Chiral Recognition by DNA-Immobilized TLC Plate
Abstract
1. Introduction
2. Experimental Section
2.1. Materials
2.2. Preparation of DNA-TLC Plate
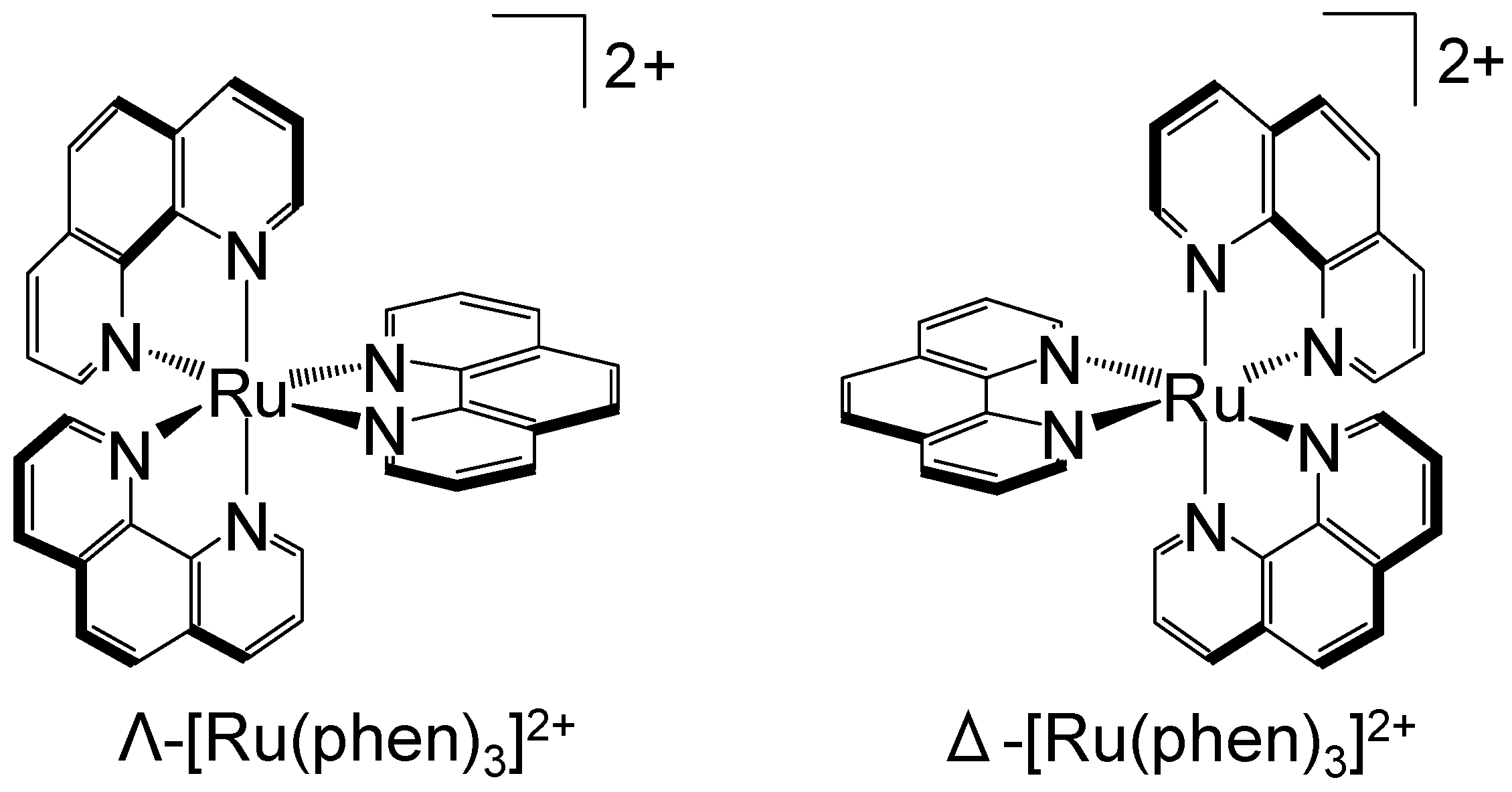
2.3. Synthesis of Tris(1,10-phenanthroline)ruthenium(II)
2.4. Separation of Amino Acid and Metal Complex on DNA-TLC Plate
2.5. IR Measurements of Amino Acid-Accumulated DNA Film
3. Results and Discussion
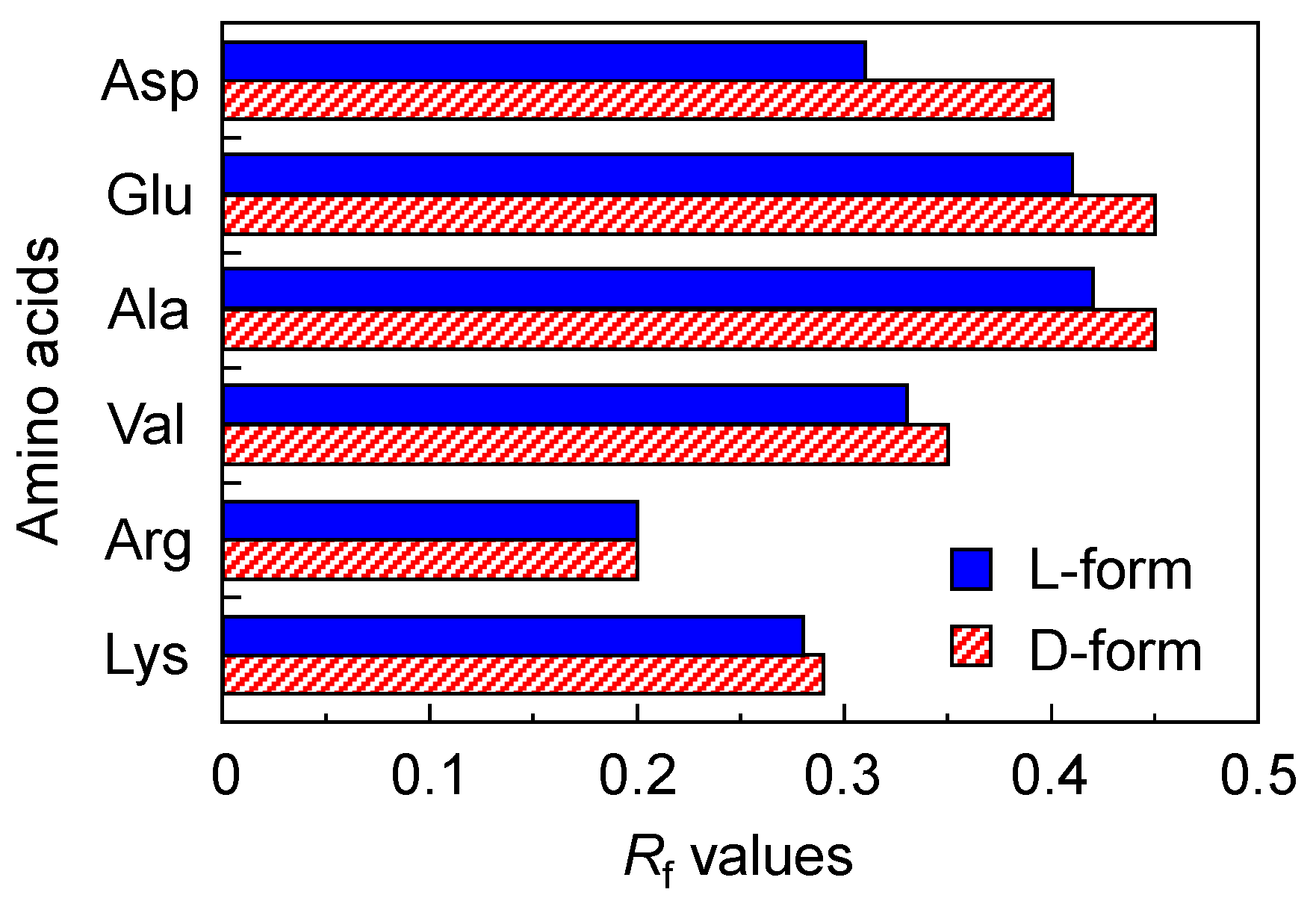
3.1. Chiral Recognition of Amino Acid on DNA-TLC Plate
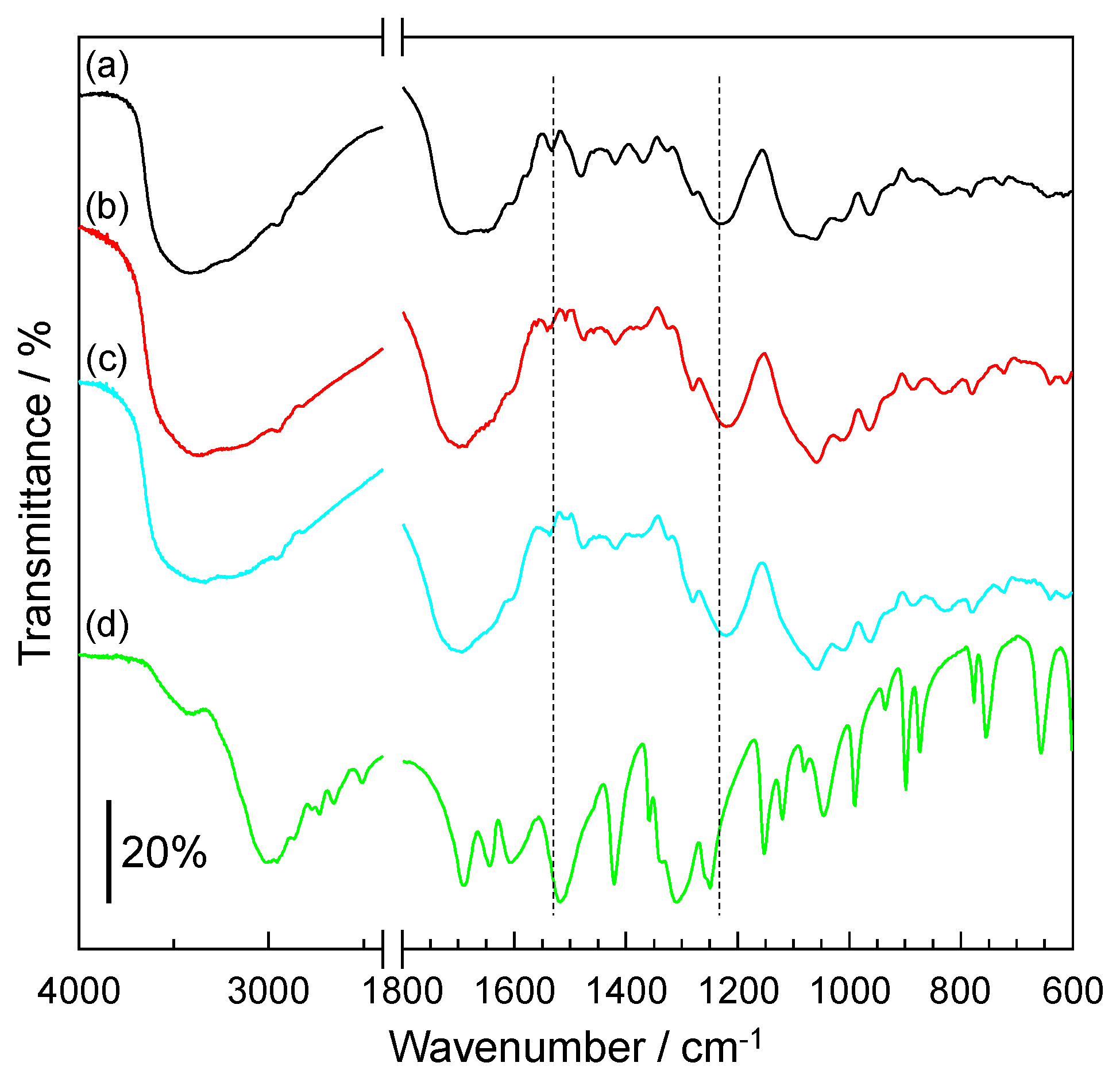
3.2. IR Spectrum of Amino Acid-Accumulated DNA Film
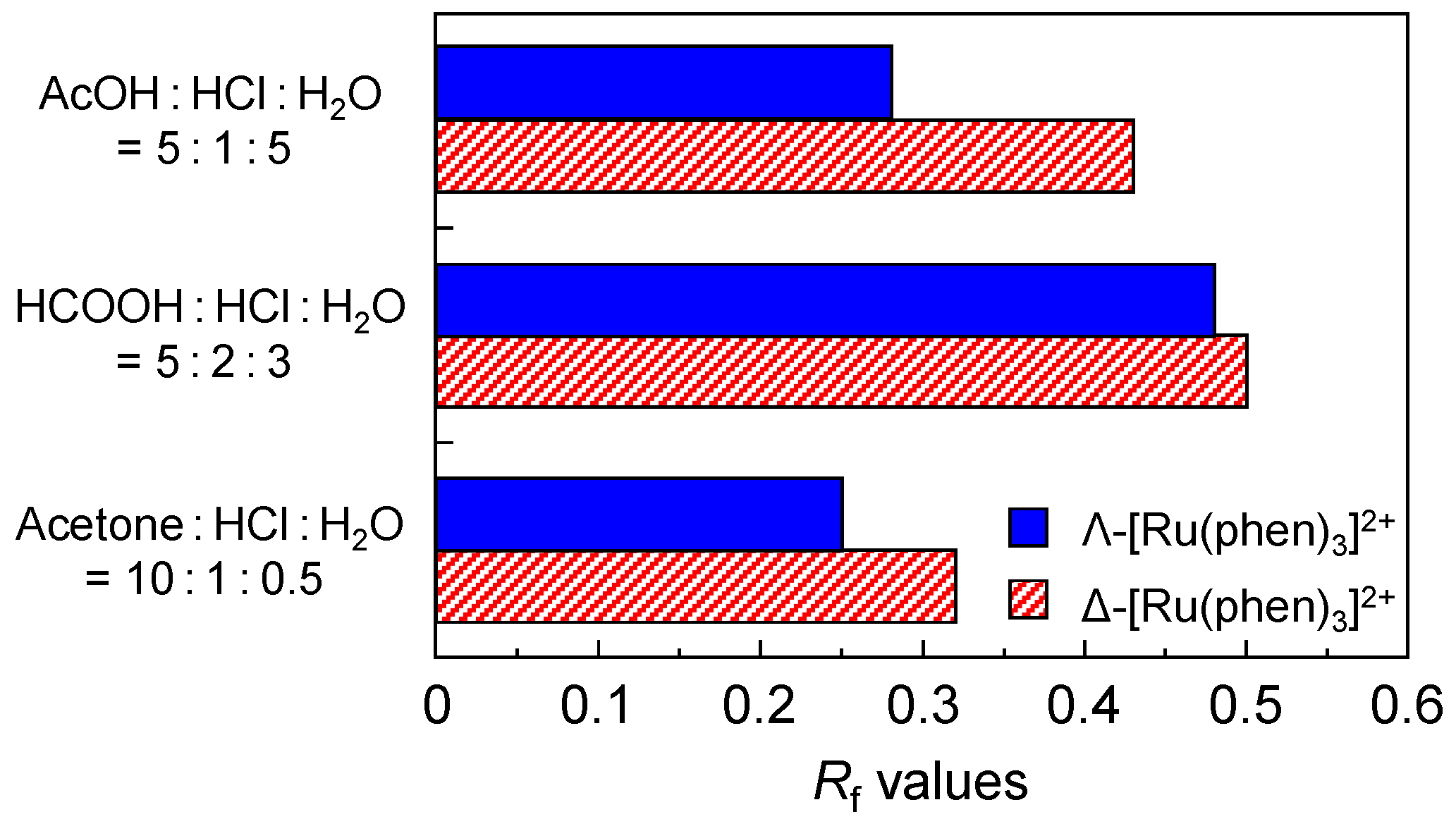
3.3. Chiral Recognition of Metal Ion Complex on DNA-TLC Plate
4. Conclusions
Supplementary Materials
Author Contributions
Conflicts of Interest
References
- Zhang, M.; Qing, G.; Sun, T. Chiral biointerface materials. Chem. Soc. Rev. 2012, 41, 1972–1984. [Google Scholar] [CrossRef] [PubMed]
- Yashima, E.; Maeda, K. Chirality-responsive helical polymers. Macromolecules 2008, 41, 3–12. [Google Scholar] [CrossRef]
- Yashima, E.; Ousaka, N.; Taura, D.; Shimomura, K.; Ikai, T.; Maeda, K. Supramolecular helical systems: Helical assemblies of small molecules, foldamers, and polymers with chiral amplification and their functions. Chem. Rev. 2016, 116, 13752–13990. [Google Scholar] [CrossRef] [PubMed]
