Machine Learning for the Classification of Alzheimer’s Disease and Its Prodromal Stage Using Brain Diffusion Tensor Imaging Data: A Systematic Review
Abstract
1. Introduction
2. Materials and Methods
3. Results
3.1. AD/HC Classification
3.1.1. DTI Analysis
3.1.2. Multimodal Analysis
3.2. AD/MCI/HC Classification
4. Discussion
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
Appendix A
List of Acronyms and Abbreviations
| AAL | Automated Anatomical Labeling atlas |
| ACC | Accuracy |
| AD | Alzheimer’s disease |
| ADNI | Alzheimer’s Disease Neuroimaging Initiative |
| AFQ | Automated fiber quantification |
| AK | Axial kurtosis |
| AdaBoost | Adaptive boosting |
| ALL-DKI | Combination of kurtosis and diffusion indices from DKI |
| BC | Betweenness centrality |
| CAD | Computer-aided diagnosis |
| cMCI | MCI patients that eventually converts to AD |
| CORR-MMSE | Correlation coefficient with the MMSE score |
| CSF | Cerebrospinal fluid |
| CV | Cross-validation |
| CWAS | Connectome-wide association |
| DA | Axial diffusivity |
| Diff-DKI | DKI diffusion indices |
| Diff-DTI | DTI diffusion indices |
| DKI | Diffusion kurtosis imaging |
| DR | Radial diffusivity |
| DTI | Diffusion tensor imaging |
| eMCI | Early MCI patient |
| FA | Fractional anisotropy |
| FD | Fiber density |
| GMC | Grey matter concentration |
| GMD | Grey matter density |
| GMV | Grey matter volume |
| HC | Healthy control patient |
| HOA | Harvard-Oxford atlas |
| ICA | Independent component analysis |
| IG | Information gain |
| ISCN | Individual structural connectivity network |
| k-NN | k-nearest neighbors algorithm |
| LDA | Linear discriminant analysis |
| LDDMM | Large deformation diffeomorphic metric mapping |
| L-MCI | Late MCI patient |
| MCI | Mild cognitive impairment |
| MD | Mean diffusivity |
| MDMR | Multivariate distance matrix regression |
| MDP | Maximum density path |
| MK | Mean kurtosis |
| MK-SVM | Multiple-kernel SVM |
| ML | Machine learning |
| MMSE | Mini-mental state examination |
| MO | Mode of anisotropy |
| MRI | Magnetic resonance imaging |
| NB | Naïve Bayes classifier |
| PCA | Principal component analysis |
| PET | Positron mission tomography |
| PLS-DA | Partial least squares discrimination analysis |
| PRISMA | Preferred Reporting Items for Systematic Reviews and Meta-Analyses |
| RA | Relative anisotropy |
| rs-fMRI | Resting-state functional MRI |
| ROI | Region of interest |
| RK | Radial kurtosis |
| SEN | Sensitivity |
| SGL | Sparse group lasso |
| SIFT | Scale invariant feature transform |
| sMRI | Structural MRI |
| SPE | Specificity |
| SURF | Speed up robust features |
| SVM | Support vector machine |
| SVM-RFE | SVM-based feature recursive elimination |
| TBSS | Tract-based special statistics |
| WM | White matter |
| WMD | White matter density |
| XGB | Extreme gradient boosting |
Appendix B
Appendix B.1. Machine Learning Overview
Appendix B.2. Support Vector Machine
Appendix B.3. Logistic Regression
Appendix B.4. Naïve Bayes Classifier
Appendix B.5. Linear Discriminant Analysis
Appendix B.6. Partial Least Squares Discriminant Analysis
Appendix B.7. K-Nearest Neighbors
Appendix B.8. Random Forest
Appendix B.9. Boosting Techniques
References
- Prince, M.; Bryce, R.; Albanese, E.; Wimo, A.; Ribeiro, W.; Ferri, C.P. The global prevalence of dementia: A systematic review and metaanalysis. Alzheimers Dement. 2013, 9, 63–75. [Google Scholar] [CrossRef]
- Collie, A.; Maruff, P. The neuropsychology of preclinical Alzheimer’s disease and mild cognitive impairment. Neurosci. Biobehav. Rev. 2000, 24, 365–374. [Google Scholar] [CrossRef]
- Alzheimer’s Association. 2019 Alzheimer’s disease facts and figures. Alzheimers Dement. 2019, 15, 321–387. [Google Scholar] [CrossRef]
- Jack, C.R.; Wiste, H.J.; Weigand, S.D.; Therneau, T.M.; Lowe, V.J.; Knopman, D.S.; Gunter, J.L.; Senjem, M.L.; Jones, D.T.; Kantarci, K.; et al. Defining imaging biomarker cut points for brain aging and Alzheimer’s disease. Alzheimers Dement. 2017, 13, 205–216. [Google Scholar] [CrossRef] [PubMed]
