An UPLC–ESI–MS/MS Bioanalytical Methodology for the Quantification of Gilteritinib in Human Liver Microsomes: Application to In Vitro and In Silico Metabolic Stability Estimation
Abstract
1. Introduction
2. Results and Discussions
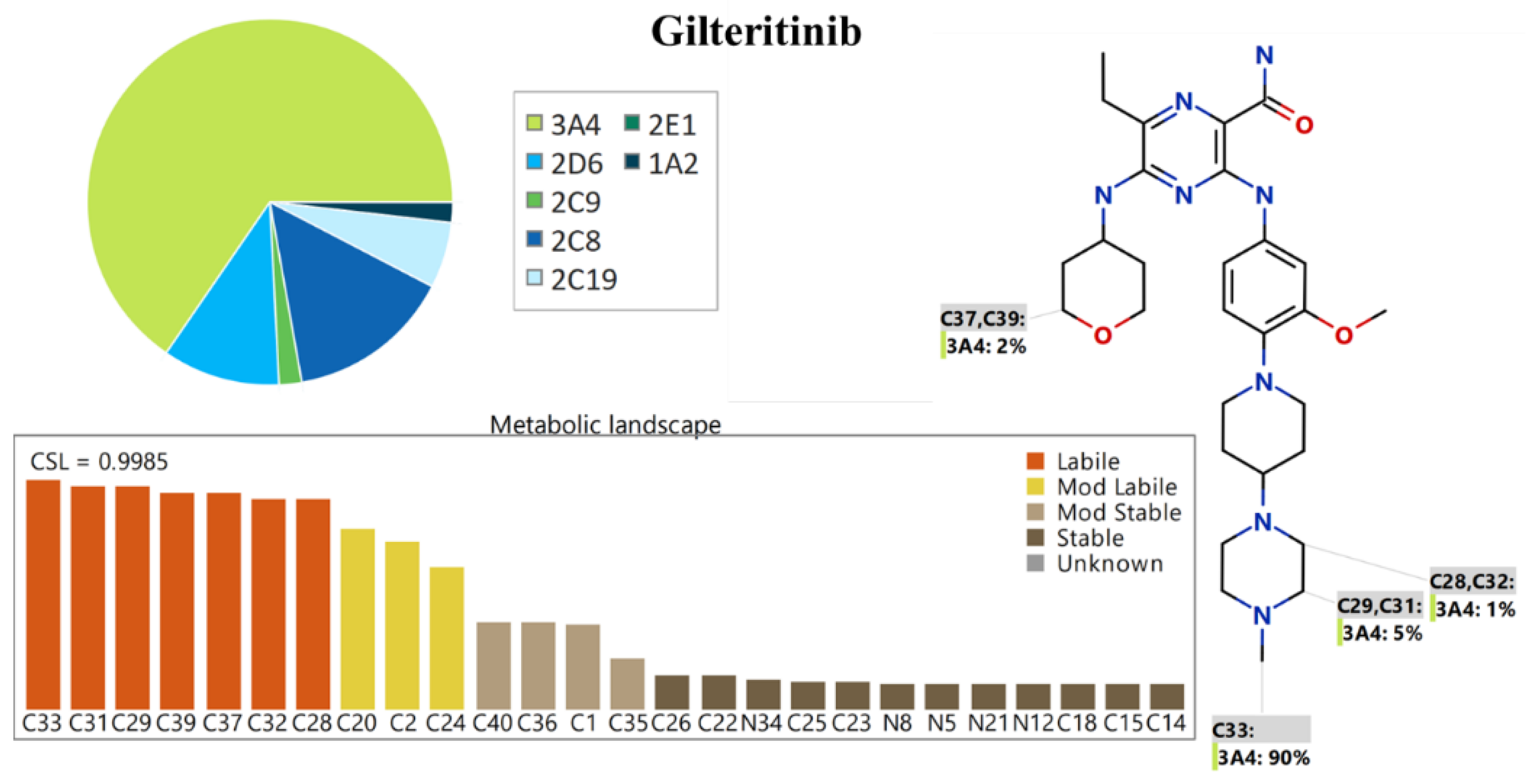
2.1. In Silico GTB Metabolic Stability
2.2. In Silico GTB Utilizing DEREK Software
2.3. UPLC–ESI–MS/MS Methodology Development
2.4. Validation Steps of the LC–ESI–MS/MS Methodology
2.4.1. Specificity
2.4.2. Sensitivity and Linearity of the UPLC-SEI-MS/MS Method
2.4.3. Precision and Accuracy of the UPLC-SEI-MS/MS Method
2.4.4. Matrix Effects of HLMs and GTB Extraction Recovery of the UPLC-SEI-MS/MS Method
2.4.5. Stability of GTB in DMSO and HLMs incubation Matrix
2.5. The In Vitro Metabolic Incubation for the Determination of GTB Metabolic Stability
3. Methodologies and Material
3.1. Instruments and Materials
3.2. In Silico Evaluation of GTB Metabolic Stability
3.3. In Silico Proposal of GTB DEREK Software
3.4. LC–ESI–MS/MS
3.4.1. LC Chromatographic Tuned Parameters
3.4.2. MS/MS Tuned Parameters
3.5. GTB Working Solutions
3.6. Calibration Curve of GTB
3.7. Extraction Methodology of GTB and EFB
3.8. Validation of the UPLC–ESI–MS/MS Methodology
3.8.1. Specificity of the UPLC–ESI–MS/MS Methodology
3.8.2. Linearity and Sensitivity of the UPLC–ESI–MS/MS Methodology
3.8.3. Accuracy and Precision of the UPLC–ESI–MS/MS Methodology
3.8.4. Extraction Recovery and Matrix Effect
3.8.5. Stability
3.9. In Vitro Assessment of GTB Metabolic Stability
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AUC | area under the curve |
| Clint | intrinsic clearance |
| DMSO | dimethyl sulfoxide |
| EFB | encorafenib |
| ESI | electrospray ionization |
| EGFR | epidermal growth factor receptor |
| GTB | gilteritinib |
| HLMs | human liver microsomes |
| IS | internal standard |
| LLOQ | lower limit of quantification |
| LC-MS/MS | liquid chromatography tandem mass spectrometry |
| MRM | multiple reaction monitoring |
| NSCLCs | Non-small cell lung cancers |
| QC | quality control |
| RSD | relative standard deviation |