- Kane-Maguire, L.A.P.; Wallace, G.G. Chiral conducting polymers. Chem. Soc. Rev. 2010, 39, 2545–2576. [Google Scholar] [CrossRef] [PubMed]
- Lv, Z.; Chen, Z.; Shao, K.; Qing, G.; Sun, T. Stimuli-directed helical chirality inversion and bio-applications. Polymers 2016, 8, 310. [Google Scholar] [CrossRef]
- Bi, C.; Zheng, X.; Azaria, S.; Beeram, S.; Li, Z.; Hage, D.S. Chromatographic studies of protein-based chiral separations. Separations 2016, 3, 27. [Google Scholar] [CrossRef] [PubMed]
- Guibal, E. Heterogeneous catalysis on chitosan-based materials: A review. Prog. Polym. Sci. 2005, 30, 71–109. [Google Scholar] [CrossRef]
- Lee, B.; Kwon, H.; Kim, S.; Rotermund, F. Natural silk protein as a new broadband nonlinear optical material. Opt. Mater. Express 2016, 6, 993–1002. [Google Scholar] [CrossRef]
- Saenger, W. Principles of Nucleic Acid Structure; Springer-Verlag: Berlin, Germany, 1987. [Google Scholar]
- Bates, A.D.; Maxwell, A. DNA Topology; Oxford University Press: New York, NY, USA, 2005. [Google Scholar]
- Roelfes, G.; Boersma, A.J.; Feringa, B.L. Highly enantioselective DNA-based catalysis. Chem. Commun. 2006, 6, 635–637. [Google Scholar] [CrossRef] [PubMed]