- Woolf, B.P. Building Intelligent Interactive Tutors: Student-Centered Strategies for Revolutionizing E-Learning; Morgan Kaufmann: Burlington, MA, USA, 2010. [Google Scholar]
- Nayak, J.; Naik, B.; Behera, H.S. A Comprehensive Survey on Support Vector Machine in Data Mining Tasks: Applications & Challenges. Int. J. Database Theory Appl. 2015, 8, 169–186. [Google Scholar] [CrossRef]
- Frisoni, G.B.; Fox, N.; Jack, C.R.; Scheltens, P.; Thompson, P. The clinical use of structural MRI in Alzheimer disease. Nat. Rev. Neurol. 2010, 6, 67–77. [Google Scholar] [CrossRef]
- Greicius, M.D.; Srivastava, G.; Reiss, A.L.; Menon, V. Default-mode network activity distinguishes Alzheimer’s disease from healthy aging: Evidence from functional MRI. Proc. Natl. Acad. Sci. USA 2004, 101, 4637–4642. [Google Scholar] [CrossRef] [PubMed]
- Fripp, J.; Bourgeat, P.; Acosta, O.; Raniga, P.; Modat, M.; Pike, K.E.; Jones, G.; O’Keefe, G.; Masters, C.L.; Ames, D.; et al. Appearance modeling of 11C PiB PET images: Characterizing amyloid deposition in Alzheimer’s disease, mild cognitive impairment and healthy aging. NeuroImage 2008, 43, 430–439. [Google Scholar] [CrossRef] [PubMed]
- Cabral, C.; Silveira, M. Classification of Alzheimer’s disease from FDG-PET images using favourite class ensembles. In Proceedings of the 2013 35th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Osaka, Japan, 3–7 July 2013; Volume 2013, pp. 2477–2480. [Google Scholar] [CrossRef]
- Szmuda, M.; Szmuda, T.; Springer, J.; Rogowska, M.; Sabisz, A.; Dubaniewicz, M.; Mazurkiewicz-Bełdzińska, M. Diffusion tensor tractography imaging in pediatric epilepsy—A systematic review. Neurologia i Neurochirurgia Polska 2016, 50, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Arab, A.; Wojna-Pelczar, A.; Khairnar, A.; Szabo, N.; Ruda-Kucerova, J. Principles of diffusion kurtosis imaging and its role in early diagnosis of neurodegenerative disorders. Brain Res. Bull. 2018, 139, 91–98. [Google Scholar] [CrossRef] [PubMed]
- Billeci, L.; Calderoni, S.; Tosetti, M.; Catani, M.; Muratori, F. White matter connectivity in children with autism spectrum disorders: A tract-based spatial statistics study. BMC Neurol. 2012, 12, 148. [Google Scholar] [CrossRef]
- Le Bihan, D.; Poupon, C.; Clark, C.A.; Pappata, S.; Molko, N.; Chabriat, H. Diffusion tensor imaging: Concepts and applications. J. Magn. Reson. Imaging 2001, 13, 534–546. [Google Scholar] [CrossRef] [PubMed]
- Pierpaoli, C.; Jezzard, P.; Basser, P.J.; Barnett, A.; Di Chiro, G. Diffusion tensor MR imaging of the human brain. Radiology 1996, 201, 637–648. [Google Scholar] [CrossRef] [PubMed]
- Alexander, A.L.; Lee, J.E.; Lazar, M.; Field, A.S. Diffusion tensor imaging of the brain. Neurotherapeutics 2007, 4, 316–329. [Google Scholar] [CrossRef] [PubMed]
- Alves, G.S.; Knöchel, V.O.; Knöchel, C.; Carvalho, A.F.; Pantel, J.; Engelhardt, E.; Laks, J. Integrating Retrogenesis Theory to Alzheimer’s Disease Pathology: Insight from DTI-TBSS Investigation of the White Matter Microstructural Integrity. BioMed Res. Int. 2015, 2015, 1–11. [Google Scholar] [CrossRef]
- Smith, S.M.; Jenkinson, M.; Johansen-Berg, H.; Rueckert, D.; Nichols, T.E.; Mackay, C.E.; Watkins, K.E.; Ciccarelli, O.; Cader, M.Z.; Matthews, P.M.; et al. Tract-based spatial statistics: Voxelwise analysis of multi-subject diffusion data. NeuroImage 2006, 31, 1487–1505. [Google Scholar] [CrossRef]
- Hagmann, P.; Cammoun, L.; Gigandet, X.; Meuli, R.; Honey, C.J.; Wedeen, V.J.; Sporns, O. Mapping the Structural Core of Human Cerebral Cortex. PLoS Boil. 2008, 6, e159. [Google Scholar] [CrossRef]
- Xie, S.; Xiao, J.X.; Gong, G.L.; Zang, Y.-F.; Wang, Y.H.; Wu, H.K.; Jiang, X.X. Voxel-based detection of white matter abnormalities in mild Alzheimer disease. Neurology 2006, 66, 1845–1849. [Google Scholar] [CrossRef]
- Ringman, J.M.; O’Neill, J.; Geschwind, D.; Medina, L.D.; Apostolova, L.G.; Rodriguez, Y.; Schaffer, B.; Varpetian, A.; Tseng, B.; Ortiz, F.; et al. Diffusion tensor imaging in preclinical and presymptomatic carriers of familial Alzheimer’s disease mutations. Brain 2007, 130, 1767–1776. [Google Scholar] [CrossRef]
