Abstract
Recent advancements in genome sequencing and bioinformatic analysis of fungal genomes have revealed that fungi possess cryptic or silent biosynthetic gene clusters (BGCs). This discovery suggests that our understanding of the universe of fungal secondary metabolomes is limited. In this review, we summarize recent strategies for activating cryptic BGCs in fungi, identify fungal secondary metabolites, and highlight their biological activities. We also briefly discuss the isolation and purification methods for these compounds. Our focus is on genetics-dependent and genetics-independent approaches for activating cryptic BGCs in fungi. Using the strategies discussed here, cryptic fungal natural products hold significant potential as a source for the discovery of new drug candidates.
1. Introduction
Natural products have been recognized as crucial sources for new drug discovery. Over the past 38 years, more than half of the clinical drugs that have been approved by the FDA were derived from natural sources, and natural products still hold promising potential for discovering novel drug candidates and bioactive chemical templates [1,2]. Fungi, in particular, offer an incredibly prolific and diverse array of bioactive secondary metabolites, making them an important natural resource for producing unique chemical compounds to combat a variety of diseases [3,4,5]. Notably, a multitude of fungal natural products exhibiting various biological effects have been discovered, suggesting that fungi play a role in communicating with other organisms and adapting to different environments [6,7]. Some of these identified fungal natural products have already been utilized in the health–functional food, agrochemical, cosmetic, and pharmaceutical industries [8].
In the early 2000s, technological advances in genome sequencing and bioinformatics on filamentous fungi began to reveal a discrepancy between the number of biosynthetic gene clusters (BGCs) encoding the biosynthesis of fungal secondary metabolites and the actual number of identified fungal compounds from the target strain [9,10]. This fact suggested that fungi have a great potential for identifying structurally and/or biologically novel secondary metabolites. However, many BGCs are not actively expressed in the normal laboratory growth environment. These are so-called cryptic or silent BGCs [11,12]. It is estimated that there are over 5 million fungal species on earth, and each of these species is capable of producing a variety of secondary metabolites, including bioactive compounds, pigments, and toxins [9,12]. These secondary metabolites are produced by specialized biosynthetic pathways, which are encoded by clusters of genes known as BGCs. Despite the availability of over 1000 fully sequenced fungal genomes and the identification of tens of thousands of BGCs, only a small fraction (<3%) of these clusters have been linked to specific secondary metabolites in part because of the cryptic BGCs of fungi [9].
Neurospora crassa, a member of the Ascomycota phylum, serves as a model organism for the study of fungal genetics, physiology, and development. It has been widely employed to investigate fundamental processes, such as circadian rhythm and gene regulation [13]. N. crassa is known to produce a variety of secondary metabolites, including carotenoids, melanins, and mycotoxin sterigmatocystin [13,14]. The sequencing information of N. crassa was completed in 2003 and has been found to contain numerous BGCs, many of which are predicted to encode secondary metabolites [15]. Recently, about 70 BGCs including polyketide synthases (PKSs), non-ribosomal peptide synthetases (NRPSs), terpene synthases, and siderophore synthetases were reported from the sequencing data of the fungus [14,15]. However, only a few of BGCs of N. crassa have been linked to specific secondary metabolites or characterized in detail. Bioinformatics-based predictions of the chemical structures based on the uncharacterized BGCs suggested that many of them were likely to have novel structures. Experimental characterization of these novel metabolites is often challenging since many BGCs are weakly expressed under laboratory conditions and may require specific environmental cues, growth conditions, and extraction and isolation techniques to induce production [16,17,18].
After the completion of genome sequencing on N. crassa, the genomes of many fungi, including those of both Ascomycota and Basidiomycota phyla, have been found to contain numerous cryptic BGCs [19]. Aspergillus nidulans is one of the most well-studied secondary metabolite producers. A. nidulans has been shown to produce a diverse array of secondary metabolites, including emericellamides, terrain, asperfuranone, fumitremorgins, gliotoxin, and aspernidine A [20]. Several studies have used computational methods to predict the number of BGCs in the A. nidulans genome. One such study, published in 2015, identified 52 BGCs in A. nidulans using a combination of genome mining and phylogenetic analysis [21]. Another study, published in 2018, identified 63 BGCs in the strain using a similar approach [21].
The discovery of cryptic BGCs in microorganisms, including fungi, has spurred the development of new experimental methodologies for identifying the secondary metabolites of these clusters, which led to the realization that they have the potential to produce novel specialized metabolites, giving rise to a new field of research called genome-guided natural product discovery [22]. Aside from pinpointing the genomics-driven approach, traditional approaches for identifying and characterizing natural products, such as fractionation and purification followed by structural elucidation using techniques such as NMR spectroscopy and mass spectrometry, can be time-consuming and require large amounts of material. To address these challenges, newer approaches such as metabolomics, transcriptomics, and proteomics have been developed to more efficiently identify and characterize natural products from cryptic BGCs [23].
In this review, we will provide an overview of recent natural product discovery strategies for activating cryptic BGCs in fungi (Figure 1). We will cover genetics-dependent and genetics-independent approaches, as well as the isolation and structural elucidation of cryptic fungal metabolites and their biological activities. Additionally, we will briefly discuss techniques used for the purification and identification of new secondary metabolites and consider future aspects of the natural product field.
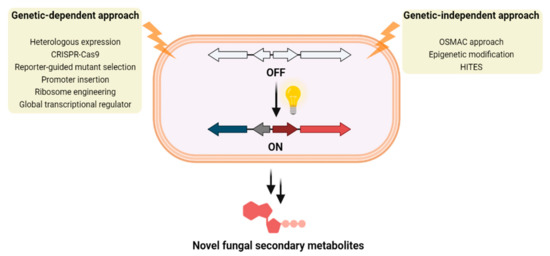
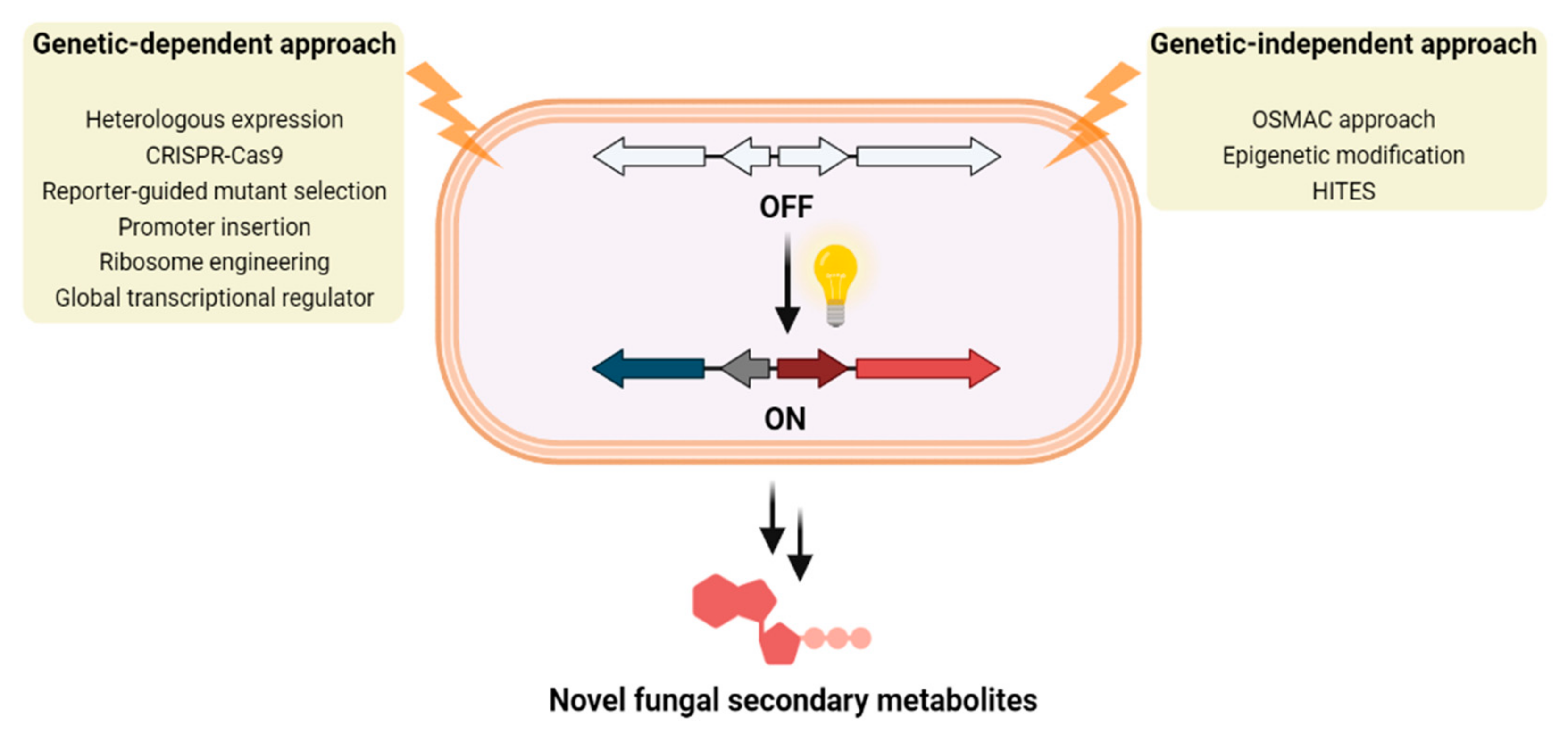
Figure 1.
Two main approaches for the identification of novel fungal secondary metabolites by the activation of cryptic fungal BGCs.
2. Organization of Biosynthetic Gene Clusters of Fungi and Their Regulation
Fungi can produce various secondary metabolites with diverse biological activities, such as antibiotics, antifungals, immunosuppressants, and anticancer agents. These secondary metabolites are often encoded by BGCs, which are physically co-localized on the fungal genome and contain all the genes necessary for the biosynthesis of the corresponding secondary metabolite [21,24]. The organization of BGCs in fungi can differ depending on the type of secondary metabolite being produced, but there are some common features. Typically, BGCs are composed of a core set of genes that encode enzymes responsible for the biosynthesis of secondary metabolites, as well as regulatory genes that control gene expression and coordinate the biosynthesis process [24,25]. In many cases, BGCs are found within mobile genetic elements such as transposable elements, plasmids, or integrative and conjugative elements, which can facilitate their transfer between different fungal strains or even different fungal species. The structure of BGCs can also be highly variable, with some BGCs containing only a few genes, while others can harbor dozens of genes that are organized into sub-clusters or modules. These sub-clusters may be responsible for the synthesis of different parts of the secondary metabolite, which are then combined to form the final product [26,27].
Fungal BGCs can be quite large, often exceeding 100 kb in size [19,28]. This fact presents a challenge for researchers who want to study the activity of these gene clusters by expressing them heterologously in a different host organism, such as E. coli or yeast. Fungal BGCs are classified based on the type of secondary metabolite they encode, including polyketides, non-ribosomal peptides, terpenoids, saccharides, and ribosomally synthesized and post-translationally modified peptides (RiPPs) [29,30,31]. The organization of BGCs in fungi is highly complex and dynamic, reflecting the diverse functions and ecological roles of the secondary metabolites they produce. Polyketide synthases (PKSs) are a class of enzymes found in fungi and other organisms that are responsible for the biosynthesis of polyketides. PKSs are modular enzymes that utilize a repeating cycle of catalytic domains to assemble complex polyketide chains from simple building blocks, such as acetate and malonate. Each module typically contains several different domains that are responsible for different steps in the biosynthesis process, such as chain initiation, chain elongation, and chain termination. Fungal NRPs utilize a repeating cycle of catalytic domains to assemble complex peptides from simple amino acid building blocks. Each module in NRPs harbors several various domains, which lead to the biosynthesis processes including amino acid activation, amino acid incorporation, and peptide bond formation.
The position of fungal BGCs is usually observed proximal to the telomeres in the genome and often within heterochromatin regions [32]. Heterochromatic regions are generally considered to be silent regions of the genome with low gene density and reduced recombination [32,33]. This may provide a more stable genomic environment for the BGCs, which are often under positive selection due to their role in fungal survival. The expression of BGCs is tightly regulated via a complex interaction of genetic, epigenetic, and environmental factors. The regulation of BGCs is important for ensuring that these clusters are expressed under appropriate conditions and that the products of biosynthesis are synthesized and utilized efficiently. A transcription factor is a protein that can bind to specific DNA sequences and activate or repress gene expression. Many BGCs are controlled by transcription factors that are specific to the biosynthetic pathway and that respond to environmental signals to activate or repress expression [19]. The structure of chromatin has a significant impact on gene expression in fungi. The presence of histone modifications such as methylation, acetylation, and phosphorylation controls the accessibility of DNA, and therefore the expression of the genes within BGCs [34]. Many BGCs are expressed in response to specific environmental triggers, such as nutrient availability or the presence of competing organisms. These signals affect transcription factors or other regulatory elements that modulate the expression of BGCs. In some cases, BGCs can be acquired through horizontal gene transfer, which involves the transfer of genetic material from one organism to another [35].
Fungi are highly adaptable organisms that live in diverse and complex natural environments, and their growth and metabolism are influenced by a variety of biotic and abiotic factors. However, laboratory growth conditions are usually simple and standardized and may not accurately reflect the physical structure, nutrient availability, and microbial diversity of actual natural environments. Additionally, laboratory conditions may not accurately mimic the environmental stresses that fungi encounter in the wild, such as changes in temperature, pH, osmotic pressure, and competition with other microorganisms. As a result, fungal fermentations in the general laboratory may not accurately represent the full range of metabolic and biosynthetic capabilities that fungi exhibit in their natural habitats [36]. Therefore, understanding the regulatory processes that modulate the growth, metabolism, and biosynthetic capabilities of fungi is critical for unlocking their full potential as sources of bioactive compounds. This requires a multidisciplinary approach that combines microbiology, biochemistry, and genetics to fully understand the complex regulation of fungi.