- Park, S.; Sugiyama, H. DNA as a chiral scaffold for asymmetric synthesis. Molecules 2012, 17, 12792–12803. [Google Scholar] [CrossRef] [PubMed]
- Higuchia, A.; Hayashi, A.; Kanda, N.; Sanui, K.; Kitamura, H. Chiral separation of amino acids in ultrafiltration through DNA-immobilized cellulose membranes. J. Mol. Struct. 2005, 739, 145–152. [Google Scholar] [CrossRef]
- Yoshikawa, M.; Maruhashi, M.; Iwamoto, Y.; Ogata, N. Chiral separation of racemic amino acids through DNA. Macromol. Symp. 2007, 249–250, 557–561. [Google Scholar] [CrossRef]
- Michaud, M.; Jourdan, E.; Villet, A.; Ravel, A.; Grosset, C.; Peyrin, E. A DNA aptamer as a new target-specific chiral selector for HPLC. J. Am. Chem. Soc. 2003, 125, 8672–8679. [Google Scholar] [CrossRef] [PubMed]
- Michaud, M.; Jourdan, E.; Ravelet, C.; Villet, A.; Ravel, A.; Grosset, C.; Peyrin, E. Immobilized DNA aptamers as target-Specific chiral stationary phases for resolution of nucleoside and amino acid derivative enantiomers. Anal. Chem. 2004, 76, 1015–1020. [Google Scholar] [CrossRef] [PubMed]
- Tohala, L.; Oukacine, F.; Ravelet, C.; Peyrin, E. Chiral Resolution Capabilities of DNA Oligonucleotides. Anal. Chem. 2015, 87, 5491–5495. [Google Scholar] [CrossRef] [PubMed]