- Cherubini, A.; Péran, P.; Spoletini, I.; Di Paola, M.; Di Iulio, F.; Hagberg, G.; Sancesario, G.; Gianni, W.; Bossù, P.; Caltagirone, C.; et al. Combined Volumetry and DTI in Subcortical Structures of Mild Cognitive Impairment and Alzheimer’s Disease Patients. J. Alzheimer’s Dis. 2010, 19, 1273–1282. [Google Scholar] [CrossRef]
- Teipel, S.J.; Grothe, M.J.; Zhou, J.; Sepulcre, J.; Dyrba, M.; Sorg, C.; Babiloni, F. Measuring Cortical Connectivity in Alzheimer’s Disease as a Brain Neural Network Pathology: Toward Clinical Applications. J. Int. Neuropsychol. Soc. 2016, 22, 138–163. [Google Scholar] [CrossRef] [PubMed]
- Naggara, O.; Oppenheim, C.; Rieu, D.; Raoux, N.; Rodrigo, S.; Barba, G.D.; Meder, J.-F. Diffusion tensor imaging in early Alzheimer’s disease. Psychiatry Res. Neuroimaging 2006, 146, 243–249. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Schuff, N.; Jahng, G.-H.; Bayne, W.; Mori, S.; Schad, L.; Mueller, S.; Du, A.-T.; Kramer, J.H.; Yaffe, K.; et al. Diffusion tensor imaging of cingulum fibers in mild cognitive impairment and Alzheimer disease. Neurology 2007, 68, 13–19. [Google Scholar] [CrossRef] [PubMed]
- Medina, D.; Detoledo-Morrell, L.; Urresta, F.; Gabrieli, J.D.; Moseley, M.; Fleischman, D.; Bennett, D.A.; Leurgans, S.; Turner, D.A.; Stebbins, G.T. White matter changes in mild cognitive impairment and AD: A diffusion tensor imaging study. Neurobiol. Aging 2006, 27, 663–672. [Google Scholar] [CrossRef]
- E Rose, S.; McMahon, K.L.; Janke, A.L.; O’Dowd, B.; De Zubicaray, G.I.; Strudwick, M.W.; Chalk, J.B. Diffusion indices on magnetic resonance imaging and neuropsychological performance in amnestic mild cognitive impairment. J. Neurol. Neurosurg. Psychiatry 2006, 77, 1122–1128. [Google Scholar] [CrossRef]
- Fellgiebel, A.; Dellani, P.R.; Greverus, D.; Scheurich, A.; Stoeter, P.; Müller, M.J. Predicting conversion to dementia in mild cognitive impairment by volumetric and diffusivity measurements of the hippocampus. Psychiatry Res. Neuroimaging 2006, 146, 283–287. [Google Scholar] [CrossRef]
- Müller, M.J.; Greverus, D.; Dellani, P.R.; Weibrich, C.; Wille, P.R.; Scheurich, A.; Stoeter, P.; Fellgiebel, A. Functional implications of hippocampal volume and diffusivity in mild cognitive impairment. NeuroImage 2005, 28, 1033–1042. [Google Scholar] [CrossRef]
- Müller, M.J.; Greverus, D.; Weibrich, C.; Dellani, P.R.; Scheurich, A.; Stoeter, P.; Fellgiebel, A. Diagnostic utility of hippocampal size and mean diffusivity in amnestic MCI. Neurobiol. Aging 2007, 28, 398–403. [Google Scholar] [CrossRef]
- Fellgiebel, A.; Müller, M.J.; Wille, P.; Dellani, P.R.; Scheurich, A.; Schmidt, L.G.; Stoeter, P. Color-coded diffusion-tensor-imaging of posterior cingulate fiber tracts in mild cognitive impairment. Neurobiol. Aging 2005, 26, 1193–1198. [Google Scholar] [CrossRef]
- Choo, I.H.; Lee, N.Y.; Oh, J.-S.; Lee, J.S.; Lee, N.S.; Song, I.C.; Youn, J.C.; Kim, S.G.; Kim, K.W.; Jhoo, J.H.; et al. Posterior cingulate cortex atrophy and regional cingulum disruption in mild cognitive impairment and Alzheimer’s disease. Neurobiol. Aging 2010, 31, 772–779. [Google Scholar] [CrossRef]
- Selnes, P.; Aarsland, D.; Bjornerud, A.; Gjerstad, L.; Wallin, A.; Hessen, E.; Reinvang, I.; Grambaite, R.; Auning, E.; Kjærvik, V.K.; et al. Diffusion Tensor Imaging Surpasses Cerebrospinal Fluid as Predictor of Cognitive Decline and Medial Temporal Lobe Atrophy in Subjective Cognitive Impairment and Mild Cognitive Impairment. J. Alzheimer’s Dis. 2013, 33, 723–736. [Google Scholar] [CrossRef] [PubMed]
- Liberati, A.; Altman, D.G.; Tetzlaff, J.; Mulrow, C.; Gøtzsche, P.C.; Ioannidis, J.P.A.; Clarke, M.; Devereaux, P.J.; Kleijnen, J.; Moher, D. The PRISMA statement for reporting systematic reviews and meta-analyses of studies that evaluate health care interventions: Explanation and elaboration. J. Clin. Epidemiol. 2009, 62, e1–e34. [Google Scholar] [CrossRef] [PubMed]
