Quantitative Analysis of Chlorogenic Acid, Rutin, and Isoquercitrin in Extracts of Cudrania tricuspidata Leaves Using HPLC-DAD
Abstract
1. Introduction
2. Materials and Methods
2.1. Equipment and Reagents
2.2. Plant Materials and Preparation of Analytical Sample
2.3. Analytical Conditions for HPLC and Mass Spectrometry
2.4. Method Validation
2.5. Calibration Curves
2.6. Data Analysis
2.7. Experimental Design for the Development of the Response Surface Methodology Model
3. Results and Discussion
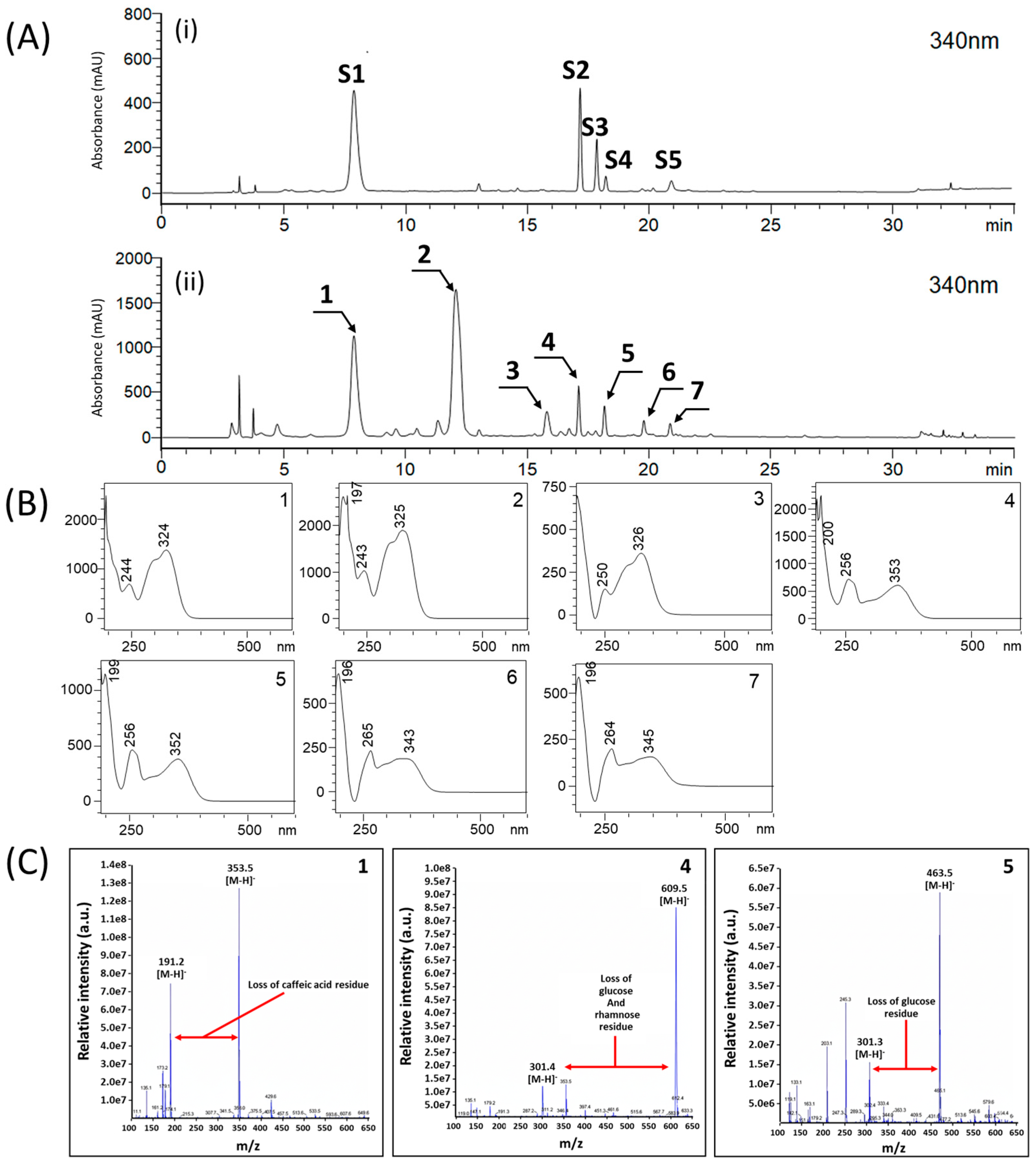
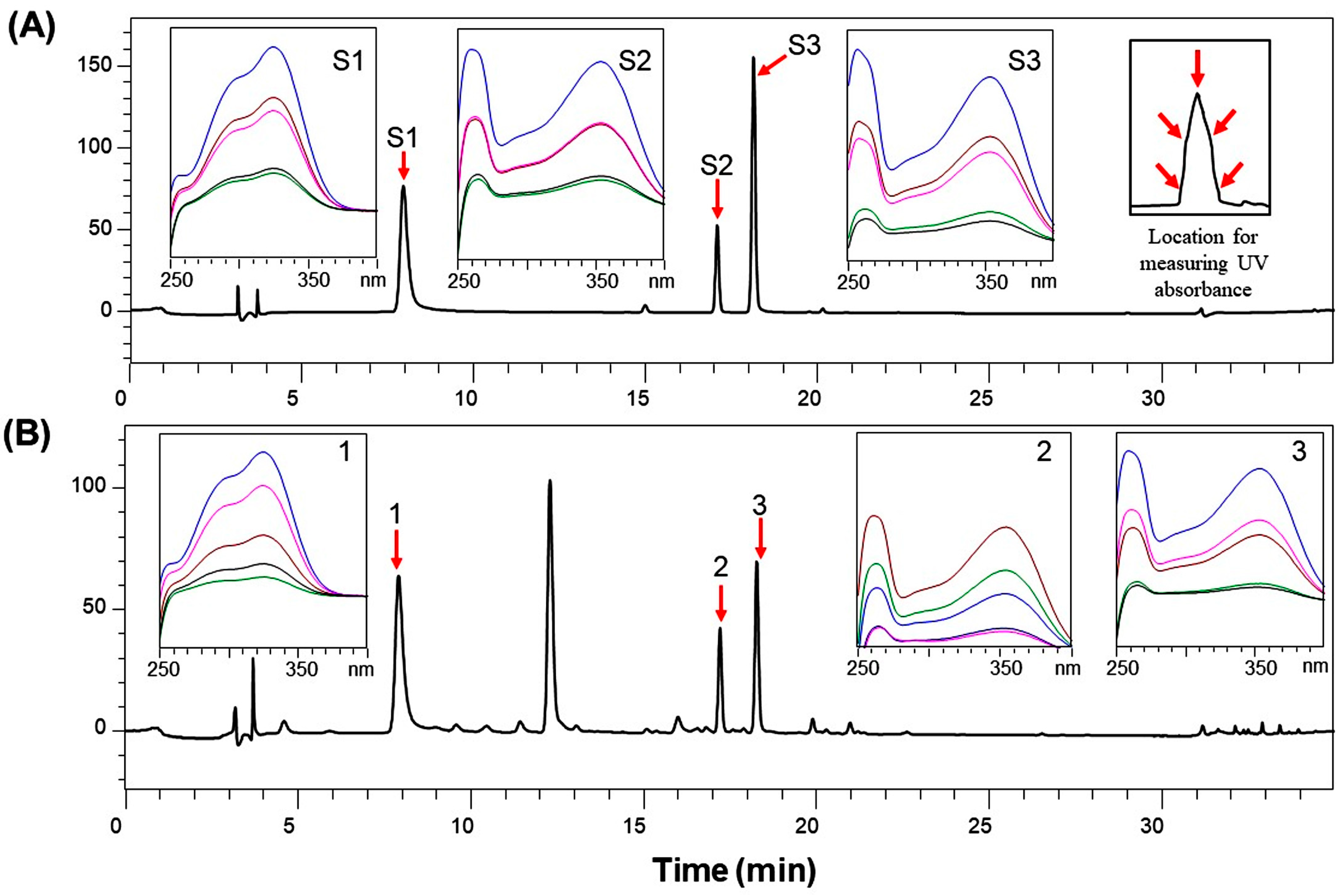
3.1. HPLC Analysis of Marker Substances in Ctl
3.2. Validation of the HPLC Analysis Method for Marker Substances from Ctl
3.3. Optimization of Extraction Conditions and Analysis of Variable Interactions for Chlorogenic Acid, Rutin, and Isoquercitrin
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
| HPLC | High-performance liquid chromatography |
| DAD | Diode array detector |
| LOD | Limit of detection |
| ROS | Reactive oxygen species |
| ATP | Adenosine triphosphate |
| Ctl | C. tricuspidata leaves |
| ACN | Acetonitrile |
| ICH | International Council for Harmonisation of Technical Requirements for Pharmaceuticals for Human Use |
| %RSD | Relative standard deviation |
References
- Choi, M.H.; Yang, S.H.; Kim, D.S.; Shin, H.S. Comparative analysis of biological activity between domestic Cudrania tricuspidata fruit extract and fermented fruit extract using Lactobacillus plantarum. KSBB J. 2021, 36, 247–253. [Google Scholar] [CrossRef]
- Działo, M.; Mierziak, J.; Korzun, U.; Preisner, M.; Wojtasik, W.; Szopa, J. The potential of plant phenolics in prevention and therapy of skin disorders. Int. J. Mol. Sci. 2016, 17, 160. [Google Scholar] [CrossRef]
- Espino, J.; Cosme, P.; Rodríguez, A.B.; Garrido, M. Plant phenolics: Bioavailability as a key determinant of their potential health-promoting applications. Antioxidants 2020, 9, 1263. [Google Scholar] [CrossRef]
- Nisa, R.U.; Nisa, A.U.; Tantray, A.Y.; Shah, A.H.; Jan, A.T. Plant phenolics with promising therapeutic applications against skin disorders: A mechanistic review. J. Agric. Food Chem. 2024, 16, 101090. [Google Scholar] [CrossRef]
- Kim, J.; Cho, N.; Kim, E.M.; Park, K.S.; Kang, Y.W.; Nam, J.H.; Nam, M.S.; Kim, K.K. Cudrania tricuspidata leaf extracts and its components, chlorogenic acid, kaempferol, and quercetin, increase claudin 1 expression in human keratinocytes, enhancing intercellular tight junction capacity. Appl. Biol. Chem. 2020, 63, 23. [Google Scholar] [CrossRef]