| RE | relative error |
| S/N | signal to noise ratio |
| SD | standard deviation |
| TKIs | tyrosine kinase inhibitors |
| t1/2 | half-life |
References
- Mattiuzzi, C.; Lippi, G. Current Cancer Epidemiology. J. Epidemiol. Glob. Health 2019, 9, 217–222. [Google Scholar] [CrossRef] [PubMed]
- Roskoski, R., Jr. Properties of FDA-approved small molecule protein kinase inhibitors: A 2020 update. Pharmacol. Res. 2020, 152, 104609. [Google Scholar] [CrossRef] [PubMed]
- Barinaga, M. From Bench Top to Bedside. Science 1997, 278, 1036–1039. [Google Scholar] [CrossRef] [PubMed]
- Kawase, T.; Nakazawa, T.; Eguchi, T.; Tsuzuki, H.; Ueno, Y.; Amano, Y.; Suzuki, T.; Mori, M.; Yoshida, T. Effect of Fms-like tyrosine kinase 3 (FLT3) ligand (FL) on antitumor activity of gilteritinib, a FLT3 inhibitor, in mice xenografted with FL-overexpressing cells. Oncotarget 2019, 10, 6111–6123. [Google Scholar] [CrossRef] [PubMed]
- Pulte, E.D.; Norsworthy, K.J.; Wang, Y.; Xu, Q.; Qosa, H.; Gudi, R.; Przepiorka, D.; Fu, W.; Okusanya, O.O.; Goldberg, K.B.; et al. FDA Approval Summary: Gilteritinib for Relapsed or Refractory Acute Myeloid Leukemia with a FLT3 Mutation. Clin. Cancer Res. 2021, 27, 3515–3521. [Google Scholar] [CrossRef]
- Dhillon, S. Gilteritinib: First Global Approval. Drugs 2019, 79, 331–339. [Google Scholar] [CrossRef]
- Bouhaddou, M.; Memon, D.; Meyer, B.; White, K.M.; Rezelj, V.V.; Correa Marrero, M.; Polacco, B.J.; Melnyk, J.E.; Ulferts, S.; Kaake, R.M.; et al. The Global Phosphorylation Landscape of SARS-CoV-2 Infection. Cell 2020, 182, 685–712.e19. [Google Scholar] [CrossRef]
- Usuki, K.; Sakura, T.; Kobayashi, Y.; Miyamoto, T.; Iida, H.; Morita, S.; Bahceci, E.; Kaneko, M.; Kusano, M.; Yamada, S.; et al. Clinical profile of gilteritinib in Japanese patients with relapsed/refractory acute myeloid leukemia: An open-label phase 1 study. Cancer Sci. 2018, 109, 3235–3244. [Google Scholar] [CrossRef]