3. Characterization of Biosynthetic Gene Clusters and Natural Product Discovery
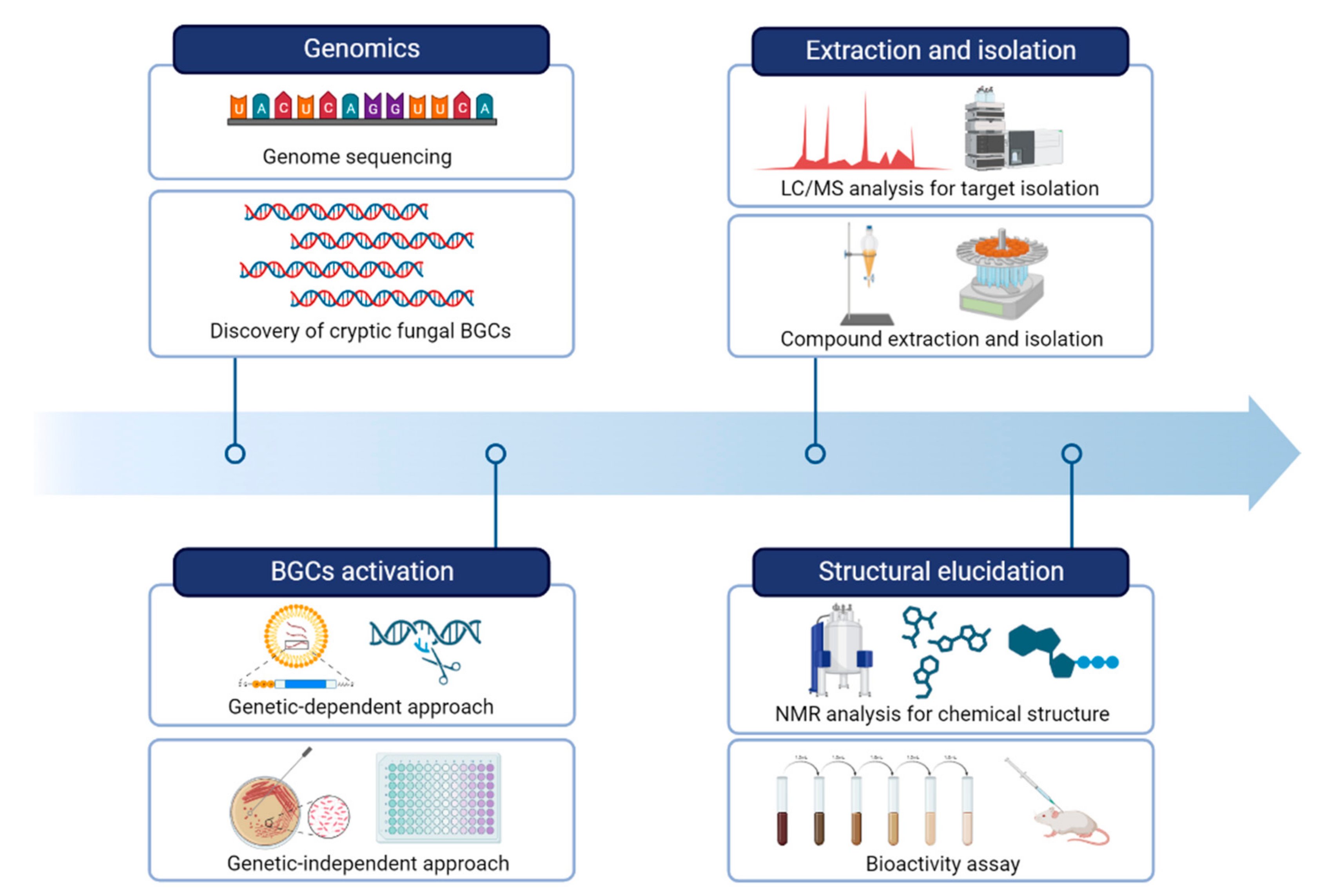
Next-generation sequencing (NGS) technologies have revolutionized the field of genomics by allowing rapid and cost-effective acquisition of genomic data. The rapid pace of technological advancements in NGS has led to an exponential increase in the amount of fungal genomic data generated, which has in turn fueled the development of new analytical tools and computational approaches to handle and analyze these data [27,37]. It is now common for researchers to identify BGCs responsible for the production of fungal secondary metabolites. By obtaining a draft genome sequence of fungi, researchers utilize a variety of bioinformatic tools to identify and analyze potential BGCs involved in the biosynthesis of a particular compound of interest (Figure 2).
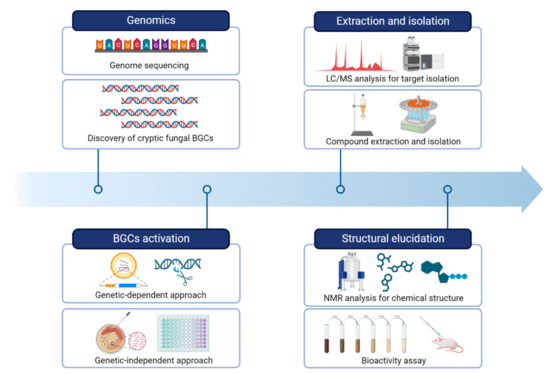
Figure 2.
A workflow of strategies for natural product research by activating fungal BGCs.
Once a draft genome sequence has been acquired, researchers start to use tools such as antiSMASH and MIBiG to identify potential BGCs within the genome [38,39]. These tools analyze the genome sequence for specific gene clusters known to be involved in the biosynthesis of secondary metabolites, such as polyketides, non-ribosomal peptides, and terpenes. By comparing the identified BGCs to known BGCs in databases, researchers can predict the structure and function of the secondary metabolites. antiSMASH is one of the most widely used BGC detection tools [38]. It is a web-based tool that allows users to input draft genome sequences and predict the location of BGCs in the genome. The tool uses a variety of algorithms to identify BGCs, including hidden Markov models (HMMs), Pfam domains, and Clusters of Orthologous Groups (COGs). The tool also provides annotations of the predicted BGCs, including predictions of the chemical structure of the metabolite produced. Alternative tools for BGCs identification are PRISM and BAGEL [40,41]. PRISM uses a machine learning algorithm to predict BGCs in microbial genomes. It integrates multiple data sources, including gene co-occurrence patterns, gene expression data, and functional annotations, to identify BGCs. PRISM also includes tools for visualizing and exploring predicted BGCs, including interactive network visualizations. BAGEL is a tool specifically designed for the identification of bacteriocin gene clusters, which are BGCs involved in the biosynthesis of antimicrobial peptides produced by bacteria. BAGEL uses a combination of HMMs and machine learning algorithms to predict bacteriocin gene clusters in microbial genomes.
When the above tools are applied to the genome sequence of a specific fungal strain, they are expected to identify cryptic BGCs that might be responsible for the production of unknown compounds and would be a promising starting point for natural product discovery. The type of NGS technology utilized impacts the capability to characterize the full complement of BGCs in a particular genome [42]. Different NGS technologies have different strengths and limitations, and the choice of technology will depend on factors such as the size and complexity of the genome, the sequencing depth needed, and the availability of bioinformatics tools and resources [43]. Some NGS technologies, such as PacBio and Oxford Nanopore, can generate long reads that can span entire BGCs and provide more complete sequence information than short-read technologies such as Illumina [44]. In addition to the choice of NGS technology, the ability to identify the full complement of BGCs in a particular genome also depends on the quality of the genome assembly, the bioinformatics tools used for BGC identification, and the expertise of the researchers involved [43,44]. Genome assembly is a critical step in NGS-based approaches for BGC identification, as errors or gaps in the assembly can lead to missed or incomplete BGCs.
Although NGS-based approaches have been utilized to identify the full complement of putative specialized metabolite BGCs, there are limitations to the application of natural product discovery. It is still challenging to predict the chemical structures of the metabolites produced by these BGCs based only on genomic and bioinformatic information [45,46]. The identification of BGCs is just the first step in the process of discovering and characterizing natural products. After a specific BGC is identified, it is necessary to express and characterize the genes involved in the biosynthesis of the secondary metabolite, to produce and purify the metabolite itself, and to test its relevant biological activity. These steps require significant resources and expertise, and may not be feasible for all BGCs identified from genomic data (Figure 2).
4. Genetics-Dependent Approach
4.1. Heterologous Expression
Fungal secondary metabolites are very valuable compounds that can be used for medical purposes. However, the majority of fungal BGCs, which produce fungal secondary metabolites, are cryptic in a general laboratory cultivation system. One of the useful approaches for finding fungal secondary metabolites is heterologous expression (Figure 3). Heterologous expression is the method of expressing a gene or gene fragment of interest using a host organism that naturally does not have the gene and is an effective way to identify the function of the gene [47]. The general procedure is transforming the specific target gene using a vector, culturing and expressing it, identifying the chemical structure, and confirming its bioactivities. Host organisms for heterologous expression are mainly Escherichia coli (bacteria), Saccharomyces cerevisiae (yeast), Aspergillus nidulans, and Aspergillus oryzae (filamentous fungi) for obtaining fungal secondary metabolites [47].
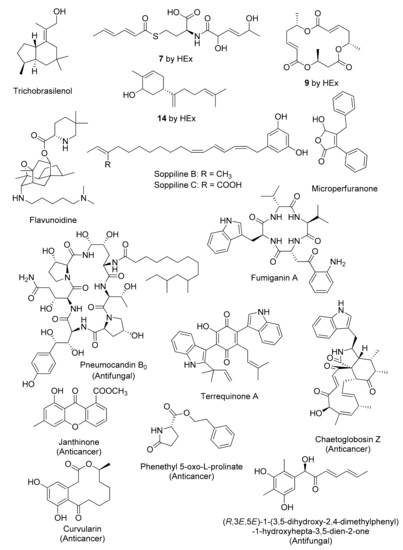
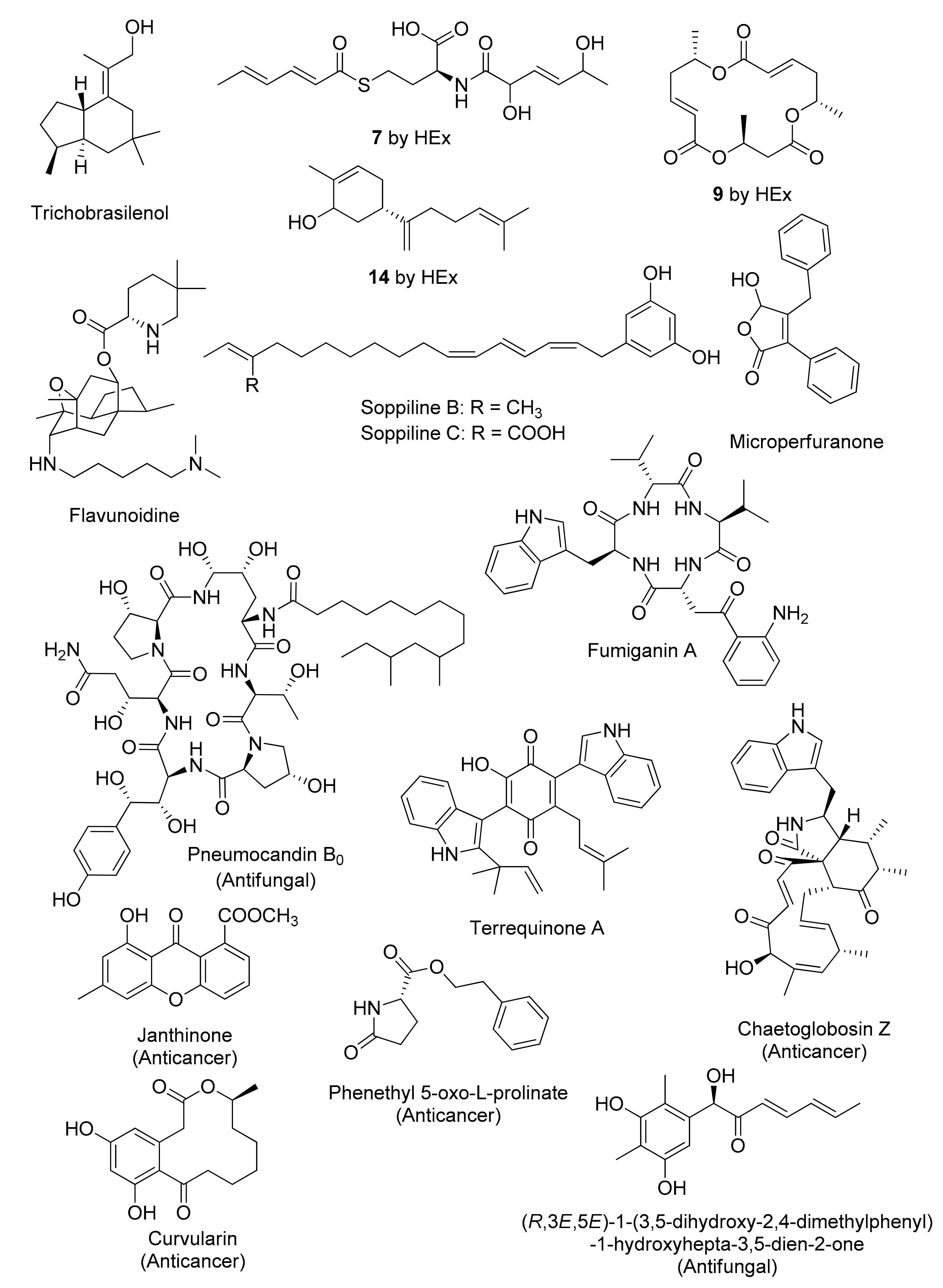
Figure 3.
Representative chemical structures of cryptic fungal metabolites obtained by genetics-dependent approach.