- Yu, C.; Wang, D.; Yang, Z.; Wang, T. Pharmacological effects of polyphenol phytochemicals on the intestinal inflammation via targeting TLR4/NF-κB signaling pathway. Int. J. Mol. Sci. 2022, 23, 6939. [Google Scholar] [CrossRef] [PubMed]
- Xin, L.T.; Yue, S.J.; Fan, Y.C.; Wu, J.S.; Yan, D.; Guan, H.S.; Wang, C.Y. Cudrania tricuspidata: An updated review on ethnomedicine, phytochemistry and pharmacology. RSC Adv. 2017, 7, 31807–31832. [Google Scholar] [CrossRef]
- Ko, W.; Kim, N.; Lee, H.; Woo, E.R.; Kim, Y.C.; Oh, H. Anti-inflammatory effects of compounds from Cudrania tricuspidata in HaCaT human keratinocytes. Int. J. Mol. Sci. 2021, 22, 7472. [Google Scholar] [CrossRef]
- Shin, J.; Oh, T.H.; Kim, J.Y.; Shim, J.J.; Lee, J.L. Efficacy and safety of the Cudrania tricuspidata extract on functional dyspepsia: A randomized double-blind placebo-controlled multicenter study. J. Clin. Med. 2021, 10, 5323. [Google Scholar] [CrossRef]
- Rhee, M.H.; Shin, J.H.; Kwon, H.W. Antiplatelet effect of cudraxanthone L isolated from Cudrania tricuspidata via inhibition of phosphoproteins. Nat. Prod. Sci. 2020, 26, 350–357. [Google Scholar] [CrossRef]
- Lee, D.H.; Son, Y.H.; Jang, J.H.; Lee, S.Y.; Kim, H.J. The growth characteristics and the active compounds of Cudrania tricuspidata fruits in different cultivation environments in South Korea. Plants 2023, 12, 2107. [Google Scholar] [CrossRef]
- Kim, H.; Chin, K.B. Evaluation of antioxidant activity of Cudrania tricuspidata leaves, fruit powder, and fruit in pork patties during storage. Food Sci. Anim. Resour. 2020, 40, 771–785. [Google Scholar] [CrossRef]
- Park, J.B.; Kim, D.W.; Lim, K.T.; Oh, S.; Lee, S.J. A 75 kDa glycoprotein isolated from Cudrania tricuspidata Bureau induces colonic epithelial proliferation and ameliorates mouse colitis. Chin. J. Nat. Med. 2021, 19, 250–260. [Google Scholar] [CrossRef]
- Jee, S.C.; Lee, K.M.; Kim, M.; Lee, Y.J.; Kim, S. Neuroprotective effect of Cudrania tricuspidata fruit extracts on scopolamine-induced learning and memory impairment. Int. J. Mol. Sci. 2020, 21, 9202. [Google Scholar] [CrossRef]
- Choi, J.H.; Kim, M.K.; Yeo, S.H.; Kim, S. Short-term Cudrania tricuspidata fruit vinegar administration attenuates obesity in high-fat diet-fed mice by improving fat accumulation and metabolic parameters. Sci. Rep. 2020, 10, 78166. [Google Scholar] [CrossRef] [PubMed]