- Su, X.; Qin, F.; Kong, L.; Ou, J.; Xie, C.; Zou, H. Characterization of enantioselective binding of racemic natural tetrahydropalmatine to DNA by chromatographic methods. J. Chromatogr. B 2007, 845, 174–179. [Google Scholar] [CrossRef] [PubMed]
- Huang, R.; Xiong, W.; Wang, D.; Guo, L.; Lin, Z.; Yu, L.; Chu, K.; Qiu, B.; Chen, G. Label-free aptamer-based partial filling technique for enantioseparation and determination of dl-tryptophan with micellar electrokinetic chromatography. Electrophoresis 2013, 34, 254–259. [Google Scholar] [CrossRef] [PubMed]
- Grote, J.G.; Hagen, J.A.; Zetts, J.S.; Nelson, R.L.; Iggs, D.E.; Stone, D.M.O.; Yaney, P.P.; Heckman, E.; Zhang, C.; Steier, W.H.; et al. Investigation of polymers and marine-derived DNA in optoelectronics. J. Phys. Chem. B 2004, 108, 8584–8591. [Google Scholar] [CrossRef]
- Yamada, M.; Hara, S.; Yamada, T.; Katagiri, F.; Hozumi, K.; Nomizu, M. Double-stranded DNA stereoselectively promotes aggregation of amyloid-like fibrils and generates peptide/DNA matrices. Biopolymers 2014, 102, 465–472. [Google Scholar] [CrossRef] [PubMed]
- Randerath, K. Thin-Layer Chromatography; Verlag Chemie Academic Press: New York, NY, USA, 1968. [Google Scholar]
- Satoh, S.; Fugetsu, B.; Nomizu, M.; Nishi, N. Functional DNA-silica composite prepared by sol-gel method. Polym. J. 2005, 37, 94–101. [Google Scholar] [CrossRef]