- Perl, A.E.; Altman, J.K.; Cortes, J.; Smith, C.; Litzow, M.; Baer, M.R.; Claxton, D.; Erba, H.P.; Gill, S.; Goldberg, S.; et al. Selective inhibition of FLT3 by gilteritinib in relapsed or refractory acute myeloid leukaemia: A multicentre, first-in-human, open-label, phase 1-2 study. Lancet Oncol. 2017, 18, 1061–1075. [Google Scholar] [CrossRef]
- Cardoso, E.; Csajka, C.; Schneider, M.P.; Widmer, N. Effect of Adherence on Pharmacokinetic/Pharmacodynamic Relationships of Oral Targeted Anticancer Drugs. Clin. Pharmacokinet. 2018, 57, 1–6. [Google Scholar] [CrossRef]
- Bryant, A.L.; LeBlanc, T.W.; Albrecht, T.; Chan, Y.N.; Richardson, J.; Foster, M.; Dang, M.; Dudley, W.; Owenby, S.; Wujcik, D. Oral adherence in adults with acute myeloid leukemia (AML): Results of a mixed methods study. Support Care Cancer 2020, 28, 5157–5164. [Google Scholar] [CrossRef]
- Houston, J.B. Utility of in vitro drug metabolism data in predicting in vivo metabolic clearance. Biochem. Pharmacol. 1994, 47, 1469–1479. [Google Scholar] [CrossRef]
- Obach, R.S.; Baxter, J.G.; Liston, T.E.; Silber, B.M.; Jones, B.C.; MacIntyre, F.; Rance, D.J.; Wastall, P. The prediction of human pharmacokinetic parameters from preclinical and in vitro metabolism data. J. Pharmacol. Exp. Ther. 1997, 283, 46–58. [Google Scholar]
- Attwa, M.W.; Kadi, A.A.; Darwish, H.W.; Amer, S.M.; Alrabiah, H. A reliable and stable method for the determination of foretinib in human plasma by LC-MS/MS: Application to metabolic stability investigation and excretion rate. Eur. J. Mass Spectrom. 2018, 24, 344–351. [Google Scholar] [CrossRef]
- Amer, S.M.; Kadi, A.A.; Darwish, H.W.; Attwa, M.W. LC–MS/MS method for the quantification of masitinib in RLMs matrix and rat urine: Application to metabolic stability and excretion rate. Chem. Cent. J. 2017, 11, 136. [Google Scholar] [CrossRef]
- Zhang, M.; Tajima, S.; Suetsugu, K.; Hirota, T.; Tsuchiya, Y.; Yamauchi, T.; Yoshimoto, G.; Miyamoto, T.; Egashira, N.; Akashi, K.; et al. Development and Validation of an LC-MS/MS Method to Quantify Gilteritinib and Its Clinical Application in Patients With FLT3 Mutation-Positive Acute Myelogenous Leukemia. Ther. Drug Monit. 2022, 44, 592–596. [Google Scholar] [CrossRef]