- Nan, L.; Nam, H.H.; Choo, B.K. Protective effect of fermented Cudrania tricuspidata fruit extracts on acute rat reflux esophagitis. Pharmacogn. Mag. 2022, 18, 435–442. [Google Scholar]
- Li, X.; Yao, Z.; Jiang, X.; Sun, J.; Ran, G.; Yang, X.; Zhao, Y.; Yan, Y.; Chen, Z.; Tian, L.; et al. Bioactive compounds from Cudrania tricuspidata: A natural anticancer source. Crit. Rev. Food Sci. Nutr. 2020, 60, 494–514. [Google Scholar] [CrossRef] [PubMed]
- Smith, J.; Doe, A. The role of chlorogenic acid in glucose metabolism and diabetes management. J. Nutr. Biochem. 2021, 34, 123–135. [Google Scholar]
- Johnson, R.; Wang, H. Rutin and cardiovascular health: Mechanistic insights and therapeutic applications. Cardiovasc. Pharmacol. 2020, 45, 210–225. [Google Scholar]
- Yang, S.C.; Chang, Z.Y.; Hsiao, C.Y.; Alshetaili, A.; Wei, S.H. Topical Anti-Inflammatory Effects of Quercetin Glycosides on Atopic Dermatitis-Like Lesions: Influence of the Glycone Type. Inflammation 2025, 48, 2236. [Google Scholar]
- Green, J.M. Peer reviewed: A practical guide to analytical method validation. Anal. Chem. 1996, 68, 305A–309A. [Google Scholar] [CrossRef]
- Shabir, G.A. Validation of high-performance liquid chromatography methods for pharmaceutical analysis: Understanding the differences and similarities between validation requirements of the US Food and Drug Administration, the US Pharmacopeia and the International Conference on Harmonization. J. Chromatogr. A 2003, 987, 57–66. [Google Scholar] [PubMed]
- Orfali, R.; Perveen, S.; Aati, H.Y.; Alam, P.; Noman, O.M.; Palacios, J.; Al-Kurbi, B.S.S.; Al-Taweel, A.M.; Khan, A.; Mehmood, R.; et al. High-performance thin-layer chromatography for rutin, chlorogenic acid, caffeic acid, ursolic acid, and stigmasterol analysis in Periploca aphylla Extracts. Separations 2021, 8, 44. [Google Scholar] [CrossRef]
- Dai, Y.; Row, K.H. Determination of rutin from Ginkgo biloba L. leaves by ultrasound-assisted extraction with natural deep eutectic solvent-based cellulose polymers and high-performance liquid chromatography (HPLC). Anal. Lett. 2021, 55, 566–579. [Google Scholar] [CrossRef]
- Himshweta; Singh, M.; Verma, N.; Trehan, N. Identification of chlorogenic acid from Morus alba leaves by UV-vis spectroscopy, FTIR, UPLC-QTOF-MS and quantification by HPTLC. Commun. Soil Sci. Plant Anal. 2022, 54, 706–722. [Google Scholar] [CrossRef]