Heterologous expression in E. coli is a good approach for characterizing cryptic fungal compounds. E. coli as a host organism for heterologous expression affords many advantages, including fast cell growth, inexpensive cultivation medium, simple cultivation, and high transformation efficiency. On the other hand, there are several limitations including the lack of post-translational modifications required for the heterologous expression of fungal BCGs and the inability to recognize the fungal promoter. Therefore, for successful heterologous expression in E. coli, it is important to consider in depth the selection of cryptic fungal BGCs [48,49]. Trichobrasilenol, an unusual sesquiterpene alcohol synthesized by a sesquiterpene cyclase from Trichoderma sp., was isolated from E. coli as a host for heterologous expression [50]. This novel compound was purified by utilizing silica column chromatography, and its absolute chemical structure was elucidated by the analysis of the NMR data. Trichobrasilenol is converted by sesquiterpene cyclase to farnesyl diphosphate via skeleton rearrangement. During the conversion, an additional methyl group is attached to trichobrasilenol. Although the function of this product has not been identified, it suggests that E. coli as the host organism for the heterologous expression of fungal metabolites would be useful [50,51].
Yeasts have been widely used in research and industrial biotechnology. Among various yeast strains, S. cerevisiae is mainly utilized for the heterologous expression of fungal BGCs. The advantage is that yeast species produce very few endogenous metabolites, which suggests that it is easy and simple to identify and purify the targeted metabolites. Recently, a heterologous expression platform (HEx) was developed that introduced S. cerevisiae as a heterologous host for the expression of fungal BGCs [52]. HEx was applied to 41 fungal BGCs of various fungal species around the world, of which 22 compounds were found to be detectable. In the HEx, the heterologous expression of a fungal BGC in the engineered host strain was performed using the adh2-like promoter of yeast as the HEx promoter. The 41 BGCs for the application of HEx were selected by developing a computational pipeline that prioritizes cryptic BGCs, including Ubia-like terpene cyclases (UTCs) or a PKS enzyme. The compounds were extracted with acetone and purified using a flash chromatography system with a gradient of hexane and acetone and semi-preparative HPLC using C18 reverse-phase column chromatography. Then, the chemical structures of isolated compounds were determined using liquid chromatography–mass spectrometry (LC-MS) and NMR experiments.
With advances in fungal genomics and related technologies, fungal hosts have been increasingly used for the production of homogeneous and heterogeneous genetic products in recent years. The main fungal host used is the Aspergillus genus, which reproduces by asexual reproduction, and because it has been used in brewing and food manufacturing since ancient times, it is considered safe as a production host for heterogeneous proteins [53]. Among the members of the genus, A. nidulans and A. oryzae have been mainly utilized for the heterologous expression of fungal secondary metabolites. Using filamentous fungi as a host led to the expression of fungal BGCs without removing introns in advance. There is also the benefit of not having to perform the post-translational modification. On the other hand, precursor competition between endogenous and heterogeneous pathways can interfere with the detection of the target compound [53].
Recently, A. nidulans was used as a heterologous host to discover flavunoidine, a new biosynthetic compound from the flv cluster of Aspergillus flavus [53]. The acetone extract of A. nidulans grown on 4 L of solid CD media was isolated by CombiFlash system (reverse-phase) and semi-preparative HPLC to afford flavunoidine. The flv cluster encodes two putative terpene cyclases (FlvE and FlvF) and the NRPS (FlvI). Flavunoidine was biosynthesized when the entire flv gene cluster was heterologously inserted into A. nidulans. In the cultivation of A. flavus under the general cultivation system, no production of flavunoidine was confirmed and no bioactivities, including cytotoxicity, antifungal activity, and antibacterial activity, were identified. Using A. oryzae as a heterologous host, new fungal compounds from the cricket-associated fungus Penicillium soppi were uncovered [54]. The new aliphatic polyketide soppiline A and the alkylresorcinols soppilines B and C were purified by using flash silica gel column chromatography and semi-preparative HPLC. Soppilines A–C contain a unique Z,E,Z-triene motif, which suggests unusual biosynthesis machinery in which a highly rehydrating PKS gene (PspA) is related to a unique biosynthetic mechanism. The highly rehydrating PKS gene (PspA) is related to positional and geometrical isomerization of double bonds during chain elongation cycles. In addition, PspB, a type III PKS, was also found to have a unique biosynthetic mechanism that intercepts the growing immature polyketide chain. This is the first study on soppiline biosynthesis to show polyketide chain transfer from PspA to PspB in fungi
4.2. CRISPR-Cas9
Recently, the CRISPR-Cas9 system has been applied for filamentous fungi to explore the production of fungal secondary metabolites (Figure 3). The CRISPR-Cas9 system consists of two components, namely Cas9 proteins and single guide RNA (sgRNA). The Cas9 protein is an important component of the CRISPR-Cas9 system that performs the endonuclease function and is approximately 1400 amino acids [55]. When the CRISPR-Cas9 system is utilized in fungi, the Cas9 protein is typically codon-optimized to match the fungal genome, which ensures efficient translation of the protein in the fungal cells. In addition, a nuclear localization signal is usually added to both ends of the Cas9 gene to facilitate its transport into the fungal nucleus, where it can carry out its gene-editing function. When the CRISPR-Cas9 system is utilized in fungi, genomes encoding the Cas9 protein are usually fungal codon-optimized, and a nuclear localization signal is added at both ends of the Cas9 gene [56,57,58].
The sgRNA used in the CRISPR-Cas9 system consists of two main components: the targeting sequence and the scaffold structure. The targeting sequence is a short RNA sequence, typically approximately 20 nucleotides in length, that is complementary to a specific region of the DNA target site [59]. This targeting sequence is designed to be specific to the target DNA sequence of interest so that it can direct the Cas9 protein to the desired location in the genome for gene editing. The scaffold structure, on the other hand, is a longer RNA sequence, typically approximately 80 nucleotides in length, that is necessary for binding to the Cas9 protein [57,58]. The scaffold structure contains sequences that are recognized by the Cas9 protein and enables it to bind to the sgRNA and form a complex that can recognize and cleave the target DNA sequence. By designing sgRNAs that are specific to the target DNA sequence and optimized for binding to the Cas9 protein, researchers can achieve highly precise and efficient gene editing using the CRISPR-Cas9 system.
CRISPR-Cas9-based genome editing in Glarea lozoyensis produces pneumocandin B0 [60]. Pneumocandins are a group of lipohexapeptides that belong to the echinocandin family of antifungal drugs and have known for their potent inhibition of fungal cell wall formation, which renders them effective against a broad range of fungal pathogens. In particular, pneumocandin B0 is a precursor to caspofungin, which is a potent antifungal drug used to treat invasive fungal infections. The resulting compound, caspofungin, is highly effective against a broad range of fungal pathogens and is generally well tolerated by patients. By utilizing a CRISPR-Cas9-based gene editing approach, significant enhancement of the accumulation of pneumocandin B0 in G. lozoyensis SIPI1208 was confirmed. The scalability of the gene editing approach was demonstrated by performing the same modifications in a large-scale industrial fermentation process, and the genetically modified strain produced higher yields of pneumocandin B0 than the wild-type strain, indicating the potential of this approach for commercial production of fungal secondary products.
CRISPR-mediated transcriptional activation (CRISPRa) is a technique that allows for the targeted activation of specific genes in cells, including those involved in the biosynthesis of secondary metabolites in fungi. This approach allows for the rapid screening of multiple gene clusters in parallel and can help to identify new compounds that may have therapeutic potential. Utilizing the CRISPRa system, the micA gene was selected for the biosynthesis of the natural product microperfuranone in the fungus A. nidulans [61]. To achieve strong activation of the micA gene, multiple CRISPR RNAs (crRNAs) targeting different regions of the micA promoter were designed. These crRNAs were expressed along with the dCas9-VP64 transcriptional activator, and the production of microperfuranone was monitored as a measure of gene activation. The results of the analysis showed that the accumulation of microperfuranone was significantly enhanced in the CRISPRa transformants compared to the control strains. Additionally, multigene CRISPRa was used to identify a new fungal secondary metabolite, dehydromicroperfuranone. Multigene CRISPRa refers to a technique that allows the simultaneous activation of multiple genes using the CRISPR-Cas9 system. For the purification of dehydromicroperfuranone from liquid culture media of genetically modified A. nidulans, Diaion HP-20, Sephadex LH-20 column, and flash chromatography system using a C18 preparative column were utilized. The CRISPR-Cas9 system has advanced genetic engineering and has already been applied to make significant advances in various fields, including medicine and agriculture. By using this technology to activate the expression of BGCs, researchers could potentially increase the production of these compounds and identify new ones from fungi.
4.3. Reporter-Guided Mutant Selection (RGMS)
The RGMS method is a genetic engineering technique used to create overproducing strains of secondary metabolites of microorganisms by inducing silent genetic clusters [62,63]. The mutagenesis step in the RGMS method involves inducing random mutations throughout the genome of the microorganism. This is done to create genetic diversity and increase the likelihood of identifying mutants that have increased expression of the silent genetic cluster. By applying a reporter gene to monitor the expression of the silent genetic cluster, it is easy to identify mutants with increased expression of the cluster and select the desired phenotype without complicated screening techniques (Figure 3).
UV mutagenesis and transposon mutagenesis were utilized in the RGMS method to obtain mutated strains [64]. UV mutagenesis involves exposing the microorganism to UV radiation, which can cause random mutations throughout the genome. This can lead to an increase in genetic diversity, which can increase the likelihood of identifying mutants with the desired phenotype. Transposon mutagenesis involves the use of transposons, which are DNA sequences that can move around the genome of the microorganism. By inserting a transposon into the genome, it is possible to disrupt the function of a specific gene or regulatory element, which can lead to changes in the production of secondary metabolites from the microorganism [65].
RGMS was developed for the first time to increase the production of lovastatin in Aspergillus terreus, a fungus that naturally produces this cholesterol-lowering drug [66]. Lovastatin is a potent inhibitor of hydroxymethylglutaryl coenzyme A (HMG-CoA) reductase, which is an enzyme involved in the synthesis of cholesterol in the liver. Inhibition of HMG-CoA reductase by lovastatin leads to a reduction in the production of cholesterol, which can lower blood cholesterol levels and reduce the risk of cardiovascular disease. The lovF gene is essential in the biosynthesis of lovastatin, and its promoter is therefore related to the production of the compound. By fusing the lovF promoter with the ble gene which encodes a protein with phleomycin resistance as the reporter gene, plasmid p3473 of lovF-ble was constructed and transformed into the wild-type A. terreus. After the plasmid was introduced into the fungus, it allowed for easy screening of mutants with increased expression of the lovastatin BGC by measuring the level of phleomycin resistance in UV irradiation. Mutants with the highest level of phleomycin resistance were then further characterized for their lovastatin production. The application of the lovF promoter-ble reporter gene construct was a key component of the RGMS method to increase lovastatin production, as it allowed for the easy identification of mutants with increased expression of the lovastatin BGC.
4.4. Promoter Insertion
If the BGCs of fungi remain silent in laboratory conditions, it is impossible to isolate secondary metabolites. Another genetic approach to solve this problem is promoter insertion technology (Figure 3). A promoter is a regulatory element in DNA sequences that is responsible for initiating transcription, which is the process by which genetic information is copied from DNA into RNA. The method of functional promoter insertion is an important technique for regulating gene expression and optimizing metabolite biosynthesis in microorganisms [67,68]. Promoter insertion has several advantages over other approaches for inducing gene expression, such as random mutagenesis or deletion of regulatory elements. By inserting a powerful exogenous promoter upstream of the BGC, it is feasible to control the level and timing of gene expression, allowing for more efficient biosynthesis of the target metabolite. Moreover, the use of functional promoter insertion can help to overcome the limitations of endogenous regulatory elements, which may not be optimized for high-level expression of the target gene cluster, and increase the expression of the BGC [69]. Among the functional promoters, glyceraldehyde-3-phosphate dehydrogenase gene promoter (PgpdA) of A. nidulans is a well-characterized promoter and is commonly used in genetic engineering and synthetic biology applications.
Fumiganins A and B are novel cyclic tetrapeptides that were identified by activating the nsc NRPS gene through the promoter insertion method [70]. The nsc NRPS is a silent BGC gene of A. fumigatus associated with resistance to oxidative stress and biosynthesis of cyclic tetrapeptides. The expression intensities of PgpdA, a well-known constitutional promoter, and PzipA and PsltA of A. nidulans were compared. The metabolomic study showed that PzipA and PsltA have strong promoter activity for the production of fumiganins A and B. In addition, PzipA was inserted in the silent nsc BGC in Neosartorya fischeri known to produce neosartoricins, which led to the production of neosartoricins with PzipA 1.46-fold higher compared to that with PgpdA. To isolate fumiganins A and B, the culture media of A. nidulans were extracted with ethyl acetate, dichloromethane, and methanol and purified by preparative HPLC. FT-IR spectrophotometry and NMR spectroscopy were used for structural identification of the cyclic tetrapeptides.
The biosynthetic pathway of sterigmatocystin toxin and at least six important secondary metabolites were activated by replacing the promoter of llmG, a gene encoding LaeA-like putative methyl transferases in A. nidulans, with a stronger promoter [71]. When the gene encoding McrA, a transcription factor that negatively regulates fungal secondary metabolites, was removed, the llmG gene as a master positive regulator was upregulated. Furthermore, by replacing the promoter of llmG with a stronger constitutive PgpdA promoter, the production of fungal secondary metabolites was increased considerably. The upregulation of LlmG by two combination methods resulted in a 460-fold improved production of various secondary metabolites including sterigmatocystin, terrequinone A, nidulanin A, cichorine, emodin/monodictyphenone, and prenyl xanthone compared to the control strain. For the extraction and purification of the compounds, extraction with EtOAc, silica gel chromatography with EtOAc and hexanes, and preparative HPLC were utilized. Promoter insertion is a powerful tool for activating silent BGCs and discovering novel secondary metabolites with potential therapeutic or industrial applications. However, it is important to carefully consider the potential risks associated with the overproduction of toxic metabolites or the unintended activation of unwanted BGCs. Therefore, it is essential to apply a comprehensive approach that integrates genetic engineering, transcriptomics, proteomics, and metabolomics to understand the complex regulatory networks that control secondary metabolism in fungi and to develop safe and effective strategies for activating silent BGCs.
