M.N.; Dulgerian, N.; Eckenrode, B.A. Odor analysis of decomposing buried human remains. J. Forensic Sci. 2008, 53, 384–391. [Google Scholar] [CrossRef]
- Dekeirsschieter, J.; Verheggen, F.; Gohy, M.; Hubrecht, F.; Bourguignon, L.; Lognay, G.; Haubruge, E. Cadaveric volatile organic compounds released by decaying pig carcasses (Sus domesticus L.) in different biotopes. Forensic Sci. Int. 2009, 189, 46–53. [Google Scholar] [CrossRef]
- Stutz, H.; Silverman, G.; Angelini, P.; Levin, R.J. Bacteria and volatile compounds associated with ground beef spoilage. J. Food Sci. 1991, 56, 1147–1153. [Google Scholar] [CrossRef]
- Armstrong, P.; Nizio, K.D.; Perrault, K.A.; Forbes, S.L. Establishing the volatile profile of pig carcasses as analogues for human decomposition during the early postmortem period. Heliyon 2016, 2, e00070. [Google Scholar] [CrossRef] [PubMed]
- Statheropoulos, M.; Spiliopoulou, C.; Agapiou, A. A study of volatile organic compounds evolved from the decaying human body. Forensic Sci. Int. 2005, 153, 147–155. [Google Scholar] [CrossRef] [PubMed]
- Hoffman, E.M.; Curran, A.M.; Dulgerian, N.; Stockham, R.A.; Eckenrode, B.A. Characterization of the volatile organic compounds present in the headspace of decomposing human remains. Forensic Sci. Int. 2009, 186, 6–13. [Google Scholar] [CrossRef]
- Edwards, H.G.; Farwell, D.W. Ivory and simulated ivory artefacts: Fourier transform Raman diagnostic study. Spectrochim. Acta A 1995, 51, 2073–2081. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).