- Kostikova, V.A.; Petrova, N.V.; Shaldaeva, T.M.; Koval, V.V.; Chernonosov, A.A. Non-targeted screening of metabolites in aqueous-ethanol extract from Spiraea hypericifolia (Rosaceae) using LC-HRMS. Int. J. Mol. Sci. 2023, 24, 13872. [Google Scholar] [CrossRef]
- Vollmannová, A.; Bojňanská, T.; Musilová, J.; Lidiková, J.; Cifrová, M. Quercetin as one of the most abundant represented biological valuable plant components with remarkable chemoprotective effects—A review. Heliyon 2024, 10, e33342. [Google Scholar] [CrossRef]
- Oziembłowski, M.; Kapusta, I.; Popiół, J.; Piorun, M.; Wójcik, J.; Kaszuba, M.; Szymczak, A. optimization of chlorogenic acid in ethanol extracts of Elderberry flowers using Response Surface Methodology. Appl. Sci. 2023, 13, 3201. [Google Scholar] [CrossRef]
- Chew, S.K.; Teoh, W.H.; Hong, S.L.; Yusoff, R. Rutin extraction from female Carica papaya Linn. using ultrasound and microwave-assisted extractive methods: Optimization and extraction efficiencies. Heliyon 2023, 9, e20260. [Google Scholar] [CrossRef]
- Lin, L.Z.; Harnly, J.; Zhang, R.W.; Fan, X.E.; Chen, H.J. Quantitation of the hydroxycinnamic acid derivatives and the glycosides of flavonols and flavones by UV absorbance after identification by LC-MS. J. Agric. Food Chem. 2012, 60, 544–553. [Google Scholar] [CrossRef]
- Belwal, T.; Dhyani, P.; Bhatt, I.D.; Rawal, R.S.; Pande, V. Optimization extraction conditions for improv-ing phenolic content and antioxidant activity in Berberis asiatica fruits using Response Surface Methodol-ogy (RSM). Food Chem. 2016, 207, 115–124. [Google Scholar] [CrossRef] [PubMed]
- Taniguchi, M.; LaRocca, C.A.; Bernat, J.D.; Lindsey, J.S. Digital database of absorption spectra of diverse flavonoids enables structural comparisons and quantitative evaluations. J. Nat. Prod. 2023, 86, 1087–1119. [Google Scholar] [CrossRef] [PubMed]
- Sova, M.; Saso, L. Natural sources, pharmacokinetics, biological activities and health benefits of hydroxycinnamic acids and their metabolites. Nutrients 2020, 12, 2190. [Google Scholar] [CrossRef] [PubMed]
- Dias, M.C.; Pinto, D.C.G.A.; Silva, A.M.S. Plant flavonoids: Chemical characteristics and biological activity. Molecules 2021, 26, 5377. [Google Scholar] [CrossRef]
- Ivanauskas, L.; Uminska, K.; Gudžinskas, Z.; Heinrich, M.; Georgiyants, V.; Kozurak, A.; Mykhailenko, O. Phenological variations in the content of polyphenols and triterpenoids in Epilobium angustifolium Herb originating from Ukraine. Plants 2024, 13, 120. [Google Scholar] [CrossRef]