4.5. Ribosome Engineering
Ribosomes are large molecular machines composed of RNA and protein molecules that work together to translate mRNA into proteins. Random mutations in RNA polymerase or ribosomes indeed occur naturally or are induced by mutagenesis, which can result in mutants with altered properties, such as higher yields of desired secondary metabolites (Figure 3). Ribosome engineering has emerged as an effective approach to improve fungal secondary metabolite production and even to enable the production of entirely new compounds [72,73]. This approach involves targeted modifications of ribosomal genes or their expression to enhance the translation efficiency of BGCs. The workflow of ribosome engineering consists of spontaneous mutagenesis using antibiotics targeting ribosomes, screening of antibiotic-resistant mutants, and selection of mutants with higher yields and/or structurally novelty of secondary metabolites. Ribosome engineering has several advantages over traditional methods for improving secondary metabolite production, including speed, cost-effectiveness, and the ability to generate large libraries of mutants with minimal effort [74]. Additionally, ribosome engineering can be combined with other approaches such as mutagenesis or promoter engineering to further enhance secondary metabolite production.
Janthinone, fructigenine A, aspterric acid methyl ester, and citrinin were isolated from a mutant marine-derived Penicillium purpurogenum G59 through ribosome engineering [75]. The gentamicin used in this study is an aminoglycoside antibiotic targeting ribosomes and is used for ribosome engineering for antibiotic production in bacteria. However, in fungi, the permeability of the fungal membrane is low, which can lower the intracellular concentration of gentamycin. To solve this problem, dimethyl-sulfoxide (DMSO) was used to improve cell membrane permeability. Newly produced secondary metabolites were isolated by TLC, HPLC, and VLC from EtOAc extracts of gentamicin-resistant mutants generated by the DMSO-mediated method, and their chemical structures were elucidated by analyzing their NMR spectroscopic data. The four newly isolated compounds showed antitumor activity displaying the inhibition of the proliferation of human cancer K562 cells, with inhibition rates of 34.6%, 60.8%, 31.7%, and 67.1% at 100 μg/mL, respectively. In addition, the compounds have not been found in any P. purpurogenum strains, indicating that the introduction of gentamicin resistance results in the production of new dormant secondary metabolites by inducing cryptic BGCs. In the subsequent study, the DMSO-mediated method using neomycin was applied to the same P. purpurogenum G59 in place of gentamycin [76]. Five secondary metabolites, curvularin, citrinin, penicitrinone A, erithro-23-O-methylneoclocitrinol, and 22E-7α-methoxy-5α,6α-epoxyergosta-8,22-dien-3β-ol, were isolated, and their cytotoxic effects on K562 cells were confirmed. All isolated compounds have not been found in other wild-type P. purpurogenum strains, indicating the expression of silent fungal BGCs by ribosome engineering.
Deep-sea fungus and neomycin-resistant Aspergillus versicolor ZBY-3 was shown to activate silent fungus BGCs to newly produce secondary metabolites [77]. Unlike previous studies that used the DMSO-mediated method to increase membrane permeability, this study applied an ultrasound-mediated method to induce a transient membrane permeability improvement. The ZBY-3 spores were treated with high concentrations of neomycin with ultrasound irradiation, resulting in mutations. HPLC–photodiode array detector–UV and HPLC–electron spray ionization–MS analyses were used to obtain six antitumor compounds, cyclo(D-Pro-D-Phe), cyclo(D-Tyr-D-Pro), phenethyl 5-oxo-L-prolinate, cyclo(L-Ile-L-Pro), cyclo(L-Leu-L-Pro), and 3β,5α,9α-trihydroxy-(22E,24R)-ergosta-7,22-dien-6-one. Ribosome engineering has revolutionized the field of natural product exploration, enabling researchers to quickly and efficiently identify new compounds with potential therapeutic or industrial applications.
4.6. Global Transcriptional Regulator
Global transcriptional regulators are proteins that control the expression of many genes across the genome. They play a critical role in regulating various cellular processes, such as metabolism, stress response, and development, by coordinating the expression of multiple genes involved in these processes. Some examples of global transcriptional regulators include cAMP receptor protein (CRP) in bacteria and nuclear factor-kappa B (NF-κB) and activator protein 1 (AP-1) in eukaryotes [78,79,80]. These proteins can bind to specific DNA sequences, often located in the promoter regions of target genes, and activate or repress their transcription. By regulating the expression of multiple genes in a coordinated manner, global transcriptional regulators make organisms respond to complex environmental cues and optimize their survival and growth under different conditions. Global transcriptional regulators play an important role in conveying environmental signals to regulate fungal secondary metabolism. For example, in response to changes in pH or carbon source, the transcription factor PacC can be activated to regulate the expression of genes involved in secondary metabolism in Aspergillus species [81,82]. Genetic manipulation of global regulators significantly affects fungal growth and the production of secondary metabolites [83]. Altering the expression or activity of these regulators results in abnormal growth and development or causes drastic changes in the metabolomics of fungi. This makes global regulators attractive targets for manipulating fungal secondary metabolism to uncover cryptic fungal metabolites (Figure 3).
The LaeA protein was first identified as a global regulator of secondary metabolism in A nidulans [84]. It was shown to affect the biosynthesis of multiple secondary metabolites in this fungus. The exact mechanism by which LaeA controls processes in fungi is still not fully understood [85]. However, it is believed that LaeA may function by regulating the expression of other transcription factors and chromatin remodeling enzymes, as well as through direct binding to DNA and chromatin [86,87]. It is also possible that LaeA may interact with other signaling pathways to coordinate fungal secondary metabolism with other cellular processes. Overexpression of the LaeA gene has been shown to enhance the production level of various secondary metabolites in different fungal species, including a new cytochalasin (chaetoglobosin Z) in Chaetomium globosum, sorbicilinoids in Penicillium dipodomyis, terrequinone A in A. nidulans, and cyclopiazonic acid in A. fumisynnematus [88,89,90,91]. Additionally, LaeA homologs have been identified in many other fungi and have been shown to have similar roles in regulating fungal secondary metabolism [92].
Talae1 is involved in the regulation of fungal secondary metabolism, and overexpression of Talae1 in Trichoderma afroharzianum led to the production of two structurally new polyketides, (R,3E,5E)-1-(3,5-dihydroxy-2,4-dimethylphenyl)-1-hydroxyhepta-3,5-dien-2-one and (R,3E,5E)-1-(3,5-dihydroxy-2,4-dimethylphenyl)-1-methoxyhepta-3,5-dien2-one [93]. Two polyketides were isolated from the EtOAc extract of the overexpression transformant by utilizing silica gel column chromatography, MPLC, and semi-preparative HPLC. Their chemical structures were elucidated by the analysis of high-resolution mass spectrometry (HRMS), NMR, and electronic circular dichroism (ECD) calculations. Genetic manipulation of global regulators has been shown to be a promising approach for activating new secondary metabolites and improving the metabolic potential of fungi.
5. Genetics-Independent Approach
5.1. OSMAC Approach
The one strain many compounds (OSMAC) framework was developed by Zeeck in the early 2000s as a way to explain the principles behind cultivation-based techniques for the discovery of new natural products [94]. The concept is based on the idea that the metabolic pathways of microorganisms can be influenced by changing the cultivation conditions, such as altering the nutrient availability or growing the organism under different environmental conditions. The culture medium plays a critical role in microbial growth and metabolism, as it provides the nutrients and environmental conditions for the microorganism to grow and produce metabolites. The composition of the culture medium has a significant impact on the metabolism of microorganisms and the production of natural products (Figure 4). The availability and concentration of nutrients, such as carbon, nitrogen, and minerals, can affect the biosynthesis of secondary metabolites [95,96]. In addition to nutrient composition, other factors such as pH, light or darkness, temperature, and aeration can also impact microbial metabolism and natural product production. For example, some microorganisms produce different types of metabolites at different temperatures or pH levels.
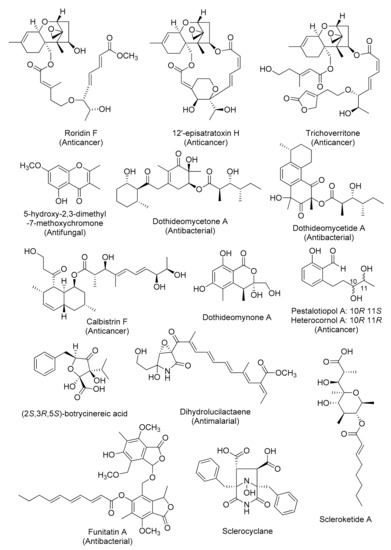
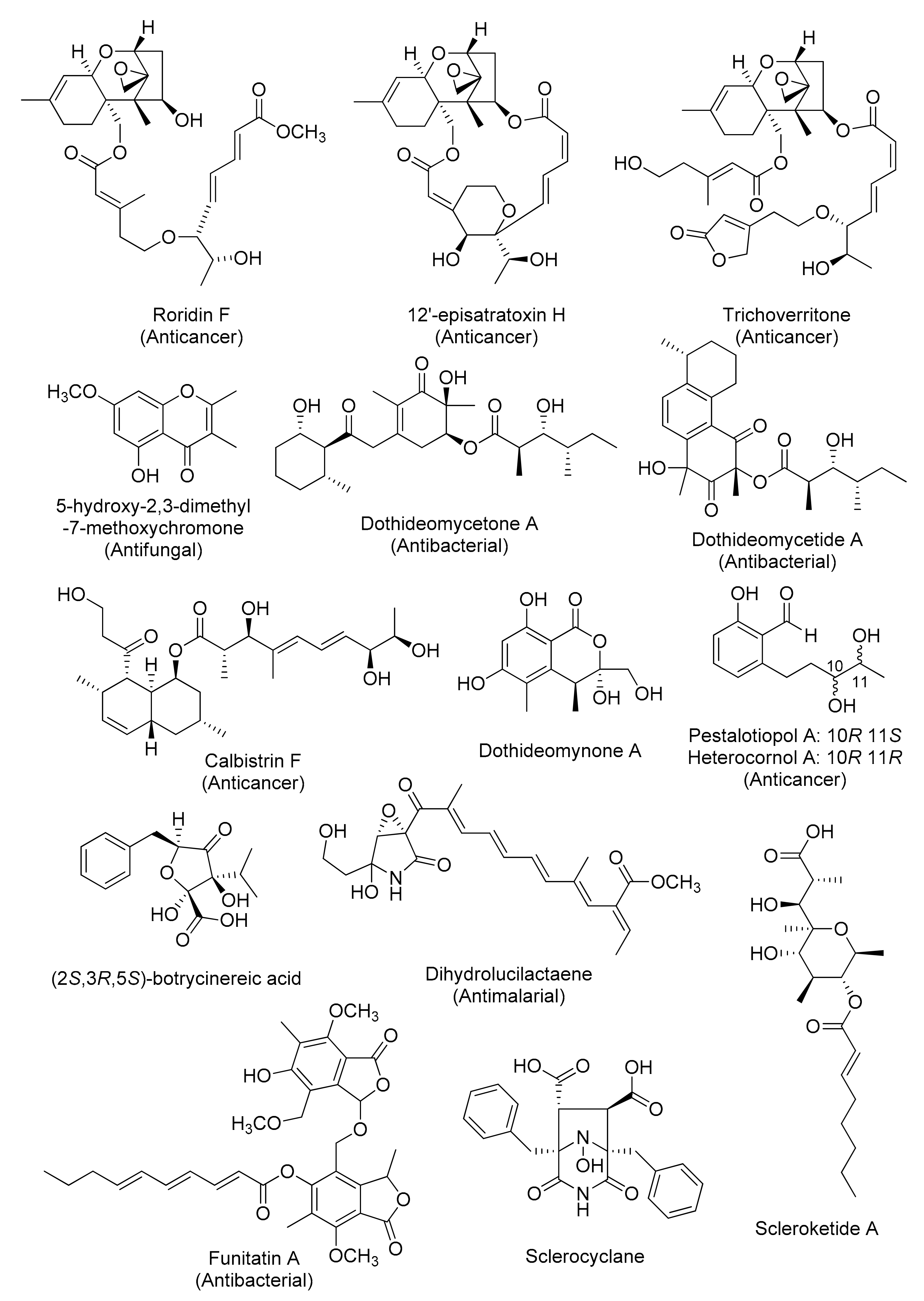
Figure 4.
Representative chemical structures of cryptic fungal metabolites obtained by genetics-independent approach.
Comparable differences in fungal metabolites were observed in the chemical investigation of Podostroma cornu-damae [97]. Macrocyclic trichothecenes were not detected in the LC/MS analysis of a MeOH extract of P. cornu-damae grown on liquid culture media. Fortunately, several macrocyclic trichothecenes were observed from a MeOH extract of P. cornu-damae grown on potato dextrose agar. Extensive chromatographic purifications including preparative and semi-preparative reverse-phase HPLC led to the isolation of eight macrocyclic trichothecenes. All identified macrocyclic trichothecenes were assessed for cytotoxicity against four human breast cancer cell lines (Bt549, HCC70, MDA-MB-231, and MDA-MB-468), and roridin E, 12′-episatratoxin H, and trichoverritone displayed significant cytotoxic activities against the breast cancer cell lines, with IC50 values in the range of 0.02–80 nM.
By employing the OSMAC strategy to search for new fungal metabolites, various liquid media (potato dextrose broth (PDB), 1/10 diluted PDB, yeast peptone glucose media, and chemically defined media) and different cultivation systems (light or dark, static or shaking) were employed to evaluate the metabolome of Trichoderma harzianum M10 [98]. A bioactive derivative of chromone, 5-hydroxy-2,3-dimethyl-7-methoxychromone, was identified when the fungal strain was grown under specific conditions using potato dextrose broth and subjected to light and shaking. The compound was isolated as a transparent crystal from ethyl acetate extract of T. harzianum by using silica gel column chromatography, and its chemical structure was characterized by X-ray diffraction, mass spectrometry, IR, and NMR. It exhibited a significant inhibitory effect against Rhizoctonia solani (showing 45% growth inhibition after 24 h of incubation at a concentration of 100 ng plug−1) and demonstrated a concentration-dependent reduction in the viability of colorectal human cancer cells.