- Graña, M.; Termenon, M.; Savio, A.; González-Pinto, A.; Echeveste, J.; Perez, J.; Besga, A. Computer Aided Diagnosis system for Alzheimer Disease using brain Diffusion Tensor Imaging features selected by Pearson’s correlation. Neurosci. Lett. 2011, 502, 225–229. [Google Scholar] [CrossRef] [PubMed]
- Patil, R.B.; Piyush, R.; Ramakrishnan, S. Identification of brain white matter regions for diagnosis of Alzheimer using Diffusion Tensor Imaging. In Proceedings of the 2013 35th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Osaka, Japan, 3–7 July 2013; pp. 6535–6538. [Google Scholar] [CrossRef]
- Patil, R.B.; Ramakrishnan, S. Analysis of sub-anatomic diffusion tensor imaging indices in white matter regions of Alzheimer with MMSE score. Comput. Methods Progr. Biomed. 2014, 117, 13–19. [Google Scholar] [CrossRef] [PubMed]
- Schouten, T.M.; Koini, M.; De Vos, F.; Seiler, S.; De Rooij, M.; Lechner, A.; Schmidt, R.; Heuvel, M.V.D.; Van Der Grond, J.; Rombouts, S.A. Individual classification of Alzheimer’s disease with diffusion magnetic resonance imaging. NeuroImage 2017, 152, 476–481. [Google Scholar] [CrossRef] [PubMed]
- Mesrob, L.; Sarazin, M.; Hahn-Barma, V.; De, S.L.C.; Dubois, B.; Gallinari, P.; Kinkingnéhun, S.; Mesrob, L.; Marie, S.; Valerie, H.-B.; et al. DTI and Structural MRI Classification in Alzheimer’s Disease. Adv. Mol. Imaging 2012, 2, 12–20. [Google Scholar] [CrossRef]
- Dyrba, M.; Ewers, M.; Wegrzyn, M.; Kilimann, I.; Plant, C.; Oswald, A.; Meindl, T.; Pievani, M.; Bokde, A.L.W.; Fellgiebel, A.; et al. Robust Automated Detection of Microstructural White Matter Degeneration in Alzheimer’s Disease Using Machine Learning Classification of Multicenter DTI Data. PLoS ONE 2013, 8, e64925. [Google Scholar] [CrossRef]
- Li, M.; Qin, Y.; Gao, F.; Zhu, W.; He, X. Discriminative analysis of multivariate features from structural MRI and diffusion tensor images. Magn. Reson. Imaging 2014, 32, 1043–1051. [Google Scholar] [CrossRef]
- Dyrba, M.; Grothe, M.J.; Kirste, T.; Teipel, S.J. Multimodal analysis of functional and structural disconnection in Alzheimer’s disease using multiple kernel SVM. Hum. Brain Mapp. 2015, 36, 2118–2131. [Google Scholar] [CrossRef]
- Chen, Y.; Sha, M.; Zhao, X.; Ma, J.; Ni, H.; Gao, W.; Ming, N. Automated detection of pathologic white matter alterations in Alzheimer’s disease using combined diffusivity and kurtosis method. Psychiatry Res. Neuroimaging 2017, 264, 35–45. [Google Scholar] [CrossRef]
- Cai, S.; Huang, K.; Kang, Y.; Jiang, Y.; Von Deneen, K.M.; Huang, L. Potential biomarkers for distinguishing people with Alzheimer’s disease from cognitively intact elderly based on the rich-club hierarchical structure of white matter networks. Neurosci. Res. 2019, 144, 56–66. [Google Scholar] [CrossRef] [PubMed]
- Tang, X.; Qin, Y.; Wu, J.; Zhang, M.; Zhu, W.; Miller, M.I. Shape and diffusion tensor imaging based integrative analysis of the hippocampus and the amygdala in Alzheimer’s disease. Magn. Reson. Imaging 2016, 34, 1087–1099. [Google Scholar] [CrossRef] [PubMed]
- Shao, J.; Myers, N.; Yang, Q.; Feng, J.; Plant, C.; Böhm, C.; Förstl, H.; Kurz, A.; Zimmer, C.; Meng, C.; et al. Prediction of Alzheimer’s disease using individual structural connectivity networks. Neurobiol. Aging 2012, 33, 2756–2765. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Nir, T.M.; Villalon-Reina, J.E.; Prasad, G.; Jahanshad, N.; Joshi, S.H.; Toga, A.W.; Bernstein, M.A.; Jack, C.R.; Weiner, M.W.; Thompson, P.; et al. Diffusion weighted imaging-based maximum density path analysis and classification of Alzheimer’s disease. Neurobiol. Aging 2015, 36, S132–S140. [Google Scholar] [CrossRef] [PubMed]
- Demirhan, A.; Nir, T.M.; Zavaliangos-Petropulu, A.; Jack, C.R.; Weiner, M.W.; Bernstein, M.A.; Thompson, P.; Jahanshad, N. Feature selection improves the accuracy of classifying Alzheimer disease using diffusion tensor images. In Proceedings of the 2015 IEEE 12th International Symposium on Biomedical Imaging (ISBI), Brooklyn, NY, USA, 16–19 April 2015; pp. 126–130. [Google Scholar] [CrossRef]