- Kumar, S.; Pandey, A.K. Chemistry and biological activities of flavonoids: An overview. Sci. World J. 2013, 162750. [Google Scholar] [CrossRef]
- da Silva, A.P.G.; Sganzerla, W.G.; John, O.D.; Marchiosi, R. A comprehensive review of the classification, sources, biosynthesis, and biological properties of hydroxybenzoic and hydroxycinnamic acids. Phytochem. Rev. 2023, 22, 1061–1090. [Google Scholar] [CrossRef]
- Fredsgaard, M.; Kaniki, S.E.K.; Antonopoulou, I.; Chaturvedi, T.; Thomsen, M.H. Phenolic compounds in Salicornia spp. and their potential therapeutic effects on H1N1, HBV, HCV, and HIV: A review. Molecules 2023, 28, 5312. [Google Scholar] [CrossRef]
- Luo, M.; Liu, X.; Zhao, Z.; Wang, F.; Shao, C. Optimization of glycerol extraction of chlorogenic acid from honeysuckle by Response Surface Methodology. Processes 2023, 11, 110. [Google Scholar] [CrossRef]
- El Maaiden, E.; Qarah, N.; Ezzariai, A.; Mazar, A.; Nasser, B.; Moustaid, K.; Boukcim, H.; Hirich, A.; Kouisni, L.; El Kharrassi, Y. Ultrasound-assisted extraction of isoquercetin from Ephedra alata (Decne): Optimization using Response Surface Methodology and in vitro bioactivities. Antioxidants 2023, 12, 725. [Google Scholar] [CrossRef]
- Goudjil, S.; Boussekine, S.; Goudjil, S.; Goudjil, H.; Yilmaz, M.A.; Ola, M.S.; Ali, A.; Cakir, O. Investigation of Algerian Crataegus monogyna Jacq phenolic compounds (using LC-ESI-MS/MS analysis, antioxidant activity, and enzyme inhibition) and their potential implications for food and nutraceutical applications. Antioxidants 2024, 13, 1350. [Google Scholar] [CrossRef]

| Compound | Repetition | Range (μg/mL) | Regression Equation | r2 |
|---|---|---|---|---|
| Chlorogenic acid | 1 | 0.1–100 | y = 9183.3x − 3035 | 0.999 |
| 2 | 0.1–100 | y = 9148.1x − 1980 | 1.000 | |
| 3 | 0.1–100 | y = 9139.8x − 1481 | 0.999 | |
| Mean of slope (S0) | 9157.07 | SD of intercept (σ) | 793.361 | |
| LOQ (10 × σ/s) | 1.25 (μg/mL) | |||
| LOD (3.3 × σ/s) | 0.29 (μg/mL) | |||
| Rutin | 1 | 0.1–100 | y = 44,092x − 12,859 | 0.999 |
| 2 | 0.1–100 | y = 45,190x − 21,972 | 0.996 | |
| 3 | 0.1–100 | y = 44,007x − 16,073 | 0.994 | |
| Mean of slope (S0) | 37,084.67 | SD of intercept (σ) | 4621.954 | |
| LOQ (10 × σ/s) | 0.87 (μg/mL) | |||
| LOD (3.3 × σ/s) | 0.41 (μg/mL) | |||
| Isoquercitrin | 1 | 0.1–100 | y = 42,269x − 11,208 | 0.999 |
| 2 | 0.1–100 | y = 42,318x − 6833 | 0.999 | |
| 3 | 0.1–100 | y = 42,143x − 6690 | 0.999 | |
| Mean of slope (S0) | 42,243.33 | SD of intercept (σ) | 2568.098 | |
| LOQ (10 × σ/s) | 0.61 (μg/mL) | |||