To discover diverse metabolites of Dothideomycete sp. CRI7, the OSMAC approach was applied by cultivating CRI7 under different media conditions [99]. Two azaphilone derivatives and one tricyclic polyketide (dothideomycetones A-B and dothideomycetide A) were identified from CRI7 grown on PDB made with potato tubers, and dothideomycetone C exhibited antibacterial effects against Staphylococcus aureus ATCC 25923 and ATCC 33591 (methicillin-resistant strain) with MIC values of 128 and 256 μg mL−1, respectively. The sensitivity of secondary metabolite production to the source of potato and malt extract used in the preparation of PDB and Czapek malt media suggests that the composition of the media plays a critical role in regulating the biosynthesis of secondary metabolites by CRI7. The fungal strain growing in PDB broth prepared from a commercial potato powder instead of fresh tubers of potato and Czapek malt medium yielded the identification of six new polyketides (calbistrins F–H and dothideomynone A–C), and Sephadex LH-20 and C18 reverse-phase HPLC were utilized for the purification of the six polyketides [99]. Calbistrin F showed weak cytotoxic activity against the MOLT-3 cell line (IC50 = 37.3 μg/mL), and calbistrin H displayed a radical scavenging effect (IC50 = 21.7 μM) as it repressed superoxide anion generation triggered by 12-O-tetradecanoylphorbol-13-acetate in differentiated HL-60 human promyelocytic leukemia cells.
5.2. Epigenetic Modification
Epigenetic modification, which involves the addition of chemicals that can alter gene expression to the culture medium, is another extension of the cultivation-dependent approach (Figure 4). Epigenetic modifications, such as DNA methylation and histone acetylation, play a role in regulating the expression of these BGCs. DNA methyl-transferase (DNMT) and histone deacetylase (HDAC) inhibitors have been shown to activate cryptic BGCs in various microorganisms by altering the epigenetic state of the genes responsible for natural product biosynthesis [100,101]. By utilizing these inhibitors to induce chromatin remodeling, it is possible to amplify the chemical diversity of natural products produced by microorganisms in the culture. DNMT inhibitors including 5-azacytidine and 5-aza-2′-deoxycytidine inhibit the activity of DNMT enzymes, which are responsible for adding methyl groups to DNA, leading to hypomethylation of genomic DNA. HDAC inhibitors such as trichostatin A and vorinostat inhibit the activity of HDAC enzymes, leading to an increase in histone acetylation and changes in gene expression [102]. The BGCs responsible for the biosynthesis of secondary metabolites are often found in regions of the genome that are transcriptionally silent or in a heterochromatin state and possibly regulated by the transcription of constitutive genes through epigenetic modification. Small-molecule epigenetic modulators have been utilized to control the expression of previously silent genes, including those involved in the biosynthesis of secondary metabolites.
Chemical modifiers of DNA methyltransferase (DNMT), including 5-azacytidine (5-AZA), 5-aza-2′-deoxycytidine (decitabine), hydralazine hydrochloride, N-acetyl-D-glucosamine (GlcNAc), procainamide, procaine, and N-phthalyl-L-tryptophan (RG-108), have been reported to induce the fungal BGCs [103]. Among them, 5-AZA is the most commonly employed DNMT inhibitor for reactivating the silenced genes through DNMT-induced methylation. 5-AZA is a cytosine analog that can be incorporated into DNA during replication, where it forms covalent bonds with DNMT, trapping the enzyme on the DNA and leading to its degradation. This results in the depletion of DNMT activity and DNA hypomethylation, enabling the reactivation of cryptic fungal BGCs. Four new polyketide derivatives, pestalotiopols A–D, were purified from Pestalotiopsis sp. cultures treated with two DNA methyltransferase modifiers, 5-aza-2′-deoxycytidine and RG-108 [104]. The EtOAc-soluble crude extract was isolated by silica gel column (Sephadex LH-20) chromatography and semi-preparative HPLC, and then its chemical structures were characterized through extensive spectroscopic analysis. Pestalotiopols A-B, and heterocornols A and E exhibited cytotoxic activity against four human cancer cell lines, namely BGC-823, SMMC-7721, Ichikawa, and 7860, with IC50 values ranging from 16.5 to 56.5 mM.
HDACs are a class of enzymes that remove acetyl groups from the lysine residues of histone and non-histone proteins [105]. The acetylation of histones is a key mechanism in the regulation of gene transcription, and HDACs play an important role in the regulation of chromatin structure and gene expression. When HDACs eliminate acetyl groups from histones, the positively charged lysine residues become more positively charged, which results in a tighter interaction between histones and DNA. This tighter interaction makes it more difficult for the transcription machinery to access and transcribe the genes located in that chromatin region, resulting in the repression of gene expression. Inhibition of HDAC activity leads to an increase in histone acetylation, which loosens the interaction between histones and DNA, allowing for greater accessibility of the transcription machinery to the genes. Suberoyl bishydroxamic acid (SBHA), suberoylanilide hydroxamic acid (SAHA), and nicotinamide are among the most commonly used HDAC inhibitors in microorganisms [106].
Botrycinereic acid is a cryptic secondary metabolite that was identified from Botrytis cinerea strain B05.10 through the treatment of SAHA, a potent HDAC inhibitor [107]. For the isolation of botrycinereic acid, silica gel column chromatography, preparative TLC, and normal phase HPLC were applied to the EtOAc-soluble extract of B05.10. In addition, overexpression of botrycinereic acid was achieved by inactivating the stc2 gene, which encodes an unidentified sesquiterpene cyclase. The fungus Talaromyces funiculosus HPU-Y01, which was isolated from the Yellow River wetland, was treated with SAHA at a concentration of 300 μM [108]. This epigenetic manipulation led to the production of a novel compound, a highly modified fatty acid ester designated funitatin A which contains a rare dimeric cyclopaldic acid structure motif. Funitatin A was purified from the EtOAc extract of the culture supernatant by using medium-pressure liquid chromatography (MPLC) and semi-preparative HPLC. Funitatin A displayed potential antimicrobial activity against Proteus species and E coli, with MIC values of 3.13 μM.
Other chemical epigenetic modifiers have been reported to be effective for inducing cryptic BGCs in fungi. Anacardic acid, BRD4770, bortezomib, and NPD938 are among the chemical epigenetic modifiers that have been successfully used in the screening of fungal secondary metabolism [100]. They target histone acetyltransferase, a histone methyltransferase, the proteasome, and unidentified mechanisms, respectively. The production of three lucilactaene analogs, dihydroNG391, dihydrolucilactaene, and 13α-hydroxylucilactaene, was induced in cultures of Fusarium sp. RK97-94 treated with NPD938 at 30 μM [109]. Silica open column chromatography, MPLC, and preparative HPLC were utilized for the purification of the above three fungal compounds. DihydroNG391 showed weak in vitro antimalarial activity (with an IC50 value of 62 μM). In contrast, dihydrolucilactaene and 13α-hydroxylucilactaene exhibited potent antimalarial activity (with IC50 values of 0.0015 and 0.68 μM, respectively) against Plasmodium falciparum. Chemical epigenetic modifiers allow for the induction of cryptic BGCs without prior knowledge of the target genome features. This approach is particularly useful in the discovery of cryptic secondary metabolites since it can lead to the production of compounds that would not have been obtained through traditional cultivation methods. In addition, the technique is relatively low-cost and easy to apply in high-throughput screens, making it a powerful tool for the identification of novel fungal compounds.
5.3. High-Throughput Elicitor Screening (HiTES)
Microorganisms produce secondary metabolites to communicate with each other and compete for resources in their environment. These compounds act as signaling small molecules that allow microorganisms to coordinate their activities, or as weapons that help them defend against other microbes. Recent studies have reported that secondary metabolites produced by microorganisms induce the expression of silent BGCs originating from other microbes, leading to the discovery of new metabolites. This approach is known as “inducing silent BGCs” and involves exposing microorganisms to chemical signals or environmental cues that trigger the expression of silent BGCs.
Recently, a method known as HiTES was developed to utilize this approach, which identifies the signals required to elicit silent BGCs [110,111,112]. In the HiTES method, microorganisms are exposed to a large library of potential elicitors, and the resulting induction of silent BGCs is monitored using various methods, such as genetic reporters, bioactivity assays, or mass spectrometry-based detection [110,111,112]. Genetic reporters are utilized to monitor the expression of BGCs in response to the elicitors. These reporters are typically fused to a promoter region that is specific to a particular silent BGC, and their expression is monitored by fluorescence or luminescence. By monitoring the expression of these reporters, it is possible to confirm which elicitors are effective in inducing the expression of specific silent BGCs. Various bioactivity assays were also applied to screen the biosynthesis of novel metabolites in response to the elicitors. These assays involve testing the extracts of the microorganisms for specific biological activities, such as antimicrobial or anticancer activity. The appearance of new bioactivity in the extracts indicates the presence of novel bioactive metabolites. Mass spectrometry-based detection was another method used to monitor the production of novel metabolites. This approach involved analyzing the metabolomics of the microorganisms using mass spectrometry, which allows for the characterization of new compounds based on their mass and fragmentation patterns.
In a recent study, HiTES coupled with MS-based read-outs was applied to offer an attractive means of inducing cryptic fungal metabolomes, since genetic manipulation of fungal strains is often challenging [113]. The HiTES approach has previously been studied only in bacteria, and the present study represents the first investigation of HiTES in fungi. Two fungal pathogens, Sclerotinia sclerotiorum and Rhizoctonia solani, were subjected to a chemical library of FDA-approved drugs as candidate elicitors. For the optimal growth of these fungal strains, HiTES was developed to perform the entire screening on agar in a 96-well microtiter format. The biosynthesis of cryptic fungal metabolites was monitored using multiplexed UPLC-Qof-MS, and the resulting metabolome was represented in a 3D plot that links each elicitor to metabolites, characterized by their m/z and MS intensity values. Agar-based HiTES on S. sclerotiorum and R. solani enabled the discovery of 13 novel natural products in 4 compound groups, including an alkaloid (sclerocyclane), modified fatty acids (tetrasclerols), polyketides (scleroketides), and siderophores (solanibactins). All identified compounds were purified by using an open silica column chromatography and preparative and semi-preparative HPLC, and their absolute structures were elucidated based on the analysis of NMR spectroscopy, ECD calculation, and organic reactions. As a result, the rapid and broad application of the HiTES approach to any microbe that can be cultured and the successful stimulation of cryptic secondary metabolites in a high-throughput manner were verified (Figure 4).
6. Techniques for the Extraction, Purification, and Identification of Secondary Metabolites
After a BGC has been expressed using one of the approaches described above, the next step is to detect any changes in the metabolite profile that may indicate the production of a new compound (Figure 2). This is typically achieved through metabolomics, which is the systematic study of small molecules, or metabolites, present in a biological system [114]. HPLC is a powerful analytical technique commonly used in metabolomics to separate and identify individual metabolites within a complex mixture. It is especially useful for the separation of non-volatile and polar metabolites, which cannot be effectively separated by gas chromatography (GC). In addition, HPLC is a highly reproducible and reliable method that can provide high-resolution separation of complex mixtures of metabolites. HPLC separates molecules based on their physical and chemical properties such as size, charge, polarity, and hydrophobicity, and it can be coupled with various detection methods such as UV-visible spectrophotometry, fluorescence spectroscopy, and mass spectrometry to identify and quantify individual metabolites [115]. In particular, HRMS is a powerful technique used in metabolomics for the identification and quantification of individual secondary metabolites in natural product discovery [115,116]. HRMS provides accurate and precise measurements of the mass-to-charge ratio (m/z) of metabolites, with high resolving power and mass accuracy, which afford the information of the molecular formula of a targeted compound. Knowing the molecular formula of a compound offers an initial assessment of the likelihood of a novel chemical structure. Many natural products have been isolated and characterized, and their chemical structures and molecular formulas are available in public databases such as PubChem, ChemSpider, and Metlin [117]. By comparing the molecular formula of unknown compounds with the molecular formulas of known natural products, it may be possible to identify the class or family of natural products to which the unknown metabolite belongs before starting the extraction and isolation steps. This information guides further analysis and characterization of the targeted compounds.
Extraction is an important process for acquiring microbial compounds from cultures and involves the separation of the target compounds from the culture medium and other unwanted components. The extraction method depends on the chemical and physical properties of the target compounds and the nature of the culture medium [118]. In organic solvent extraction, an organic solvent such as methanol, ethyl acetate, or chloroform is utilized to extract the target compounds from the culture medium. It is necessary to consider the polarity of targeted compounds in the culture medium to select an organic solvent. For example, polar metabolites such as RiPPs, aminoglycosides, and alkaloids are not extracted from the culture medium efficiently. Another method for extraction is solid-phase extraction, which involves the use of a solid-phase material such as a resin or silica gel to selectively adsorb the target compounds from the culture medium. The culture medium is passed through a column containing the solid-phase material, and the target compounds are retained on the material while unwanted components are washed away. The target compounds are subsequently eluted from the column with a solvent and collected for further analysis.
Open-column chromatography is a common technique used in the isolation and purification of natural products from complex mixtures. It is a type of liquid chromatography that uses a column packed with a stationary phase and a mobile phase to separate and purify the target compounds based on their chemical and physical properties. The sample is dissolved in a suitable solvent and applied to the top of the column. The column is then eluted with a solvent system that is carefully selected based on the polarity of the target compounds. The mobile phase gradually moves down the column, and the target compounds are selectively adsorbed and retained on the stationary phase based on their polarity, size, and other properties. The usage of open-column chromatography is an iterative process where fractions are collected at specific time intervals or absorbance readings, and each fraction is analyzed for the presence of the target compounds using various analytical techniques such as thin-layer chromatography (TLC) or mass spectrometry. However, it can be time-consuming and may require multiple rounds of purification to achieve the desired purity and yield. To overcome this, other chromatographic techniques such as flash chromatography or preparative HPLC can be used for larger-scale purification of natural products [118].