- Prasad, G.; Joshi, S.H.; Nir, T.M.; Toga, A.W.; Thompson, P.M.; Alzheimer’s Disease Neuroimaging Initiative (ADNI). Brain connectivity and novel network measures for Alzheimer’s disease classification. Neurobiol. Aging 2015, 36, S121–S131. [Google Scholar] [CrossRef] [PubMed]
- Ebadi, A.; Da Rocha, J.L.D.; Nagaraju, D.B.; Tovar-Moll, F.; Bramati, I.; Coutinho, G.; Sitaram, R.; Rashidi, P. Ensemble Classification of Alzheimer’s Disease and Mild Cognitive Impairment Based on Complex Graph Measures from Diffusion Tensor Images. Front. Mol. Neurosci. 2017, 11. [Google Scholar] [CrossRef] [PubMed]
- Maggipinto, T.; Bellotti, R.; Amoroso, N.; Diacono, D.; Donvito, G.; Lella, E.; Monaco, A.; Scelsi, M.A.; Tangaro, S.; Initiative, A.D.N. DTI measurements for Alzheimer’s classification. Phys. Med. Boil. 2017, 62, 2361–2375. [Google Scholar] [CrossRef] [PubMed]
- Eldeeb, G.W.; Zayed, N.; Yassine, I.A. Alzheimer’S Disease Classification Using Bag-Of-Words Based on Visual Pattern of Diffusion Anisotropy for DTI Imaging. In Proceedings of the 2018 40th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Honolulu, HI, USA, 17–21 July 2018; pp. 57–60. [Google Scholar]
- Ye, C.; Mori, S.; Chan, P.; Ma, T. Connectome-wide network analysis of white matter connectivity in Alzheimer’s disease. NeuroImage Clin. 2019, 22, 101690. [Google Scholar] [CrossRef]
- Da Rocha, J.L.D.; Bramati, I.E.; Coutinho, G.; Moll, F.T.; Sitaram, R. Fractional Anisotropy changes in Parahippocampal Cingulum due to Alzheimer’s Disease. Sci. Rep. 2020, 10, 1–8. [Google Scholar] [CrossRef]
- Dou, X.; Yao, H.; Feng, F.; Wang, P.; Zhou, B.; Jin, D.; Yang, Z.; Li, J.; Zhao, C.; Wang, L.; et al. Characterizing white matter connectivity in Alzheimer’s disease and mild cognitive impairment: An automated fiber quantification analysis with two independent datasets. Cortex 2020, 129, 390–405. [Google Scholar] [CrossRef]
- Weston, P.S.; Simpson, I.J.; Ryan, N.S.; Ourselin, S.; Fox, N. Diffusion imaging changes in grey matter in Alzheimer’s disease: A potential marker of early neurodegeneration. Alzheimer’s Res. Ther. 2015, 7, 47. [Google Scholar] [CrossRef] [PubMed]
- Misra, C.; Fan, Y.; Davatzikos, C. Baseline and longitudinal patterns of brain atrophy in MCI patients, and their use in prediction of short-term conversion to AD: Results from ADNI. NeuroImage 2009, 44, 1415–1422. [Google Scholar] [CrossRef] [PubMed]
- A Phelps, E.; Phelps, E. Human emotion and memory: Interactions of the amygdala and hippocampal complex. Curr. Opin. Neurobiol. 2004, 14, 198–202. [Google Scholar] [CrossRef] [PubMed]
- Laakso, M.; Soininen, H.; Partanen, K.; Helkala, E.-L.; Hartikainen, P.; Vainio, P.; Hallikainen, M.; Hänninen, T.; Sr, P.J.R. Volumes of hippocampus, amygdala and frontal lobes in the MRI-based diagnosis of early Alzheimer’s disease: Correlation with memory functions. J. Neural Transm. 1995, 9, 73–86. [Google Scholar] [CrossRef] [PubMed]
- Lehéricy, S.; Baulac, M.; Chiras, J.; Piérot, L.; Martin, N.; Pillon, B.; Deweer, B.; Dubois, B.; Marsault, C. Amygdalohippocampal MR volume measurements in the early stages of Alzheimer disease. Am. J. Neuroradiol. 1994, 15, 929–937. [Google Scholar]
- Chu, C.; Hsu, A.-L.; Chou, K.-H.; Bandettini, P.; Lin, C. Does feature selection improve classification accuracy? Impact of sample size and feature selection on classification using anatomical magnetic resonance images. NeuroImage 2012, 60, 59–70. [Google Scholar] [CrossRef] [PubMed]
- Teipel, S.J.; Reuter, S.; Stieltjes, B.; Acosta-Cabronero, J.; Ernemann, U.; Fellgiebel, A.; Filippi, M.; Frisoni, G.B.; Hentschel, F.; Jessen, F.; et al. Multicenter stability of diffusion tensor imaging measures: A European clinical and physical phantom study. Psychiatry Res. Neuroimaging 2011, 194, 363–371. [Google Scholar] [CrossRef]
- Woo, C.-W.; Chang, L.J.; A Lindquist, M.; Wager, T.D. Building better biomarkers: Brain models in translational neuroimaging. Nat. Neurosci. 2017, 20, 365–377. [Google Scholar] [CrossRef]