| LOD (3.3 × σ/s) | 0.20 (μg/mL) | |||
| (A) Chlorogenic acid | ||||
| Repetition | Range(μg/mL) | Regression Equation | r2 | 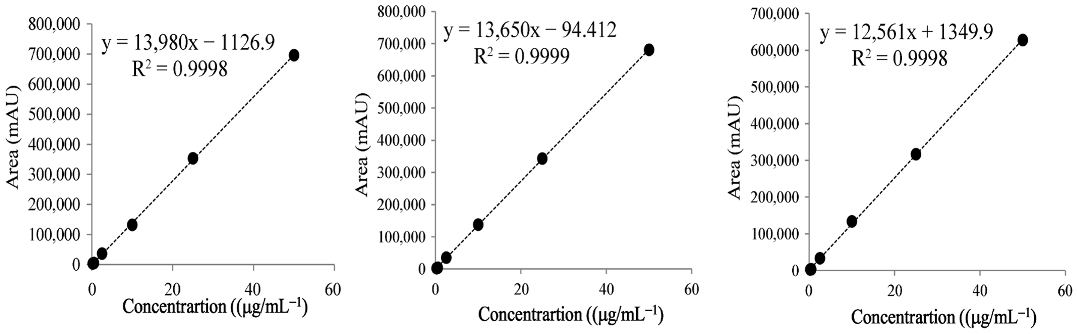 |
| 1 | 0.25–50 | y = 13,980x − 1127 | 0.999 | |
| 2 | 0.25–50 | y = 13,650x − 94 | 0.999 | |
| 3 | 0.25–50 | y = 12,561x + 1350 | 0.999 | |
| (B) Rutin | ||||
| Repetition | Range(μg/mL) | Regression Equation | r2 | 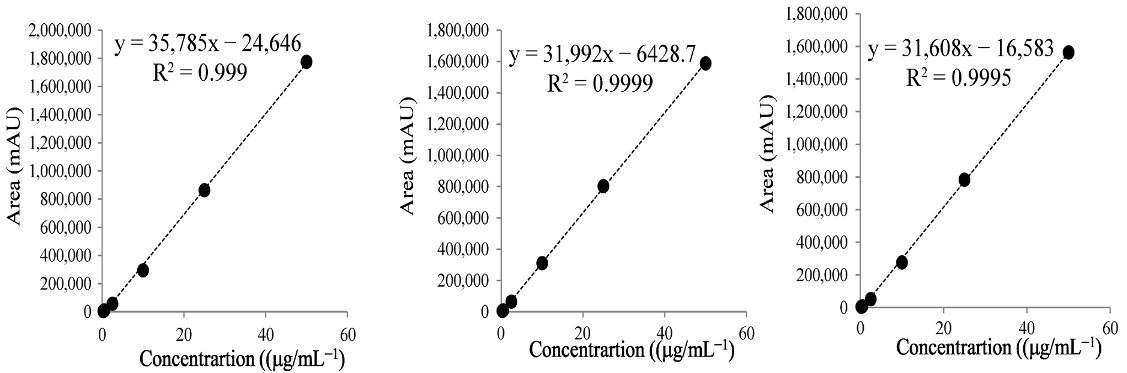 |
| 1 | 0.25–50 | y = 35,785x − 24,646 | 0.999 | |
| 2 | 0.25–50 | y = 31,992x − 6429 | 0.999 | |
| 3 | 0.25–50 | y = 31,608x − 16,583 | 0.999 | |
| (C) Isoquercitirn | ||||
| Repetition | Range(μg/mL) | Regression Equation | r2 | 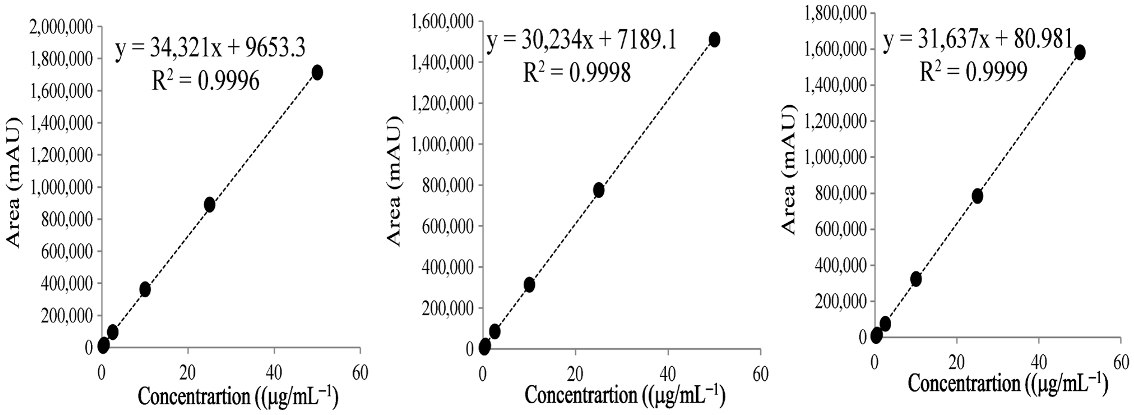 |
| 1 | 0.25–50 | y = 34,321x + 9653 | 0.999 | |
| 2 | 0.25–50 | y = 30,234x + 7189 | 0.999 | |
| 3 | 0.25–50 | y = 31,637x + 81 | 0.999 | |
| Compound | Repetition | Spiked Amounts (μg/mL) | |||
|---|---|---|---|---|---|
| 1.0 | 2.0 | 3.0 | 4.0 | ||
| Chlorogenic acid | 1 | 101.34 | 100.60 | 102.68 | 102.00 |
| 2 | 101.60 | 100.52 | 102.60 | 102.04 | |
| 3 | 101.04 | 100.54 | 102.61 | 102.16 | |
| 4 | 101.14 | 100.92 | 102.49 | 102.11 | |
| 5 | 101.28 | 100.69 | 102.49 | 101.74 | |
| Mean recovery rate (%) | 101.28 | 100.65 | 102.57 | 102.01 | |
| Net recovery rate (%) | 101.63 | ||||
| Range of recovery rate (%) | 101.04–102.68 | ||||
| Rutin | 1 | 102.39 | 100.88 | 102.50 | 101.99 |
| 2 | 101.90 | 100.28 | 102.36 | 102.14 | |
| 3 | 102.05 | 100.57 | 102.76 | 101.96 | |
| 101.36 | 100.23 | 102.89 | 102.08 | ||
| 101.95 | 100.48 | 102.22 | 102.16 | ||
| Mean recovery rate (%) | 101.93 | 100.49 | 102.55 | 102.07 | |
| Net recovery rate (%) | 101.76 | ||||
| Range of recovery rate (%) | 100.23–102.89 | ||||
| Isoquercitrin | 1 | 100.81 | 100.32 | 103.70 | 104.15 |
| 2 | 100.67 | 100.37 | 103.61 | 104.30 | |
| 3 | 100.73 | 100.31 | 103.39 | 104.22 | |
| 4 | 100.71 | 100.30 | 103.27 | 104.22 | |
| 5 | 100.71 | 100.32 | 103.14 | 104.43 | |
| Mean recovery rate (%) | 100.73 | 100.32 | 103.42 | 104.26 | |
| Net recovery rate (%) | 102.18 | ||||
| Range of recovery rate (%) | 100.30–104.43 | ||||
| Amount of Ethanolic Extract of Ctl | |||
|---|---|---|---|
| 1.0 g | 2.0 g | 3.0 g | |
| Repetition | Chlorogenic acid (μg/g) | Chlorogenic acid (μg/g) | Chlorogenic acid (μg/g) |
| 1 | 715.62 | 721.01 | 724.12 |
| 2 | 711.22 | 722.12 | 729.13 |
| 3 | 716.06 | 720.69 | 730.22 |
| 4 | 710.68 | 721.12 | 724.15 |
| 5 | 718.25 | 721.03 | 729.54 |