- Yamada, M.; Aono, H. DNA-inorganic hybrid material as selective absorbent for harmful compounds. Polymer 2008, 49, 4658–4665. [Google Scholar] [CrossRef]
- Dwyer, F.P.; Humpolets, J.E.; Nyholm, R.S. The chemistry of ruthenium. Part I. The redox potential of the tris-orthophenanthroline ruthenous ion. J. Proc. R. Soc. N. S. W. 1946, 80, 212–216. [Google Scholar]
- Dwyer, F.P.; Gyarfas, E.C. The chemistry of ruthenium. Part VI. The existence of the tris-o-phenanthroline ruthenium II and the tris-o-phenanthroline ruthenium III ions in entantiomorphous forms. J. Proc. R. Soc. N. S. W. 1949, 83, 170–173. [Google Scholar]
- Yamada, M.; Kato, K.; Nomizu, M.; Sakairi, N.; Ohkawa, K.; Yamamoto, H.; Nishi, N. Preparation and characterization of DNA films induced by UV irradiation. Chem. Eur. J. 2002, 8, 1407–1412. [Google Scholar] [CrossRef]
- Liu, X.D.; Kubo, T.; Diao, H.Y.; Benjamas, J.; Yonemichi, T.; Nishi, N. DNA/polyvinyl alcohol interpenetrating polymer network as stationary phase for thin layer chromatography. Anal. Biochem. 2009, 393, 67–72. [Google Scholar] [CrossRef] [PubMed]
- Tajmir-Riahi, H.A.; Naoui, M.; Ahimad, R. The effects of Cu2+ and Pb2+ on the solution structure of calf thymus DNA: DNA condensation and denaturation studied by Fourier-transform IR difference spectroscopy. Biopolymers 1993, 33, 1819–1827. [Google Scholar] [CrossRef] [PubMed]