Semi-preparative high-performance liquid chromatography (semi-HPLC) is typically utilized for the final purification step of natural products. In semi-HPLC, the stationary phase is typically a reverse-phase resin, and the mobile phase is a solvent or a mixture of solvents that are carefully selected based on the polarity and chemical properties of the target compounds [115,118]. The sample is dissolved in a suitable solvent and injected into the column, and the column is then eluted with a solvent gradient that is optimized to selectively separate the target compounds. Semi-HPLC is a powerful tool for the isolation and purification of natural products, particularly those that are present in low concentrations or are difficult to separate by other techniques from the culture medium.
After the targeted compounds were isolated by semi-HPLC, they are analyzed by various analytical techniques, such as NMR spectroscopy and mass spectrometry, to confirm their chemical structures (Figure 2). NMR spectroscopy is a powerful technique for the determination of the planar structure, relative configuration, and functional group assignments in natural products. It is particularly important for identifying and characterizing new and structurally complex natural products. Several types of NMR experiments can be used to analyze natural products, including one-dimensional (1D) and two-dimensional (2D) NMR experiments. Common 1D NMR experiments include proton NMR (1H NMR) and carbon NMR (13C NMR), which provide information on the number and type of hydrogen and carbon atoms in a molecule. Two-dimensional NMR experiments, such as correlation spectroscopy (COSY), heteronuclear single quantum coherence (HSQC), and heteronuclear multiple bond correlation (HMBC), provide information on the connectivity of atoms within a molecule and the relative orientation of functional groups.
Determination of the absolute configuration of natural products is a crucial step in understanding their biological activity and potential use as a drug [119]. Circular dichroism (CD) spectroscopy is a specifically designed technique for the determination of the absolute configuration of chiral molecules [119]. This method depends on the interactions between circularly polarized light and chiral molecules. Recently, computational ECD calculation that predicts the ECD spectra of chiral molecules has been widely applied for the characterization of the absolute configuration of natural products [120]. Vibrational circular dichroism (VCD) is a spectroscopic technique employed in the investigation of molecular chirality and structural characteristics, particularly in the case of natural products [121]. VCD relies on the detection of variances in the absorption of left- and right-circularly polarized infrared (IR) light by chiral molecules. The analysis of VCD spectra entails the utilization of theoretical calculations and a comparison with reference spectra. By comparing experimental VCD data with computational models, researchers are able to attribute specific vibrational modes to distinct regions of the molecule, thereby gaining valuable insights into its three-dimensional structure [122,123]. Several organic reactions can be used to determine the absolute configuration of natural products and involve the use of chiral reagents or catalysts to induce stereochemistry in the reaction products.
X-ray crystallography is a powerful technique used to determine the three-dimensional structure and is also useful for the determination of the absolute stereochemistry of natural products [124,125]. Crystals of the molecules are mounted on a goniometer and exposed to a beam of X-rays. The diffraction pattern produced by the X-rays is recorded and processed to create an electron density map, which can be used to determine the positions of the atoms in the molecule. Once the electron density map has been generated, the structure of the molecule can be refined using various computational tools and software. Similarly, micro-electron diffraction (Micro-ED) is a new technique for determining the crystal structure of small molecules, including natural products [126]. This method involves the collection of electron diffraction data from very small crystals, typically less than 1 micron in size. The advantage of Micro-ED is that it allows the determination of crystal structures from very small samples, which is often the case for natural products. It also allows the determination of crystal structures from samples that are difficult or impossible to crystallize using traditional methods.
7. Conclusions
Activating cryptic BGCs in fungi has become an important method for discovering new secondary metabolites that could have potential uses in biotechnology and pharmaceuticals. Despite the vast biosynthetic potential of fungi, many of their gene clusters remain silent or poorly expressed under typical laboratory conditions. While various strategies have been developed to activate cryptic fungal secondary metabolites, no single method has been universally accepted, as each approach has its own advantages and limitations. Therefore, the choice of the appropriate approach often depends on the specific research question and the characteristics of the fungal species being studied.
This review emphasizes the significance of activating cryptic fungal BGCs, as it enables the production of novel fungal metabolites that may have important biological and biotechnological applications. We discuss two main approaches for activating these gene clusters: genetics-dependent and genetics-independent. Genetics-dependent approaches are highly targeted and specific, but they require a deep understanding of the fungus’s genetic makeup and may have unintended effects on other biological processes. Genetics-independent approaches are simpler to implement and provide a more natural approach to activate gene clusters, but they can result in the production of complex mixtures of unintended compounds and require optimization of culture conditions. The choice of approach often depends on the specific research question and the characteristics of the fungal species being investigated.
Overall, a combination of both genetics-dependent and genetics-independent approaches may provide the best opportunities for the successful activation of cryptic BGCs and the discovery of novel fungal natural products. Future studies on the induction of cryptic BGCs in fungi are likely to continue to explore and develop a variety of methods and strategies. Advances in microbial ecology and understanding real microbial interactions in the environment may provide new insights into the environmental and ecological factors that influence the activation of cryptic fungal BGCs. Therefore, future studies are likely to be interdisciplinary, combining approaches from molecular biology, genomics, natural product chemistry, and microbial ecology to further advance our understanding and ability to activate cryptic fungal BGCs.
Author Contributions
Conceptualization, S.R.L.; formal analysis, J.Y.H., E.J. and Y.C.K.; investigation, J.Y.H., E.J. and Y.C.K.; writing—original draft preparation, J.Y.H., E.J., Y.C.K. and S.R.L.; writing—review and editing, S.R.L.; visualization, J.Y.H., E.J., Y.C.K. and S.R.L.; supervision, S.R.L.; project administration, S.R.L.; funding acquisition, S.R.L. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (2020R1A6A3A03037782).
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Newman, D.J.; Cragg, G.M. Natural products as sources of new drugs over the nearly four decades from 01/1981 to 09/2019. J. Nat. Prod. 2020, 83, 770–803. [Google Scholar] [CrossRef] [PubMed]
- Newman, D.J.; Cragg, G.M. Natural products as sources of new drugs from 1981 to 2014. J. Nat. Prod. 2016, 79, 629–661. [Google Scholar] [CrossRef] [PubMed]
- Helaly, S.E.; Thongbai, B.; Stadler, M. Diversity of biologically active secondary metabolites from endophytic and saprotrophic fungi of the ascomycete order Xylariales. Nat. Prod. Rep. 2018, 35, 992–1014. [Google Scholar] [CrossRef] [PubMed]
- Demain, A.L. Valuable secondary metabolites from fungi. In Biosynthesis and Molecular Genetics of Fungal Secondary Metabolites; Springer: New York, NY, USA, 2014; pp. 1–15. [Google Scholar]
- Bills, G.F.; Gloer, J.B. Biologically active secondary metabolites from the fungi. Microbiol. Spectr. 2017, 4, 1087–1119. [Google Scholar]
- Atanasov, A.G.; Zotchev, S.B.; Dirsch, V.M.; Supuran, C.T. Natural products in drug discovery: Advances and opportunities. Nat. Rev. Drug Discov. 2021, 20, 200–216. [Google Scholar] [CrossRef]
- Fischer, M.S.; Glass, N.L. Communicate and fuse: How filamentous fungi establish and maintain an interconnected mycelial network. Front. Microbiol. 2019, 10, 619. [Google Scholar] [CrossRef]
- Dias, D.A.; Urban, S.; Roessner, U. A historical overview of natural products in drug discovery. Metabolites 2012, 2, 303–336. [Google Scholar] [CrossRef]
- Robey, M.T.; Caesar, L.K.; Drott, M.T.; Keller, N.P.; Kelleher, N.L. An interpreted atlas of biosynthetic gene clusters from 1000 fungal genomes. Proc. Natl. Acad. Sci. USA 2021, 118, e2020230118. [Google Scholar] [CrossRef]
- Clevenger, K.D.; Bok, J.W.; Ye, R.; Miley, G.P.; Verdan, M.H.; Velk, T.; Chen, C.; Yang, K.; Robey, M.T.; Gao, P. A scalable platform to identify fungal secondary metabolites and their gene clusters. Nat. Chem. Biol. 2017, 13, 895–901. [Google Scholar] [CrossRef]
- Hoskisson, P.A.; Seipke, R.F. Cryptic or silent? The known unknowns, unknown knowns, and unknown unknowns of secondary metabolism. MBio 2020, 11, e02642-20. [Google Scholar] [CrossRef]
- Amos, G.C.; Awakawa, T.; Tuttle, R.N.; Letzel, A.-C.; Kim, M.C.; Kudo, Y.; Fenical, W.; Moore, B.S.; Jensen, P.R. Comparative transcriptomics as a guide to natural product discovery and biosynthetic gene cluster functionality. Proc. Natl. Acad. Sci. USA 2017, 114, E11121–E11130. [Google Scholar] [PubMed]
- Honda, S.; Eusebio-Cope, A.; Miyashita, S.; Yokoyama, A.; Aulia, A.; Shahi, S.; Kondo, H.; Suzuki, N. Establishment of Neurospora crassa as a model organism for fungal virology. Nat. Commun. 2020, 11, 5627. [Google Scholar]
- Keller, N.P. Fungal secondary metabolism: Regulation, function and drug discovery. Nat. Rev. Microbiol. 2019, 17, 167–180. [Google Scholar]
- Dunlap, J.C.; Borkovich, K.A.; Henn, M.R.; Turner, G.E.; Sachs, M.S.; Glass, N.L.; McCluskey, K.; Plamann, M.; Galagan, J.E.; Birren, B.W. Enabling a community to dissect an organism: Overview of the Neurospora functional genomics project. Adv. Genet. 2007, 57, 49–96. [Google Scholar]
- Liu, Z.; Lin, Z.; Nielsen, J. Expression of fungal biosynthetic gene clusters in S. cerevisiae for natural product discovery. Synth. Syst. Biotechnol. 2021, 6, 20–22. [Google Scholar] [CrossRef]
- Reen, F.J.; Romano, S.; Dobson, A.D.; O’Gara, F. The sound of silence: Activating silent biosynthetic gene clusters in marine microorganisms. Mar. Drugs 2015, 13, 4754–4783. [Google Scholar]
- Almeida, H.; Tsang, A.; Diallo, A.B. Improving candidate Biosynthetic Gene Clusters in fungi through reinforcement learning. Bioinformatics 2022, 38, 3984–3991. [Google Scholar] [CrossRef]
- Mózsik, L.; Iacovelli, R.; Bovenberg, R.A.; Driessen, A.J. Transcriptional activation of biosynthetic gene clusters in filamentous fungi. Front. Bioeng. Biotechnol. 2022, 10, 1199. [Google Scholar]
- Chiang, Y.-M.; Szewczyk, E.; Nayak, T.; Davidson, A.D.; Sanchez, J.F.; Lo, H.-C.; Ho, W.-Y.; Simityan, H.; Kuo, E.; Praseuth, A. Molecular genetic mining of the Aspergillus secondary metabolome: Discovery of the emericellamide biosynthetic pathway. Chem. Biol. 2008, 15, 527–532. [Google Scholar] [CrossRef]
- Drott, M.; Bastos, R.; Rokas, A.; Ries, L.; Gabaldón, T.; Goldman, G.; Keller, N.; Greco, C. Diversity of secondary metabolism in Aspergillus nidulans clinical isolates. Msphere 2020, 5, e00156-20. [Google Scholar] [CrossRef] [PubMed]