| Mean | 714.37 | 721.19 | 727.43 |
| SD | 3.28 | 0.54 | 3.03 |
| RSD | 0.46 | 0.08 | 0.42 |
| RSD (%) 0.08–0.46 | |||
| Repetition | Rutin (μg/g) | Rutin (μg/g) | Rutin (μg/g) |
| 1 | 319.62 | 323.13 | 328.65 |
| 2 | 316.56 | 326.23 | 329.63 |
| 3 | 319.55 | 329.46 | 328.46 |
| 4 | 318.42 | 326.59 | 326.71 |
| 5 | 315.48 | 324.88 | 328.10 |
| Mean | 317.93 | 326.06 | 328.32 |
| SD | 1.84 | 2.34 | 1.06 |
| RSD | 0.58 | 0.72 | 0.32 |
| RSD (%): 0.32–0.72 | |||
| Repetition | Isoquercitrin (μg/g) | Isoquercitrin (μg/g) | Isoquercitrin (μg/g) |
| 1 | 532.62 | 541.05 | 546.54 |
| 2 | 539.23 | 542.21 | 548.97 |
| 3 | 537.89 | 544.22 | 549.32 |
| 4 | 537.66 | 546.47 | 546.42 |
| 5 | 539.14 | 546.99 | 549.77 |
| Mean | 537.31 | 544.19 | 548.21 |
| SD | 2.72 | 2.59 | 1.60 |
| RSD | 0.51 | 0.48 | 0.29 |
| RSD (%) 0.29–0.51 | |||
| Amount of Ethanolic Extract of Ctl | |||||||
| Chlorogenic acid (μg/g) | Rutin (μg/g) | Isoquercitrin (μg/g) | |||||
| Day | Repetition | 1.5 g | 3.0 g | 1.5 g | 3.0 g | 1.5 g | 3.0 g |
| 1 | 1 | 723.67 | 724.30 | 324.12 | 327.72 | 544.37 | 546.62 |
| 2 | 727.78 | 725.16 | 322.58 | 326.85 | 541.95 | 547.27 | |
| 3 | 726.66 | 725.00 | 322.32 | 325.72 | 543.66 | 544.11 | |
| 4 | 724.09 | 726.82 | 322.63 | 326.15 | 541.96 | 555.42 | |
| 5 | 723.66 | 726.98 | 322.94 | 330.01 | 543.39 | 547.83 | |
| 2 | 1 | 727.72 | 724.82 | 326.06 | 320.14 | 543.95 | 542.92 |
| 2 | 727.17 | 729.88 | 326.12 | 329.64 | 544.89 | 543.94 | |
| 3 | 727.08 | 723.68 | 325.00 | 323.45 | 545.06 | 548.61 | |
| 4 | 726.98 | 721.11 | 325.24 | 328.54 | 545.93 | 547.18 | |
| 5 | 726.98 | 729.63 | 325.95 | 326.57 | 546.28 | 544.73 | |
| 3 | 1 | 723.32 | 725.80 | 324.81 | 328.89 | 541.34 | 542.33 |
| 2 | 723.29 | 724.65 | 324.29 | 326.54 | 542.73 | 549.67 | |
| 3 | 724.63 | 729.87 | 324.26 | 329.75 | 541.37 | 547.81 | |
| 4 | 724.24 | 723.65 | 325.03 | 328.04 | 542.39 | 541.11 | |
| 5 | 724.39 | 726.99 | 324.90 | 328.01 | 543.56 | 542.01 | |
| 4 | 1 | 723.75 | 729.22 | 329.26 | 328.85 | 542.67 | 548.11 |
| 2 | 724.02 | 724.68 | 327.68 | 327.41 | 542.35 | 546.83 | |
| 3 | 723.49 | 725.58 | 327.13 | 328.79 | 543.13 | 544.71 | |
| 4 | 723.38 | 729.46 | 324.55 | 325.54 | 542.05 | 549.56 | |
| 5 | 723.31 | 720.68 | 326.72 | 327.75 | 542.20 | 549.67 | |
| 5 | 1 | 727.16 | 728.89 | 328.20 | 328.56 | 541.43 | 545.80 |
| 2 | 724.94 | 729.63 | 327.56 | 326.98 | 541.84 | 544.74 | |
| 3 | 726.48 | 722.56 | 325.93 | 326.87 | 543.35 | 542.93 | |