- Garrison, D.A.; Jin, Y.; Uddin, M.E.; Sparreboom, A.; Baker, S.D. Development, validation, and application of an LC-MS/MS method for the determination of the AXL/FLT3 inhibitor gilteritinib in mouse plasma. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2021, 1179, 122882. [Google Scholar] [CrossRef]
- Wang, Q.; Chen, Z.; Chen, D.; Ye, X.Y. An LC-MS/MS Bioanalytical Assay for the Determination of Gilteritinib in Rat Plasma and Application to a Drug-Drug Interaction Study. Drug Des. Devel Ther. 2020, 14, 2061–2067. [Google Scholar] [CrossRef]
- Tyzack, J.D.; Kirchmair, J. Computational methods and tools to predict cytochrome P450 metabolism for drug discovery. Chem. Biol. Drug Des. 2019, 93, 377–386. [Google Scholar] [CrossRef]
- Marothu Vamsi, K.; Kantamaneni, P.; Gorrepati, M. In vitro Metabolic Stability of Drugs and Applications of LC-MS in Metabolite Profiling. In Drug Metabolism; Katherine, D., Ed.; IntechOpen: Rijeka, Croatia, 2021; p. 77. [Google Scholar]
- Kazmi, S.R.; Jun, R.; Yu, M.S.; Jung, C.; Na, D. In silico approaches and tools for the prediction of drug metabolism and fate: A review. Comput. Biol. Med. 2019, 106, 54–64. [Google Scholar] [CrossRef]
- Hunt, P.A.; Segall, M.D.; Tyzack, J.D. WhichP450: A multi-class categorical model to predict the major metabolising CYP450 isoform for a compound. J. Comput. Aided Mol. Des. 2018, 32, 537–546. [Google Scholar] [CrossRef] [PubMed]
- Shin, Y.G.; Le, H.; Khojasteh, C.; Hop, C.E.C.A. Comparison of metabolic soft spot predictions of CYP3A4, CYP2C9 and CYP2D6 substrates using MetaSite and StarDrop. Comb. Chem. High Throughput Screen. 2011, 14, 811–823. [Google Scholar] [CrossRef]
- Attwa, M.W.; Darwish, H.W.; Al-Shakliah, N.S.; Kadi, A.A. A validated lc–ms/ms assay for the simultaneous quantification of the fda-approved anticancer mixture (Encorafenib and binimetinib): Metabolic stability estimation. Molecules 2021, 26, 2717. [Google Scholar] [CrossRef] [PubMed]
- McNaney, C.A.; Drexler, D.M.; Hnatyshyn, S.Y.; Zvyaga, T.A.; Knipe, J.O.; Belcastro, J.V.; Sanders, M. An automated liquid chromatography-mass spectrometry process to determine metabolic stability half-life and intrinsic clearance of drug candidates by substrate depletion. Assay Drug Dev. Technol. 2008, 6, 121–129. [Google Scholar] [CrossRef]