- Banyay, M.; Sarkaräslund, A. A library of IR bands of nucleic acids in solution. Biophys. Chem. 2003, 104, 477–488. [Google Scholar] [CrossRef]
- Silverstein, R.M.; Webster, F.X. Spectrometric Identification of Organic Compounds; John Wiley & Sons: New York, NY, USA, 1998. [Google Scholar]
- Yamada, M.; Goto, A. Proton conduction of DNA-imidazole composite material under anhydrous condition. Polym. J. 2012, 44, 415–420. [Google Scholar] [CrossRef]
- Yamada, M.; Hashimoto, K. DNA-cyclodextrin composite material for environmental applications. Biomacromolecules 2008, 9, 3341–3345. [Google Scholar] [CrossRef] [PubMed]
- Barton, J.K.; Danishefsky, A.T.; Goldberg, J.M. Tris(phenanthroline)ruthenium(II): Stereoselectivity in binding to DNA. J. Am. Chem. Soc. 1984, 106, 2172–2176. [Google Scholar] [CrossRef]
- Yamagishi, A. Electric dichroism evidence for stereospecific binding of optically active tris chelated complexes to DNA. J. Phys. Chem. 1984, 88, 5709–5713. [Google Scholar] [CrossRef]
- Eriksson, M.; Leijon, M.; Hiort, C.; Norden, B.; Graeslund, A. Binding of Δ- and Λ-[Ru(phen)3]2+ to [d(CGCGATCGCG)]2 Studied by NMR. Biochemistry 1994, 33, 5031–5040. [Google Scholar] [CrossRef] [PubMed]