| 4 | 724.55 | 728.57 | 324.59 | 324.82 | 541.89 | 547.95 | |
| 5 | 722.41 | 724.48 | 327.15 | 324.33 | 542.43 | 549.18 | |
| Mean | 725.01 | 726.08 | 325.40 | 327.03 | 543.05 | 546.42 | |
| SD | 1.71 | 2.76 | 1.86 | 2.22 | 1.39 | 3.19 | |
| RSD (%) | 0.24 | 0.38 | 0.57 | 0.68 | 0.26 | 0.58 | |
| Net RSD (%) 0.38 | Net RSD (%) = 0.68 | Net RSD (%) = 0.58 | |||||
| Experiment Number | Extraction Temperature (°C) | Extraction Concentration (%) | Extraction Time (Min) | Chlorogenic Acid (μg/g) | Rutin (μg/g) | Isoquercitrin (μg/g) |
|---|---|---|---|---|---|---|
| 1 | 60 | 20 | 120 | 696.64 | 316.75 | 535.36 |
| 2 | 80 | 20 | 120 | 718.94 | 309.43 | 553.75 |
| 3 | 60 | 60 | 120 | 690.80 | 310.27 | 550.16 |
| 4 | 80 | 60 | 120 | 736.22 | 310.90 | 573.56 |
| 5 | 60 | 40 | 90 | 715.10 | 319.15 | 536.49 |
| 6 | 80 | 40 | 90 | 735.46 | 328.18 | 550.84 |
| 7 | 60 | 40 | 150 | 737.91 | 333.02 | 552.21 |
| 8 | 80 | 40 | 150 | 736.81 | 311.85 | 556.96 |
| 9 | 70 | 20 | 90 | 844.28 | 328.15 | 573.10 |
| 10 | 70 | 60 | 90 | 847.45 | 328.33 | 578.28 |
| 11 | 70 | 20 | 150 | 799.77 | 337.96 | 572.83 |
| 12 | 70 | 60 | 150 | 843.64 | 328.92 | 571.42 |
| 13 | 70 | 40 | 120 | 908.30 | 348.44 | 654.57 |
| 14 | 70 | 40 | 120 | 908.41 | 348.25 | 654.01 |
| 15 | 70 | 40 | 120 | 912.53 | 347.48 | 652.14 |
| 16 | 70 | 40 | 120 | 911.52 | 348.30 | 653.72 |
| 17 | 70 | 40 | 120 | 907.92 | 348.69 | 657.57 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kang, J.-Y.; Noh, H.-R.; Yoone, Y.; Kim, B.-G. Quantitative Analysis of Chlorogenic Acid, Rutin, and Isoquercitrin in Extracts of Cudrania tricuspidata Leaves Using HPLC-DAD. Separations 2025, 12, 298. https://doi.org/10.3390/separations12110298
Kang J-Y, Noh H-R, Yoone Y, Kim B-G. Quantitative Analysis of Chlorogenic Acid, Rutin, and Isoquercitrin in Extracts of Cudrania tricuspidata Leaves Using HPLC-DAD. Separations. 2025; 12(11):298. https://doi.org/10.3390/separations12110298
Chicago/Turabian StyleKang, Ju-Yeong, Hye-Ryeong Noh, Youngdae Yoone, and Bong-Gyu Kim. 2025. "Quantitative Analysis of Chlorogenic Acid, Rutin, and Isoquercitrin in Extracts of Cudrania tricuspidata Leaves Using HPLC-DAD" Separations 12, no. 11: 298. https://doi.org/10.3390/separations12110298
APA StyleKang, J.-Y., Noh, H.-R., Yoone, Y., & Kim, B.-G. (2025). Quantitative Analysis of Chlorogenic Acid, Rutin, and Isoquercitrin in Extracts of Cudrania tricuspidata Leaves Using HPLC-DAD. Separations, 12(11), 298. https://doi.org/10.3390/separations12110298