- Scherlach, K.; Hertweck, C. Mining and unearthing hidden biosynthetic potential. Nat Commun. 2021, 12, 3864. [Google Scholar]
- Machado, H.; Tuttle, R.N.; Jensen, P.R. Omics-based natural product discovery and the lexicon of genome mining. Curr. Opin. Microbiol. 2017, 39, 136–142. [Google Scholar]
- Kwon, M.J.; Steiniger, C.; Cairns, T.C.; Wisecaver, J.H.; Lind, A.L.; Pohl, C.; Regner, C.; Rokas, A.; Meyer, V. Beyond the biosynthetic gene cluster paradigm: Genome-wide coexpression networks connect clustered and unclustered transcription factors to secondary metabolic pathways. Microbiol. Spectr. 2021, 9, e00898-21. [Google Scholar] [CrossRef] [PubMed]
- Medema, M.H.; Cimermancic, P.; Sali, A.; Takano, E.; Fischbach, M.A. A systematic computational analysis of biosynthetic gene cluster evolution: Lessons for engineering biosynthesis. PLoS Comput. Biol. 2014, 10, e1004016. [Google Scholar] [CrossRef]
- Calvo, A.M.; Wilson, R.A.; Bok, J.W.; Keller, N.P. Relationship between secondary metabolism and fungal development. Microbiol. Mol. Biol. Rev. 2002, 66, 447–459. [Google Scholar]
- Manzoni, M.; Rollini, M. Biosynthesis and biotechnological production of statins by filamentous fungi and application of these cholesterol-lowering drugs. Appl. Microbiol. Biotechnol. 2002, 58, 555–564. [Google Scholar]
- Gluck-Thaler, E.; Haridas, S.; Binder, M.; Grigoriev, I.V.; Crous, P.W.; Spatafora, J.W.; Bushley, K.; Slot, J.C. The architecture of metabolism maximizes biosynthetic diversity in the largest class of fungi. Mol. Biol. Evol. 2020, 37, 2838–2856. [Google Scholar]
- Arnison, P.G.; Bibb, M.J.; Bierbaum, G.; Bowers, A.A.; Bugni, T.S.; Bulaj, G.; Camarero, J.A.; Campopiano, D.J.; Challis, G.L.; Clardy, J. Ribosomally synthesized and post-translationally modified peptide natural products: Overview and recommendations for a universal nomenclature. Nat. Prod. Rep. 2013, 30, 108–160. [Google Scholar]
- Le Govic, Y.; Papon, N.; Le Gal, S.; Bouchara, J.-P.; Vandeputte, P. Non-ribosomal peptide synthetase gene clusters in the human pathogenic fungus Scedosporium apiospermum. Front. Microbiol. 2019, 10, 2062. [Google Scholar] [CrossRef] [PubMed]
- Cox, R.J.; Simpson, T.J. Fungal type I polyketide synthases. Methods Enzymol. 2009, 459, 49–78. [Google Scholar] [PubMed]
- Palmer, J.M.; Keller, N.P. Secondary metabolism in fungi: Does chromosomal location matter? Curr. Opin. Microbiol. 2010, 13, 431–436. [Google Scholar] [CrossRef] [PubMed]
- Tamaru, H. Confining euchromatin/heterochromatin territory: Jumonji crosses the line. Genes Dev. 2010, 24, 1465–1478. [Google Scholar] [CrossRef] [PubMed]
- Lai, Y.; Wang, L.; Zheng, W.; Wang, S. Regulatory roles of histone modifications in filamentous fungal pathogens. J. Fungi 2022, 8, 565. [Google Scholar] [CrossRef] [PubMed]
- Tran, P.N.; Yen, M.-R.; Chiang, C.-Y.; Lin, H.-C.; Chen, P.-Y. Detecting and prioritizing biosynthetic gene clusters for bioactive compounds in bacteria and fungi. Appl. Microbiol. Biotechnol. 2019, 103, 3277–3287. [Google Scholar] [CrossRef]
- Jouhten, P.; Ponomarova, O.; Gonzalez, R.; Patil, K.R. Saccharomyces cerevisiae metabolism in ecological context. FEMS Yeast Res. 2016, 16, fow080. [Google Scholar] [CrossRef]
- Salem-Bango, Z.; Price, T.K.; Chan, J.L.; Chandrasekaran, S.; Garner, O.B.; Yang, S. Fungal whole-genome sequencing for species identification: From test development to clinical utilization. J. Fungi 2023, 9, 183. [Google Scholar]
- Blin, K.; Shaw, S.; Kloosterman, A.M.; Charlop-Powers, Z.; Van Wezel, G.P.; Medema, M.H.; Weber, T. antiSMASH 6.0: Improving cluster detection and comparison capabilities. Nucleic Acids Res. 2021, 49, W29–W35. [Google Scholar] [CrossRef]
- Kautsar, S.A.; Blin, K.; Shaw, S.; Navarro-Muñoz, J.C.; Terlouw, B.R.; Van Der Hooft, J.J.; Van Santen, J.A.; Tracanna, V.; Suarez Duran, H.G.; Pascal Andreu, V. MIBiG 2.0: A repository for biosynthetic gene clusters of known function. Nucleic Acids Res. 2020, 48, D454–D458. [Google Scholar] [CrossRef]
- Russell, A.H.; Truman, A.W. Genome mining strategies for ribosomally synthesised and post-translationally modified peptides. Comput. Struct. Biotechnol. J. 2020, 18, 1838–1851. [Google Scholar]
- Alam, K.; Hao, J.; Zhong, L.; Fan, G.; Ouyang, Q.; Islam, M.; Islam, S.; Sun, H.; Zhang, Y.; Li, R. Complete genome sequencing and in silico genome mining reveal the promising metabolic potential in Streptomyces strain CS-7. Front. Microbiol. 2022, 13, 3751. [Google Scholar] [CrossRef]
- Koboldt, D.C.; Steinberg, K.M.; Larson, D.E.; Wilson, R.K.; Mardis, E.R. The next-generation sequencing revolution and its impact on genomics. Cell 2013, 155, 27–38. [Google Scholar] [PubMed]
- Slatko, B.E.; Gardner, A.F.; Ausubel, F.M. Overview of next-generation sequencing technologies. Curr. Protoc. Mol. Biol. 2018, 122, e59. [Google Scholar] [CrossRef]
- Amarasinghe, S.L.; Su, S.; Dong, X.; Zappia, L.; Ritchie, M.E.; Gouil, Q. Opportunities and challenges in long-read sequencing data analysis. Genome Biol. 2020, 21, 30. [Google Scholar]
- Louwen, J.J.; Medema, M.H.; van der Hooft, J.J. Enhanced correlation-based linking of biosynthetic gene clusters to their metabolic products through chemical class matching. Microbiome 2023, 11, 13. [Google Scholar] [CrossRef] [PubMed]
- Van Der Hooft, J.J.; Mohimani, H.; Bauermeister, A.; Dorrestein, P.C.; Duncan, K.R.; Medema, M.H. Linking genomics and metabolomics to chart specialized metabolic diversity. Chem. Soc. Rev. 2020, 49, 3297–3314. [Google Scholar]
- Nevalainen, K.H.; Te’o, V.S.; Bergquist, P.L. Heterologous protein expression in filamentous fungi. Trends Biotechnol. 2005, 23, 468–474. [Google Scholar]
- Qiao, Y.-M.; Yu, R.-L.; Zhu, P. Advances in targeting and heterologous expression of genes involved in the synthesis of fungal secondary metabolites. RSC Adv. 2019, 9, 35124–35134. [Google Scholar] [CrossRef]
- Ning, Y.; Xu, Y.; Jiao, B.; Lu, X. Application of gene knockout and heterologous expression strategy in fungal secondary metabolites biosynthesis. Mar. Drugs 2022, 20, 705. [Google Scholar]
- Murai, K.; Lauterbach, L.; Teramoto, K.; Quan, Z.; Barra, L.; Yamamoto, T.; Nonaka, K.; Shiomi, K.; Nishiyama, M.; Kuzuyama, T. An unusual skeletal rearrangement in the biosynthesis of the sesquiterpene trichobrasilenol from Trichoderma. Angew. Chem. Int. Ed. 2019, 58, 15046–15050. [Google Scholar] [CrossRef]
- Feng, J.; Surup, F.; Hauser, M.; Miller, A.; Wennrich, J.-P.; Stadler, M.; Cox, R.J.; Kuhnert, E. Biosynthesis of oxygenated brasilane terpene glycosides involves a promiscuous N-acetylglucosamine transferase. Chem. Comm. 2020, 56, 12419–12422. [Google Scholar] [CrossRef] [PubMed]
- Harvey, C.J.; Tang, M.; Schlecht, U.; Horecka, J.; Fischer, C.R.; Lin, H.-C.; Li, J.; Naughton, B.; Cherry, J.; Miranda, M. HEx: A heterologous expression platform for the discovery of fungal natural products. Sci. Adv. 2018, 4, eaar5459. [Google Scholar] [CrossRef]
- Meng, X.; Fang, Y.; Ding, M.; Zhang, Y.; Jia, K.; Li, Z.; Collemare, J.; Liu, W. Developing fungal heterologous expression platforms to explore and improve the production of natural products from fungal biodiversity. Biotechnol. Adv. 2022, 54, 107866. [Google Scholar]
- Kaneko, A.; Morishita, Y.; Tsukada, K.; Taniguchi, T.; Asai, T. Post-genomic approach based discovery of alkylresorcinols from a cricket-associated fungus, Penicillium soppi. Org. Biomol. Chem. 2019, 17, 5239–5243. [Google Scholar] [CrossRef]
- Song, R.; Zhai, Q.; Sun, L.; Huang, E.; Zhang, Y.; Zhu, Y.; Guo, Q.; Tian, Y.; Zhao, B.; Lu, H. CRISPR/Cas9 genome editing technology in filamentous fungi: Progress and perspective. Appl. Microbiol. Biotechnol. 2019, 103, 6919–6932. [Google Scholar]
- Nødvig, C.S.; Nielsen, J.B.; Kogle, M.E.; Mortensen, U.H. A CRISPR-Cas9 system for genetic engineering of filamentous fungi. PLoS ONE 2015, 10, e0133085. [Google Scholar] [CrossRef]
- Nødvig, C.S.; Hoof, J.B.; Kogle, M.E.; Jarczynska, Z.D.; Lehmbeck, J.; Klitgaard, D.K.; Mortensen, U.H. Efficient oligo nucleotide mediated CRISPR-Cas9 gene editing in Aspergilli. Fungal Genet. Biol. 2018, 115, 78–89. [Google Scholar] [CrossRef]
- Generoso, W.C.; Gottardi, M.; Oreb, M.; Boles, E. Simplified CRISPR-Cas genome editing for Saccharomyces cerevisiae. J. Microbiol. 2016, 127, 203–205. [Google Scholar] [CrossRef] [PubMed]
- Schuster, M.; Kahmann, R. CRISPR-Cas9 genome editing approaches in filamentous fungi and oomycetes. Fungal Genet. Biol. 2019, 130, 43–53. [Google Scholar] [CrossRef]
- Jiang, C.; Lv, G.; Tu, Y.; Cheng, X.; Duan, Y.; Zeng, B.; He, B. Applications of CRISPR/Cas9 in the synthesis of secondary metabolites in filamentous fungi. Front. Microbiol. 2021, 12, 638096. [Google Scholar] [PubMed]
- Roux, I.; Woodcraft, C.; Hu, J.; Wolters, R.; Gilchrist, C.L.; Chooi, Y.-H. CRISPR-mediated activation of biosynthetic gene clusters for bioactive molecule discovery in filamentous fungi. ACS Synth. Biol. 2020, 9, 1843–1854. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Zhang, J.; Ma, Z.; Zhang, Y.; Bechthold, A.; Yu, X. Double-reporter-guided targeted activation of the oxytetracycline silent gene cluster in Streptomyces rimosus M527. Biotechnol. Bioeng. 2023, 120, 1411–1422. [Google Scholar] [CrossRef] [PubMed]
- El-Hawary, S.S.; Hassan, M.H.; Hudhud, A.O.; Abdelmohsen, U.R.; Mohammed, R. Elicitation for activation of the actinomycete genome’s cryptic secondary metabolite gene clusters. RSC Adv. 2023, 13, 5778–5795. [Google Scholar] [CrossRef]
- Mao, D.; Yoshimura, A.; Wang, R.; Seyedsayamdost, M.R. Reporter-Guided transposon mutant selection for activation of silent gene clusters in Burkholderia thailandensis. ChemBioChem 2020, 21, 1826–1831. [Google Scholar] [CrossRef] [PubMed]
- Mao, D.; Okada, B.K.; Wu, Y.; Xu, F.; Seyedsayamdost, M.R. Recent advances in activating silent biosynthetic gene clusters in bacteria. Curr. Opin. Microbiol. 2018, 45, 156–163. [Google Scholar] [CrossRef]
- Askenazi, M.; Driggers, E.M.; Holtzman, D.A.; Norman, T.C.; Iverson, S.; Zimmer, D.P.; Boers, M.-E.; Blomquist, P.R.; Martinez, E.J.; Monreal, A.W. Integrating transcriptional and metabolite profiles to direct the engineering of lovastatin-producing fungal strains. Nat. Biotechnol. 2003, 21, 150–156. [Google Scholar] [CrossRef]
- Moore, L.D.; Le, T.; Fan, G. DNA methylation and its basic function. Neuropsychopharmacology 2013, 38, 23–38. [Google Scholar] [PubMed]
- Kandel, E.S.; Lu, T.; Wan, Y.; Agarwal, M.K.; Jackson, M.W.; Stark, G.R. Mutagenesis by reversible promoter insertion to study the activation of NF-κB. Proc. Natl. Acad. Sci. USA 2005, 102, 6425–6430. [Google Scholar] [CrossRef]
- Zheng, W.; Wang, X.; Zhou, H.; Zhang, Y.; Li, A.; Bian, X. Establishment of recombineering genome editing system in Paraburkholderia megapolitana empowers activation of silent biosynthetic gene clusters. Microb. Biotechnol. 2020, 13, 397–405. [Google Scholar] [CrossRef]
- Wei, P.-L.; Fan, J.; Yu, J.; Ma, Z.; Guo, X.; Keller, N.P.; Li, E.; Lou, C.; Yin, W.-B. Quantitative characterization of filamentous fungal promoters on a single-cell resolution to discover cryptic natural products. Sci. China Life Sci. 2022, 66, 848–860. [Google Scholar] [CrossRef]
- Grau, M.F.; Entwistle, R.; Oakley, C.E.; Wang, C.C.; Oakley, B.R. Overexpression of an LaeA-like methyltransferase upregulates secondary metabolite production in Aspergillus nidulans. ACS Chem. Biol. 2019, 14, 1643–1651. [Google Scholar] [CrossRef]
- Shentu, X.; Liu, N.; Tang, G.; Tanaka, Y.; Ochi, K.; Xu, J.; Yu, X. Improved antibiotic production and silent gene activation in Streptomyces diastatochromogenes by ribosome engineering. J. Antibiot. 2016, 69, 406–410. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Zhong, C.; Xiao, H. Genetic engineering of filamentous fungi for efficient protein expression and secretion. Front. Bioeng. Biotechnol. 2020, 8, 293. [Google Scholar] [CrossRef] [PubMed]
- Zhu, S.; Duan, Y.; Huang, Y. The application of ribosome engineering to natural product discovery and yield improvement in Streptomyces. Antibiotics 2019, 8, 133. [Google Scholar] [CrossRef]