- Leahy, D.E. Integrating invitro ADMET data through generic physiologically based pharmacokinetic models. Expert Opin. Drug Metab. Toxicol. 2006, 2, 619–628. [Google Scholar] [CrossRef]
- Alrabiah, H.; Kadi, A.A.; Attwa, M.W.; Abdelhameed, A.S. A simple liquid chromatography-tandem mass spectrometry method to accurately determine the novel third-generation EGFR-TKI naquotinib with its applicability to metabolic stability assessment. RSC Adv. 2019, 9, 4862–4869. [Google Scholar] [CrossRef] [PubMed]
- Kadi, A.A.; Darwish, H.W.; Abuelizz, H.A.; Alsubi, T.A.; Attwa, M.W. Identification of reactive intermediate formation and bioactivation pathways in Abemaciclib metabolism by LC-MS/MS: In vitro metabolic investigation. R. Soc. Open Sci. 2019, 6, 181714. [Google Scholar] [CrossRef]
- Busby, W.F., Jr.; Ackermann, J.M.; Crespi, C.L. Effect of methanol, ethanol, dimethyl sulfoxide, and acetonitrile on in vitro activities of cDNA-expressed human cytochromes P-450. Drug Metab. Dispos. 1999, 27, 246–249. [Google Scholar]
- Störmer, E.; Roots, I.; Brockmöller, J. Benzydamine N-oxidation as an index reaction reflecting FMO activity in human liver microsomes and impact of FMO3 polymorphisms on enzyme activity. Br. J. Clin. Pharmacol. 2000, 50, 553–561. [Google Scholar] [CrossRef]
- Fouin-Fortunet, H.; Tinel, M.; Descatoire, V.; Letteron, P.; Larrey, D.; Geneve, J.; Pessayre, D. Inactivation of cytochrome P-450 by the drug methoxsalen. J. Pharmacol. Exp. Ther. 1986, 236, 237–247. [Google Scholar]
- Attwa, M.W.; AlRabiah, H.; Mostafa, G.A.E.; Kadi, A.A. Development of an LC-MS/MS Method for Quantification of Sapitinib in Human Liver Microsomes: In Silico and In Vitro Metabolic Stability Evaluation. Molecules 2023, 28, 2322. [Google Scholar] [CrossRef]
- United State of America–Food and Drug Administration. Bioanalytical Method Validation Guidance for Industry. 2018. Available online: https://www.fda.gov/ucm/groups/fdagov-public/@fdagov-drugsgen/documents/document/ucm070107.pdf (accessed on 10 February 2022).