- Yamada, M.; Nomizu, M.; Satoh, S.; Ohkawa, K.; Yamamoto, H.; Nishi, N. DNA with γ-Aminopropyltriethoxysilane switches the B- and C-form structures by thermal control. ChemBioChem 2003, 4, 232–234. [Google Scholar] [CrossRef] [PubMed]
| Entry | Mobile Phases | Ratio/v/v |
|---|---|---|
| 1 | 1-BuOH:AcOH:H2O | 4:4:1 |
| 2 | 1-BuOH:AcOH:H2O | 4:2:1 |
| 3 | (CH3)2CO:AcOH:H2O | 70:30:7 |
| 4 | 1-BuOH:EtOH:H2O | 5:4:3 |
| 5 | EtOH:H2O | 1:1 |
| 6 | AcOEt:MeOH | 4:1 |
| 7 | MeOH:CHCl3:H2O | 12:5:3 |
| 8 | (CH3)2CO:1-BuOH:NH3(aq):H2O | 65:20:10:5 |
| 9 | NH3(aq):H2O | 17:3 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yamada, M.; Inoue, M. Chiral Recognition by DNA-Immobilized TLC Plate. Separations 2018, 5, 3. https://doi.org/10.3390/separations5010003
Yamada M, Inoue M. Chiral Recognition by DNA-Immobilized TLC Plate. Separations. 2018; 5(1):3. https://doi.org/10.3390/separations5010003
Chicago/Turabian StyleYamada, Masanori, and Mami Inoue. 2018. "Chiral Recognition by DNA-Immobilized TLC Plate" Separations 5, no. 1: 3. https://doi.org/10.3390/separations5010003
APA StyleYamada, M., & Inoue, M. (2018). Chiral Recognition by DNA-Immobilized TLC Plate. Separations, 5(1), 3. https://doi.org/10.3390/separations5010003