- Cohen, D.S.; Carpenter, K.A.; Jarrell, J.T.; Huang, X. Deep learning-based classification of multi-categorical Alzheimer’s disease data. Curr. Neurobiol. 2019, 10, 141–147. [Google Scholar]
- Rana, M.; Gupta, N.; Da Rocha, J.L.D.; Lee, S.; Sitaram, R. A toolbox for real-time subject-independent and subject-dependent classification of brain states from fMRI signals. Front. Mol. Neurosci. 2013, 7. [Google Scholar] [CrossRef]
- Liberati, G.; Da Rocha, J.L.D.; Van Der Heiden, L.; Raffone, A.; Birbaumer, N.; Belardinelli, M.O.; Sitaram, R. Toward a Brain-Computer Interface for Alzheimer’s Disease Patients by Combining Classical Conditioning and Brain State Classification. J. Alzheimer’s Dis. 2012, 31, S211–S220. [Google Scholar] [CrossRef] [PubMed]
- Vieira, S.; Pinaya, W.H.L.; Mechelli, A. Using deep learning to investigate the neuroimaging correlates of psychiatric and neurological disorders: Methods and applications. Neurosci. Biobehav. Rev. 2017, 74, 58–75. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Li, Z.; Ge, Q.; Lin, N.; Xiong, M. Deep Feature Selection and Causal Analysis of Alzheimer’s Disease. Front. Mol. Neurosci. 2019, 13. [Google Scholar] [CrossRef] [PubMed]
- Marzban, E.N.; Eldeib, A.M.; Yassine, I.A.; Kadah, Y.M.; Initiative, F.T.A.D.N. Alzheimer’s disease diagnosis from diffusion tensor images using convolutional neural networks. PLoS ONE 2020, 15, e0230409. [Google Scholar] [CrossRef]
- Bishop, C. Pattern Recognition and Machine Learning; Springer-Verlag: New York, NY, USA, 2006. [Google Scholar]
- Hastie, T.; Tibshirani, R.; Friedman, J. The Elements of Statistical Learning: Data Mining, Inference, and Prediction, 2nd ed.; Springer: Berlin/Heidelberg, Germany, 2009. [Google Scholar]
- Ballabio, D.; Consonni, V. Classification tools in chemistry. Part 1: Linear models. PLS-DA. Anal. Methods 2013, 5, 3790–3798. [Google Scholar] [CrossRef]
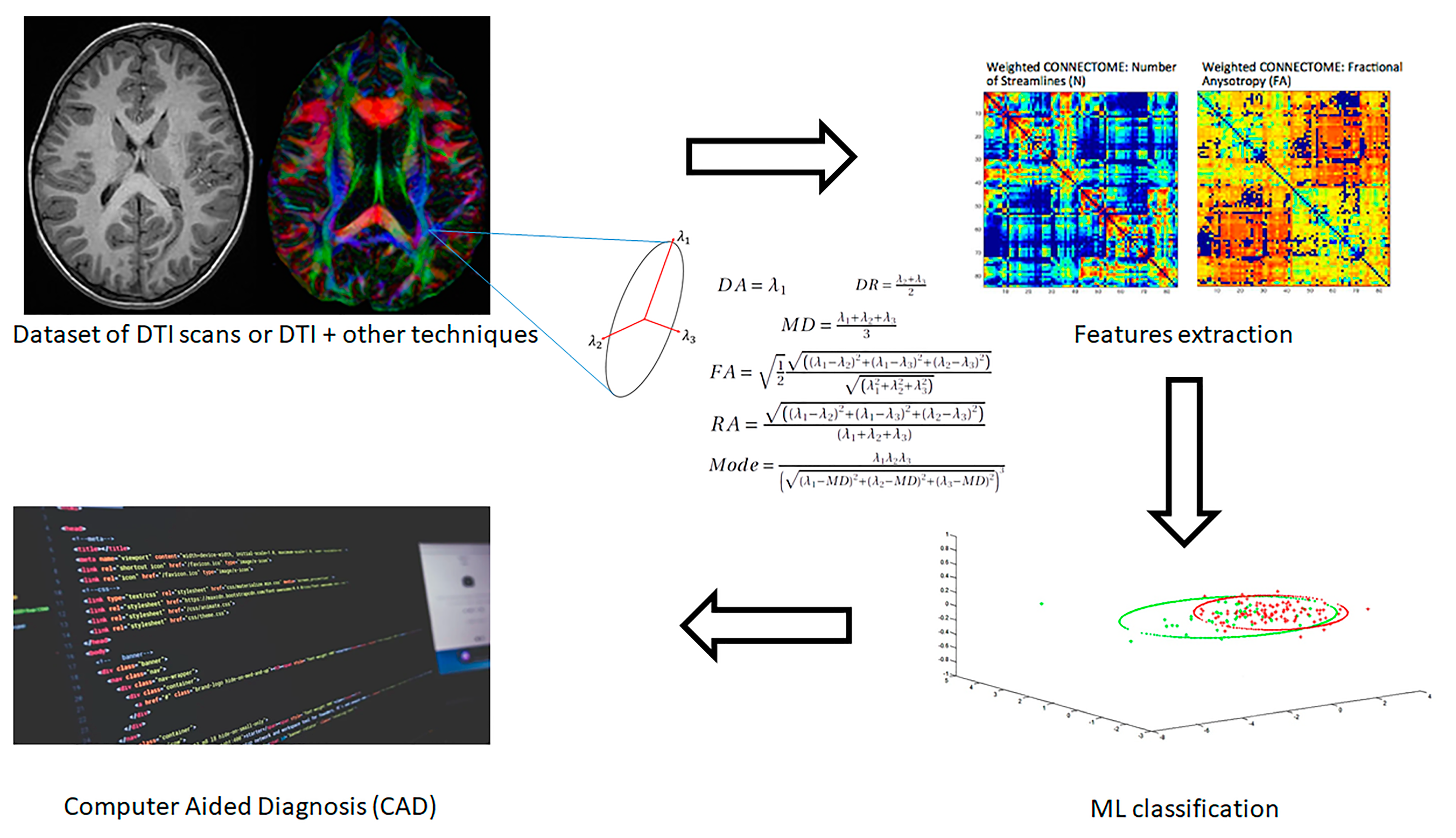


| Article | Neuroimaging Technique | Subjects | Measures | Classifier | Classification Results | ||||
|---|---|---|---|---|---|---|---|---|---|
| Feature Set/Method | ACC% | SEN% | SPE% | ||||||
| DTI analysis | |||||||||
| Graña et al., 2011 [35] | DTI | 20 AD, 25 HC | FA, MD | SVM | FA | 100.0 | 100.0 | 100.0 | |
| MD | ~99.0 | ~97.9 | ~98.1 | ||||||
| Patil et al., 2013 [36] | DTI | 34 AD, 58 HC | FA | AdaBoost | FA (10 features) | 84.5 | 80.2 | 85.2 | |
| FA (all features) | 75.3 | 71.0 | 76.7 | ||||||
| Patil and Ramakrishnan, 2014 [37] | DTI | 37 AD, 50 HC | FA, MD, DR, DA | SVM, decision stumps, simple logistic | FA (SVM) | MMSE | 94.2 | 94.4 | 93.0 |
| No MMSE | 81.6 | 81.8 | 81.4 | ||||||
| MD (SVM) | MMSE | 89.7 | 88.9 | 90.1 | |||||
| No MMSE | 87.4 | 88.2 | 86.7 | ||||||
| DR (SVM) | MMSE | 91.9 | 96.8 | 89.0 | |||||
| No MMSE | 83.9 | 89.6 | 81.0 | ||||||
| DA (SVM) | MMSE | 93.4 | 95.1 | 93.2 | |||||
| No MMSE | 81.6 | 86.2 | 79.3 | ||||||
| Schouten et al., 2017 [38] | DTI | 77 AD, 173 HC | FA, MD, DA, DR | Logistic elastic net regression | FA-TBSS | 82.6 | 83.8 | 82.1 | |
| MD-TBSS | 80.8 | 84.4 | 79.2 | ||||||
| DA-TBSS | 81.8 | 84.9 | 80.4 | ||||||
| DR-TBSS | 84.8 | 79.1 | 87.3 | ||||||
| FA-ICA | 85.1 | 86.8 | 84.4 | ||||||
| MD-ICA | 84.3 | 84.2 | 84.3 | ||||||
| DA-ICA | 83.4 | 89.7 | 80.6 | ||||||
| DR-ICA | 84.0 | 83.2 | 84.4 | ||||||
| Connectivity graph | 85.0 | 80.3 | 87.1 | ||||||
| Degree | 75.8 | 79.9 | 74.0 | ||||||
| Strength | 79.6 | 79.9 | 80.9 | ||||||
| Clustering | 75.6 | 76.6 | 79.5 | ||||||
| Betw.centrality | 64.6 | 66.9 | 66.8 | ||||||
| Path length | 69.6 | 59.5 | 72.7 | ||||||
| Transitivity | 64.9 | 62.5 | 77.2 | ||||||
| Sparse Group Lasso | 80.8 | 37.3 | 77.4 | ||||||
| Multimodal analysis | |||||||||
| Mesrob et al., 2012 [39] | DTI, sMRI | 15 AD, 16 HC | Diff: FA, MD sMRI: GMC | Non-linear SVM | MD/GMC (15 multivariate) | 99.6 | 99.2 | 99.9 | |
| MD/GMC (15 univariate) | 72.1 | 53.6 | 90.6 | ||||||
| MD/GMC (73 ROIs) | 72.4 | 62.4 | 82.4 | ||||||
| MD (73 ROIs) | 65.2 | 60.8 | 69.5 | ||||||