- Słoczyńska, K.; Gunia-Krzyżak, A.; Koczurkiewicz, P.; Wójcik-Pszczoła, K.; Żelaszczyk, D.; Popiół, J.; Pękala, E. Metabolic stability and its role in the discovery of new chemical entities. Acta Pharm. 2019, 69, 345–361. [Google Scholar] [CrossRef]
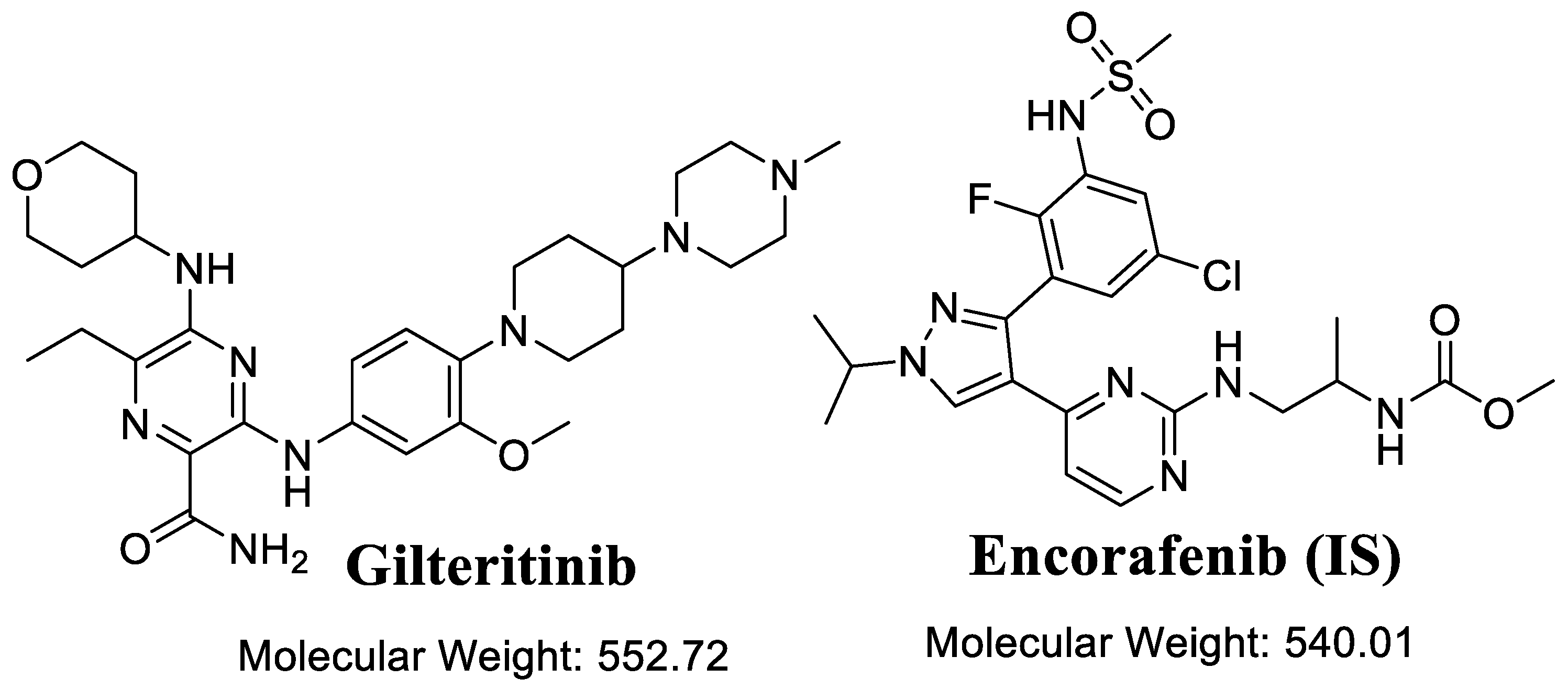





| Analyte | ESI | Rt | Parent (m/z) | Daughter Ion (1) | Daughter Ion (2) | CE, eV | Cone Voltage (V) |
|---|---|---|---|---|---|---|---|
| GTB | +ve | 0.56 | 553.0 | 436 | 453 | 30/24 | 38 |
| EFB (IS) | +ve | 1.16 | 540.0 | 116 | 359 | 32/36 | 56 |
| GTB (ng/mL) | Average | SD | RSD (%) | Accuracy (%) | Recovery |
|---|---|---|---|---|---|
| 1 (LLOQ) | 1.10 | 0.01 | 1.33 | 10.10 | 110.10 |
| 3 (LQC) | 3.06 | 0.13 | 4.32 | 2.10 | 102.10 |
| 15 | 14.81 | 0.20 | 1.38 | −1.28 | 98.72 |
| 150 | 146.81 | 2.29 | 1.56 | −2.13 | 97.87 |
| 300 | 305.01 | 2.62 | 0.86 | 1.67 | 101.67 |
| 500 | 508.11 | 4.69 | 0.92 | 1.62 | 101.62 |
| 900 (MQC) | 905.53 | 4.61 | 0.51 | 0.61 | 100.61 |
| 1500 | 1485.84 | 5.67 | 0.38 | −0.94 | 99.06 |
| 2400 (HQC) | 2430.93 | 14.01 | 0.58 | 1.29 | 101.29 |
| 3000 | 3039.44 | 20.48 | 0.67 | 1.31 | 101.31 |
| % Recovery | 101.43 ± 3.37 |
| GTB in HLMs Matrix (ng/mL) | Intra-Day Assay (Twelve Replicates in One Day) | Inter-Day Assay (Six Replicates in Three Days) | ||||||
|---|---|---|---|---|---|---|---|---|
| 1 (LLOQ) | 3 (LQC) | 900 (MQC) | 2400 (HQC) | 1 (LLOQ) | 3 (LQC) | 900 (MQC) | 2400 (HQC) | |
| Mean | 1.10 | 3.06 | 905.53 | 2430.93 | 1.11 | 3.11 | 910.30 | 2408.49 |
| SD | 0.01 | 0.13 | 4.61 | 14.01 | 0.02 | 0.10 | 4.04 | 6.52 |
| Precision (% RSD) | 1.33 | 4.32 | 0.51 | 0.58 | 2.02 | 3.24 | 0.44 | 0.27 |
| % Accuracy | 10.10 | 2.10 | 0.61 | 1.29 | 11.39 | 3.69 | 1.14 | 0.35 |
| Recovery (%) | 110.10 | 102.10 | 100.61 | 101.29 | 111.39 | 103.69 | 101.14 | 100.35 |
| GTB Concentration (ng/mL) | Short-Term Stability | Freeze–Thaw Stability | Autosampler Stability | Long-Term Stability |
|---|---|---|---|---|
| LQC (3) | 99.7 ± 3.21 | 99.24 ± 3.46 | 99.52 ± 3.08 | 98.73 ± 4.18 |