- Chai, Y.-J.; Cui, C.-B.; Li, C.-W.; Wu, C.-J.; Tian, C.-K.; Hua, W. Activation of the dormant secondary metabolite production by introducing gentamicin-resistance in a marine-derived Penicillium purpurogenum G59. Mar. Drugs 2012, 10, 559–582. [Google Scholar] [CrossRef]
- Wu, C.-J.; Yi, L.; Cui, C.-B.; Li, C.-W.; Wang, N.; Han, X. Activation of the silent secondary metabolite production by introducing neomycin-resistance in a marine-derived Penicillium purpurogenum G59. Mar. Drugs 2015, 13, 2465–2487. [Google Scholar] [CrossRef]
- Dong, Y.; Cui, C.-B.; Li, C.-W.; Hua, W.; Wu, C.-J.; Zhu, T.-J.; Gu, Q.-Q. Activation of dormant secondary metabolite production by introducing neomycin resistance into the deep-sea fungus, Aspergillus versicolor ZBY-3. Mar. Drugs 2014, 12, 4326–4352. [Google Scholar] [CrossRef]
- Perrenoud, A.; Sauer, U. Impact of global transcriptional regulation by ArcA, ArcB, Cra, Crp, Cya, Fnr, and Mlc on glucose catabolism in Escherichia coli. J. Bacteriol. 2005, 187, 3171–3179. [Google Scholar] [CrossRef]
- Glauser, D.A.; Schlegel, W. Mechanisms of transcriptional regulation underlying temporal integration of signals. Nucleic Acids Res. 2006, 34, 5175–5183. [Google Scholar] [CrossRef] [PubMed]
- Varfolomeev, E.; Goncharov, T.; Maecker, H.; Zobel, K.; Kömüves, L.G.; Deshayes, K.; Vucic, D. Cellular inhibitors of apoptosis are global regulators of NF-κB and MAPK activation by members of the TNF family of receptors. Sci. Signal. 2012, 5, ra22. [Google Scholar] [CrossRef]
- Peñalva, M.A.; Arst, J.; Herbert, N. Recent advances in the characterization of ambient pH regulation of gene expression in filamentous fungi and yeasts. Annu. Rev. Microbiol. 2004, 58, 425–451. [Google Scholar] [CrossRef]
- Merhej, J.; Richard-Forget, F.; Barreau, C. The pH regulatory factor Pac1 regulates Tri gene expression and trichothecene production in Fusarium graminearum. Fungal Genet. Biol. 2011, 48, 275–284. [Google Scholar] [CrossRef]
- Lyu, H.-N.; Liu, H.-W.; Keller, N.P.; Yin, W.-B. Harnessing diverse transcriptional regulators for natural product discovery in fungi. Nat. Prod. Rep. 2020, 37, 6–16. [Google Scholar] [CrossRef]
- Bok, J.W.; Keller, N.P. LaeA, a regulator of secondary metabolism in Aspergillus spp. Eukaryot. Cell 2004, 3, 527–535. [Google Scholar] [CrossRef] [PubMed]
- Niehaus, E.-M.; Rindermann, L.; Janevska, S.; Münsterkötter, M.; Güldener, U.; Tudzynski, B. Analysis of the global regulator Lae1 uncovers a connection between Lae1 and the histone acetyltransferase HAT1 in Fusarium fujikuroi. Appl. Microbiol. Biotechnol. 2018, 102, 279–295. [Google Scholar] [CrossRef] [PubMed]
- Brakhage, A.A. Regulation of fungal secondary metabolism. Nat. Rev. Microbiol. 2013, 11, 21–32. [Google Scholar] [CrossRef]
- Strauss, J.; Reyes-Dominguez, Y. Regulation of secondary metabolism by chromatin structure and epigenetic codes. Fungal Genet. Biol. 2011, 48, 62–69. [Google Scholar] [CrossRef] [PubMed]
- Jiang, T.; Wang, M.; Li, L.; Si, J.; Song, B.; Zhou, C.; Yu, M.; Wang, X.; Zhang, Y.; Ding, G. Overexpression of the global regulator LaeA in Chaetomium globosum leads to the biosynthesis of chaetoglobosin Z. J. Nat. Prod. 2016, 79, 2487–2494. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Han, H.; Zhang, X.; Ma, C.; Sun, C.; Che, Q.; Gu, Q.; Zhu, T.; Zhang, G.; Li, D. Discovery of two new sorbicillinoids by overexpression of the global regulator LaeA in a marine-derived fungus Penicillium dipodomyis YJ-11. Mar. Drugs 2019, 17, 446. [Google Scholar] [CrossRef]
- Bok, J.W.; Hoffmeister, D.; Maggio-Hall, L.A.; Murillo, R.; Glasner, J.D.; Keller, N.P. Genomic mining for Aspergillus natural products. Chem. Biol. 2006, 13, 31–37. [Google Scholar] [CrossRef]
- Hong, E.J.; Kim, N.K.; Lee, D.; Kim, W.G.; Lee, I. Overexpression of the laeA gene leads to increased production of cyclopiazonic acid in Aspergillus fumisynnematus. Fungal Biol. 2015, 119, 973–983. [Google Scholar] [CrossRef]
- Wang, L.; Zhang, X.; Zhang, K.; Zhang, X.; Zhu, T.; Che, Q.; Zhang, G.; Li, D. Overexpression of global regulator PbrlaeA leads to the discovery of new polyketide in fungus Penicillium brocae HDN-12-143. Front. Chem. 2020, 8, 270. [Google Scholar] [CrossRef]
- Ding, Z.; Wang, X.; Kong, F.-D.; Huang, H.-M.; Zhao, Y.-N.; Liu, M.; Wang, Z.-P.; Han, J. Overexpression of global regulator Talae1 leads to the discovery of new antifungal polyketides from endophytic fungus Trichoderma afroharzianum. Front. Microbiol. 2020, 11, 622785. [Google Scholar] [CrossRef]
- Bode, H.B.; Bethe, B.; Höfs, R.; Zeeck, A. Big effects from small changes: Possible ways to explore nature’s chemical diversity. ChemBioChem 2002, 3, 619–627. [Google Scholar] [CrossRef] [PubMed]
- Pan, R.; Bai, X.; Chen, J.; Zhang, H.; Wang, H. Exploring structural diversity of microbe secondary metabolites using OSMAC strategy: A literature review. Front. Microbiol. 2019, 10, 294. [Google Scholar] [CrossRef] [PubMed]
- Romano, S.; Jackson, S.A.; Patry, S.; Dobson, A.D. Extending the “one strain many compounds”(OSMAC) principle to marine microorganisms. Mar. Drugs 2018, 16, 244. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.R.; Seok, S.; Ryoo, R.; Choi, S.U.; Kim, K.H. Macrocyclic trichothecene mycotoxins from a deadly poisonous mushroom, Podostroma cornu-damae. J. Nat. Prod. 2018, 82, 122–128. [Google Scholar] [CrossRef]
- Staropoli, A.; Iacomino, G.; De Cicco, P.; Woo, S.L.; Di Costanzo, L.; Vinale, F. Induced secondary metabolites of the beneficial fungus Trichoderma harzianum M10 through OSMAC approach. Chem. Biol. Technol. Agric. 2023, 10, 28. [Google Scholar] [CrossRef]
- Hewage, R.T.; Aree, T.; Mahidol, C.; Ruchirawat, S.; Kittakoop, P. One strain-many compounds (OSMAC) method for production of polyketides, azaphilones, and an isochromanone using the endophytic fungus Dothideomycete sp. Phytochemistry 2014, 108, 87–94. [Google Scholar] [CrossRef]
- Xue, M.; Hou, X.; Fu, J.; Zhang, J.; Wang, J.; Zhao, Z.; Xu, D.; Lai, D.; Zhou, L. Recent advances in search of bioactive secondary metabolites from fungi triggered by chemical epigenetic modifiers. J. Fungi 2023, 9, 172. [Google Scholar] [CrossRef]
- Bind, S.; Bind, S.; Sharma, A.; Chaturvedi, P. Epigenetic modification: A key tool for secondary metabolite production in microorganisms. Front. Microbiol. 2022, 13, 230. [Google Scholar] [CrossRef]
- Bondarev, A.D.; Attwood, M.M.; Jonsson, J.; Chubarev, V.N.; Tarasov, V.V.; Schiöth, H.B. Recent developments of HDAC inhibitors: Emerging indications and novel molecules. Br. J. Clin. Pharmacol. 2021, 87, 4577–4597. [Google Scholar] [CrossRef]
- Anjum, K.; Xuewei, Y. Epigenetic strategies to discover novel fungal secondary metabolites. J. ISSN 2022, 2766, 2276. [Google Scholar] [CrossRef]
- Lei, H.; Zhang, D.; Ding, N.; Chen, S.; Song, C.; Luo, Y.; Fu, X.; Bi, X.; Niu, H. New cytotoxic natural products from the marine sponge-derived fungus Pestalotiopsis sp. by epigenetic modification. RSC Adv. 2020, 10, 37982–37988. [Google Scholar] [CrossRef]
- Seto, E.; Yoshida, M. Erasers of histone acetylation: The histone deacetylase enzymes. Cold Spring Harb. Perspect. Biol. 2014, 6, a018713. [Google Scholar] [CrossRef]
- Yang, X.; Zhang, N.; Shi, Z.; Yang, Z.; Hu, X. Histone deacetylase inhibitor suberoyl bis-hydroxamic acid suppresses cell proliferation and induces apoptosis in breast cancer cells. Mol. Med. Rep. 2015, 11, 2908–2912. [Google Scholar] [CrossRef]
- Pinto, A.A.; Barúa, J.E.; Almeida, M.O.; Viaud, M.; Zorrilla, D.; Collado, I.G.; Macías-Sánchez, A.J.; Durán-Patrón, R. Structural and biosynthetic studies of botrycinereic acid, a new cryptic metabolite from the fungus Botrytis cinerea. Bioorg. Chem. 2022, 127, 105979. [Google Scholar] [CrossRef]
- Wang, Z.; He, X.; Niu, C.; Zhou, J.; Zhang, Z. Induction of funitatin A, a new polyketide from the Yellow River wetland-derived fungus Talaromyces funiculosus. Phytochem. Lett. 2022, 47, 42–45. [Google Scholar] [CrossRef]
- Abdelhakim, I.A.; Bin Mahmud, F.; Motoyama, T.; Futamura, Y.; Takahashi, S.; Osada, H. Dihydrolucilactaene, a potent antimalarial compound from Fusarium sp. RK97-94. J. Nat. Prod. 2021, 85, 63–69. [Google Scholar] [CrossRef]
- Xu, F.; Nazari, B.; Moon, K.; Bushin, L.B.; Seyedsayamdost, M.R. Discovery of a cryptic antifungal compound from Streptomyces albus J1074 using high-throughput elicitor screens. J. Am. Chem. Soc. 2017, 139, 9203–9212. [Google Scholar] [CrossRef] [PubMed]
- Xu, F.; Wu, Y.; Zhang, C.; Davis, K.M.; Moon, K.; Bushin, L.B.; Seyedsayamdost, M.R. A genetics-free method for high-throughput discovery of cryptic microbial metabolites. Nat. Chem. Biol. 2019, 15, 161–168. [Google Scholar] [CrossRef]
- Moon, K.; Xu, F.; Zhang, C.; Seyedsayamdost, M.R. Bioactivity-HiTES unveils cryptic antibiotics encoded in actinomycete bacteria. ACS Chem. Biol. 2019, 14, 767–774. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.R.; Seyedsayamdost, M.R. Induction of diverse cryptic fungal metabolites by steroids and channel blockers. Angew. Chem. Int. Ed. 2022, 61, e202204519. [Google Scholar] [CrossRef]
- Qiu, S.; Cai, Y.; Yao, H.; Lin, C.; Xie, Y.; Tang, S.; Zhang, A. Small molecule metabolites: Discovery of biomarkers and therapeutic targets. Signal Transduct. Target. Ther. 2023, 8, 132. [Google Scholar] [CrossRef]
- Soares, J.X.; Santos, Á.; Fernandes, C.; Pinto, M.M. Liquid chromatography on the different methods for the determination of lipophilicity: An essential analytical tool in medicinal chemistry. Chemosensors 2022, 10, 340. [Google Scholar] [CrossRef]
- Xiao, J.F.; Zhou, B.; Ressom, H.W. Metabolite identification and quantitation in LC-MS/MS-based metabolomics. TrAC Trends Anal. Chem. 2012, 32, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Marco-Ramell, A.; Palau-Rodriguez, M.; Alay, A.; Tulipani, S.; Urpi-Sarda, M.; Sanchez-Pla, A.; Andres-Lacueva, C. Evaluation and comparison of bioinformatic tools for the enrichment analysis of metabolomics data. BMC Bioinform. 2018, 19, 1–11. [Google Scholar] [CrossRef]
- Zhang, Q.-W.; Lin, L.-G.; Ye, W.-C. Techniques for extraction and isolation of natural products: A comprehensive review. Chin. Med. 2018, 13, 20. [Google Scholar] [CrossRef]
- Li, X.-C.; Ferreira, D.; Ding, Y. Determination of absolute configuration of natural products: Theoretical calculation of electronic circular dichroism as a tool. Curr. Org. Chem. 2010, 14, 1678–1697. [Google Scholar] [CrossRef]
- Fleming, A.M.; Alshykhly, O.; Orendt, A.M.; Burrows, C.J. Computational studies of electronic circular dichroism spectra predict absolute configuration assignments for the guanine oxidation product 5-carboxamido-5-formamido-2-iminohydantoin. Tetrahedron Lett. 2015, 56, 3191–3196. [Google Scholar] [CrossRef]
- Koenis, M.A.; Chibueze, C.; Jinks, M.A.; Nicu, V.P.; Visscher, L.; Goldup, S.; Buma, W.J. Vibrational circular dichroism spectroscopy for probing the expression of chirality in mechanically planar chiral rotaxanes. Chem. Sci. 2020, 11, 8469–8475. [Google Scholar] [CrossRef]
- Felippe, L.G.; Batista, J.M., Jr.; Baldoqui, D.C.; Nascimento, I.R.; Kato, M.J.; He, Y.; Nafie, L.A.; Furlan, M. VCD to determine absolute configuration of natural product molecules: Secolignans from Peperomia blanda. Org. Biomol. Chem. 2012, 10, 4208–4214. [Google Scholar] [CrossRef]
- Grassin, C.; Santoro, E.; Merten, C. 7-Azaindole breaks carboxylic acid dimers and simplifies VCD spectra analyses of natural products. Chem. Comm. 2022, 58, 11527–11530. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.W.; Wang, J.W. How cryo-electron microscopy and X-ray crystallography complement each other. Protein Sci. 2017, 26, 32–39. [Google Scholar] [CrossRef]
- Guss, J.M.; King, G.F. Macromolecular structure determination: Comparison of crystallography and NMR. Encycl. Life Sci. 2002, 11, 1–5. [Google Scholar]
- Danelius, E.; Halaby, S.; van der Donk, W.A.; Gonen, T. MicroED in natural product and small molecule research. Nat. Prod. Rep. 2021, 38, 423–431. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).