| FA/MD (73 ROIs) | 68.6 | 73.4 | 63.8 | ||||||
| GMC (73 ROIs) | 76.5 | 78.7 | 74.3 | ||||||
| Dyrba et al., 2013 [40] | DTI, sMRI | 137 AD, 143 HC | Diff: FA, MD sMRI: GMD, WMD | Multivariate SVM NB | GMD (SVM) | 89.3 | 87.4 | 91.2 | |
| FA (SVM) | 80.3 | 78.8 | 81.9 | ||||||
| MD (SVM) | 83.3 | 79.6 | 86.9 | ||||||
| WMD (SVM) | 82.7 | 77.9 | 87.4 | ||||||
| Li et al., 2014 [41] | DTI, sMRI | 21 AD, 15 HC | Diff: FA sMRI: GMV | SVM | Tract-Based FA + GMV | 94.3 | 95.0 | 93.3 | |
| Tract-based FA | ~89.0 | 90.5 | 86.7 | ||||||
| Voxel-based FA | ~83.0 | 90.5 | 80.0 | ||||||
| GMV | ~88.0 | 85.0 | 93.0 | ||||||
| Dyrba et al., 2015 [42] | DTI, sMRI, rs-fMRI | 28 AD, 25 HC | Diff. FA, MD, MO sMRI: GMV Rs-fMRI: local clustering coefficient, shortest path length | SVM MK-SVM | DTI measures (SVM) | 85.0 | 86.0 | 84.0 | |
| Rs-fMRI measures (SM) | 74.0 | 82.0 | 64.0 | ||||||
| GMV (SVM) | 81.0 | 82.0 | 80.0 | ||||||
| Rs-fMRI + DTI + GMV (SVM) | 79.0 | 82.0 | 86.0 | ||||||
| DTI + GMV (SVM) | 85.0 | 79.0 | 92.0 | ||||||
| Chen et al., 2017 [43] | DTI, DKI | 27 AD, 26 HC | Diff: FA, MD, DA, DR Kur: MK, AK, RK | SVM | ALL-DKI | RFE | 96.2 | 100 | 92.8 |
| MMSE | 90.6 | 100 | 83.9 | ||||||
| Diff-DKI | RFE | 92.5 | 100 | 86.7 | |||||
| MMSE | 90.6 | 100 | 83.3 | ||||||
| Diff-DTI | RFE | 81.1 | 72.9 | 100 | |||||
| MMSE | 86.8 | 81.3 | 95.2 | ||||||
| Kur-DKI | RFE | 86.8 | 83.3 | 91.3 | |||||
| MMSE | 83.0 | 79.3 | 86.9 | ||||||
| Cai et al., 2019 [44] | DTI, sMRI | 165 AD, 165 HC | BC, connection strength | LDA | BC (AAI) | 84.6 | - | - | |
| CN (AAI) | 73.0 | - | - | ||||||
| BC + CN (AAI) | 79.8 | - | - | ||||||
| Hippocampal volume (AAI) | 68.1 | - | - | ||||||
| MMSE (AAI) | 70.2 | - | - | ||||||
| Hippocampal volume + MMSE (AAI) | 71.1 | - | - | ||||||
| BC (HOA) | 75.0 | - | - | ||||||
| CN (HOA) | 71.1 | - | - | ||||||
| BC + CN (HOA) | 72.2 | - | - | ||||||
| Hippocampal volume (HOA) | 61.5 | - | - | ||||||
| MMSE (HOA) | 70.2 | - | - | ||||||
| Hippocampal volume + MMSE (HOA) | 66.6 | - | - | ||||||
| Tang et al., 2016 [45] | DTI, sMRI | 29 AD, 23 HC | Volume, deformation, FA, MD | LDA, SVM | Results reported for Right hippocampus with SVM Volume | 78.4 | 63.6 | 100.0 | |
| Shape | |||||||||
| original | 78.4 | 72.7 | 76.7 | ||||||
| PCA | 70.3 | 63.6 | 80.0 | ||||||
| PCA + ttest | 86.5 | 81.8 | 93.3 | ||||||
| DTI | 83.8 | 86.4 | 80.0 | ||||||
| Volume + Shape | |||||||||
| original | 78.4 | 72.7 | 86.7 | ||||||
| PCA | 73.0 | 68.2 | 80.0 | ||||||
| PCA + ttest | 89.2 | 86.4 | 93.3 | ||||||
| DTI + Shape | |||||||||
| original | 81.8 | 72.7 | 93.3 | ||||||
| PCA | 83.8 | 86.4 | 80.0 | ||||||
| PCA + ttest | 94.6 | 95.5 | 93.3 | ||||||
| Article | Neuroimaging Technique | Subjects | Measures | Classifier | Classification Results | ||||
|---|---|---|---|---|---|---|---|---|---|
| Task | Feature Set/Method | ACC% | SEN% | SPE% | |||||
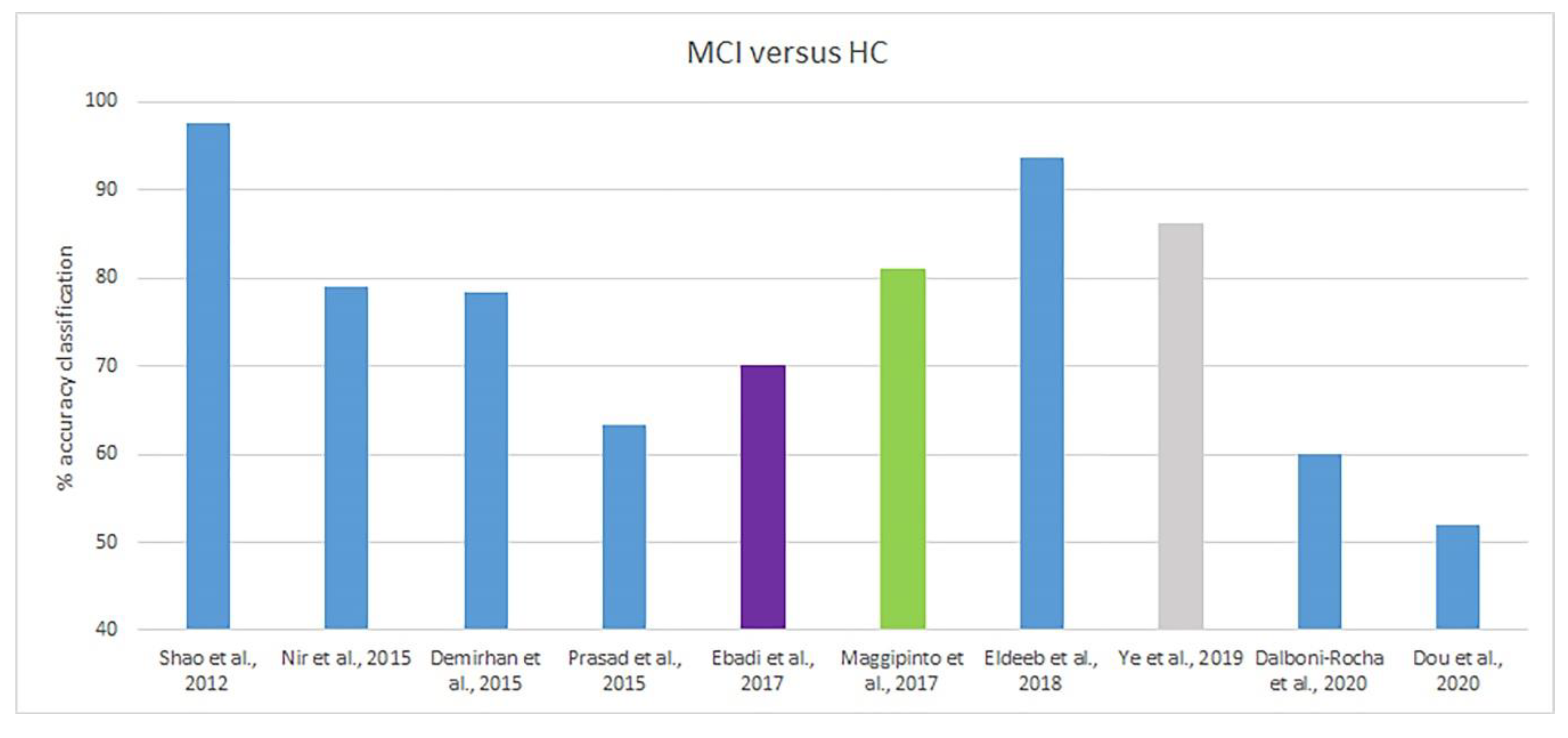
| Shao et al., 2012 [46] | DTI | 17 AD, 21 HC, 23 MCI | FA, MD, FD | SVM, k-NN, NB | AD/HC (SVM) | FD | 100.0 | - | - |
| FA | 92.1 | - | - | ||||||
| MD | 100.0 | - | - | ||||||
| MCI/HC (SVM) | FD | 97.7 | - | - | |||||
| FA | 84.1 | - | - | ||||||
| MD | 93.2 | - | - | ||||||
| MCI/AD (SVM) | FD | 85.0 | - | - | |||||
| FA | 82.5 | - | - | ||||||
| MD | 85.0 | - | - | ||||||
| Nir et al., 2015 [47] | DTI | 37 AD, 50 HC, 113 MCI | FA, MD | SVM | AD/HC | MD-fdr cva (n = 641) | 84.9 | 84.4 | 85.7 |
| FA-fdr cva (n = 214) | 77.8 | 78.2 | 77.3 | ||||||
| FA (n = 1080) | 74.5 | 75.0 | 73.9 | ||||||
| MD (n = 1080) | 80.6 | 79..2 | 82.4 | ||||||
| MCI/HC | MD-fdr cvl (n = 12) | 79.0 | 76.9 | 81.5 | |||||
| MD (n = 1080) | 68.3 | 69.8 | 66.4 | ||||||
| Demirhan et al., 2015 [48] | DTI | 43 AD, 70 HC, 114 MCI | FA | SVM | AD/HC | Whole WM Relieff1500 | 80.8 87.8 | - - | - - |
| MCI/HC | Whole WM Relieff1500 | 63.6 78.5 | - - | - - | |||||
| AD/MCI | Whole WM ReliefF1500 | 73.9 85.3 | - - | - - | |||||
| Prasad et al., 2015 [49] | DTI | 38 AD, 50 HC, 38 lMCI, 74 eMCI | Measures of connectivity | SVM | AD/HC | FI(N) + FL(N) | 78.2 | - | - |