| HQC (2400) | 101.24 ± 3.62 | 100.82 ± 3.48 | 100.68 ± 2.82 | 99.85 ± 3.28 |
| Time of Quenching (min) | Average a (ng/mL) | X b | LN X | The Linearity |
|---|---|---|---|---|
| 0.00 | 672.00 | 100.00 | 4.61 | Regression equation: y = −0.04842x + 4.659 |
| 2.50 | 626.57 | 93.24 | 4.54 | |
| 5.00 | 561.59 | 83.57 | 4.43 | R2 = 0.985 |
| 7.50 | 512.40 | 76.25 | 4.33 | |
| 15.00 | 365.03 | 54.32 | 3.99 | Slope: −0.04842 |
| 20.00 | 254.35 | 37.85 | 3.63 | |
| 25.00 | 197.90 | 29.45 | 3.38 | t1/2: 14.32 min and |
| 30.00 | 187.15 | 27.85 | 3.33 | Clint: 56.64 mL/min/kg |
| 40.00 | 166.32 | 24.75 | 3.21 | |
| 50.00 | 146.16 | 21.75 | 3.08 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Attwa, M.W.; AlRabiah, H.; Alsibaee, A.M.; Abdelhameed, A.S.; Kadi, A.A. An UPLC–ESI–MS/MS Bioanalytical Methodology for the Quantification of Gilteritinib in Human Liver Microsomes: Application to In Vitro and In Silico Metabolic Stability Estimation. Separations 2023, 10, 278. https://doi.org/10.3390/separations10050278
Attwa MW, AlRabiah H, Alsibaee AM, Abdelhameed AS, Kadi AA. An UPLC–ESI–MS/MS Bioanalytical Methodology for the Quantification of Gilteritinib in Human Liver Microsomes: Application to In Vitro and In Silico Metabolic Stability Estimation. Separations. 2023; 10(5):278. https://doi.org/10.3390/separations10050278
Chicago/Turabian StyleAttwa, Mohamed W., Haitham AlRabiah, Aishah M. Alsibaee, Ali S. Abdelhameed, and Adnan A. Kadi. 2023. "An UPLC–ESI–MS/MS Bioanalytical Methodology for the Quantification of Gilteritinib in Human Liver Microsomes: Application to In Vitro and In Silico Metabolic Stability Estimation" Separations 10, no. 5: 278. https://doi.org/10.3390/separations10050278
APA StyleAttwa, M. W., AlRabiah, H., Alsibaee, A. M., Abdelhameed, A. S., & Kadi, A. A. (2023). An UPLC–ESI–MS/MS Bioanalytical Methodology for the Quantification of Gilteritinib in Human Liver Microsomes: Application to In Vitro and In Silico Metabolic Stability Estimation. Separations, 10(5), 278. https://doi.org/10.3390/separations10050278