| eMCI/HC | FI(N+M) | 59.2 | - | - | |||||
| lMCI/HC | FL(N) | 62.8 | - | - | |||||
| eMCI/lMCI | FI(N)+ FL(N) | 63.4 | - | - | |||||
| Ebadi et al., 2017 [50] | DTI | 15 AD, 15 HC, 15 MCI | FA | Logistic regression, random forest, NB, k-NN and SVM, ensemble | AD/HC | No Feat. selection | 73.3 | - | - |
| (Ensemble) | Feat. selection | 80.0 | - | - | |||||
| MCI/HC | No Feat. selection | 50.0 | - | - | |||||
| (Ensemble) | Feat. selection | 70.0 | - | - | |||||
| AD/MCI | No Feat. selection | 73.3 | - | - | |||||
| (Ensemble) | Feat. selection | 80.0 | - | - | |||||
| Maggipinto et al., 2017 [51] | DTI | 89 AD, 90 HCI, 90 MCI | FA, MD | Random forest | AD/HC | FA, non-nested | 87.0 | - | - |
| FA, nested | 75.0 | - | - | ||||||
| MD, non- nested | 83.0 | - | - | ||||||
| MD, nested | 76.0 | - | - | ||||||
| MCI/HC | FA, non-nested | 81.0 | - | - | |||||
| FA, nested | 59.0 | - | - | ||||||
| MD, non-nested | 79.0 | - | - | ||||||
| MD, nested | 60.0 | - | - | ||||||
| Eldeeb et al., 2018 [52] | DTI | 35 AD, 31 HC, 30 MCI | FA, MD | SVM | AD/HC | MD-SIFT | 98.3 | 97.0 | 100.0 |
| MD-SURF | 74.3 | 100 | 55.0 | ||||||
| FA-SIFT | 95.5 | 98.0 | 95.0 | ||||||
| FA-SURF | 62.0 | 92.0 | 20.0 | ||||||
| MCI/HC | MD-SIFT | 93.6 | 89.0 | 97.0 | |||||
| MD-SURF | 83.0 | 82.3 | 92.0 | ||||||
| FA-SIFT | 92.0 | 95.0 | 87.08 | ||||||
| FA-SURF | 58.0 | 49.0 | 77.0 | ||||||
| AD/MCI | MD-SIFT | 92.0 | 98.0 | 91.0 | |||||
| MD-SURF | 58.0 | 94.0 | 41.0 | ||||||
| FA-SIFT | 92.0 | 98.0 | 87.0 | ||||||
| FA-SURF | 56.0 | 100.0 | 20.0 | ||||||
| Multiclass | MD-SIFT | 89.0 | - | - | |||||
| MD-SURF | 55.0 | - | - | ||||||
| FA-SIFT | 87.0 | - | - | ||||||
| FA-SURF | 43.0 | - | - | ||||||
| Ye et al., 2019 [53] | DTI | 40 AD, 27 cMCI, 48 sMCI, 46 HC | Connectivity strength | PLS-DA | AD/HC | Whole-brain | 78.5 * | 71.9 | 70.1 |
| MDMR selected | 81.7 * | 67.0 | 76.2 | ||||||
| cMCI/HC | Whole-brain | 78.3 * | 54.7 | 85.0 | |||||
| MDMR selected | 86.2 * | 71.3 | 79.3 | ||||||
| Dalboni da Rocha et al., 2020 [54] | DTI | 15 AD, 15 MCI, 15 HC | FA | SVM | AD/HC | Whole-brain | 80 | - | - |
| Hippocampal Cingulum | 87 | - | - | ||||||
| Parahippocampal Gyrus | 83 | - | - | ||||||
| MCI/HC | Whole-brain | 60 | - | - | |||||
| Parahippocampal Cingulum | 57 | - | - | ||||||
| Parahippocampal Gyrus | 47 | - | - | ||||||
| AD/MCI | Whole-brain | 77 | - | - | |||||
| Hippocampal Cingulum | 83 | - | - | ||||||
| Parahippocampal Gyrus | 67 | - | - | ||||||
| Dou et al., 2020 [55] | DTI | 89 AD, 71 aMCI, 82 HC | FA, MD, DR, DA | SVM, LDA, XGB | AD/HC (SVM) | Dataset 1 | 82.5 | 85.1 | 79.4 |
| Dataset 2 | 82.3 | 80.9 | 82.3 | ||||||
| aMCI/HC (SVM) | Dataset 1 | 52.0 | 24.7 | 74.6 | |||||
| Dataset 2 | 51.2 | 24.3 | 74.4 | ||||||
| AD/aMCI (SVM) | Dataset 1 | 77.7 | 89.3 | 61.7 | |||||
| Dataset 2 | 82.2 | 83.3 | 81.0 | ||||||
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Billeci, L.; Badolato, A.; Bachi, L.; Tonacci, A. Machine Learning for the Classification of Alzheimer’s Disease and Its Prodromal Stage Using Brain Diffusion Tensor Imaging Data: A Systematic Review. Processes 2020, 8, 1071. https://doi.org/10.3390/pr8091071
Billeci L, Badolato A, Bachi L, Tonacci A. Machine Learning for the Classification of Alzheimer’s Disease and Its Prodromal Stage Using Brain Diffusion Tensor Imaging Data: A Systematic Review. Processes. 2020; 8(9):1071. https://doi.org/10.3390/pr8091071
Chicago/Turabian StyleBilleci, Lucia, Asia Badolato, Lorenzo Bachi, and Alessandro Tonacci. 2020. "Machine Learning for the Classification of Alzheimer’s Disease and Its Prodromal Stage Using Brain Diffusion Tensor Imaging Data: A Systematic Review" Processes 8, no. 9: 1071. https://doi.org/10.3390/pr8091071
APA StyleBilleci, L., Badolato, A., Bachi, L., & Tonacci, A. (2020). Machine Learning for the Classification of Alzheimer’s Disease and Its Prodromal Stage Using Brain Diffusion Tensor Imaging Data: A Systematic Review. Processes, 8(9), 1071. https://doi.org/10.3390/pr8091071







