1. Introduction
Artificial intelligence (AI) and machine learning (ML) hold great promise for advancing biomedicine [
1,
2,
3]. In Alzheimer’s disease (AD), as in most other areas of biomedicine, AI and ML have been deployed mainly in automatic diagnosis on the basis of imaging data, augmented in some cases with biomarker or omics data, clinical findings, or demographics [
4,
5]. Accuracies in excess of 95% correct AD diagnosis are commonly reported. Despite the success in automatic diagnosis, the use of AI and ML for the purpose of drug discovery in AD is still an emerging area.
AI and ML have been combined with more traditional techniques (e.g., quantitative structure activity relationship, or QSAR) for the identification of new compounds potentially active against targets implicated in neurodegenerative diseases, including AD [
6]. Although the overwhelming majority of these studies were focused on single targets, the development of compounds capable of binding multiple targets has become a “major goal” [
6]. AI and ML have also been applied in “virtual screening” for novel compounds against specific targets. Such use against AD targets has been suggested, but here again the focus is on single drugs against single targets [
7].
The repurposing of old drugs for treatment of neurodegenerative diseases is another emerging effort [
8,
9]. The main benefit of the repurposing approach is the prior approval for safety of the candidate drugs and the consequent accessibility “to academic institutions, government and research council programmes, charities and not-for-profit organizations”, which complement “the work of pharmaceutical and biotechnology companies” and substantially reduce “the time and cost involved in progressing the potential treatment into clinical trials” [
9]. Drug candidates for AD repurposing have been identified on the basis of epidemiological and/or empirical (in vitro/in vivo) evidence, or by panels of experts. Though the emphasis of most repurposing efforts has been on single drugs, interest is growing in the AD community for using combinations of repurposed drugs, reflecting the now common understanding of AD as a multifactorial disorder [
10,
11,
12,
13,
14]. The main challenge remains to identify the best candidates from among the many possible combinations of repurposed drugs.
The identification of potentially effective AD treatments is urgently needed. The development of treatments for slowing the progression of AD and other neurodegenerative disorders is lagging far behind the improvements in treatments for other major diseases [
15,
16,
17]. Consequently, increasing numbers of people in developed countries are living long enough to become demented. No reasonable AD treatment approach should go unexplored. Considering the rapidity with which already approved and widely used drugs can be repurposed, the effort to identify potentially effective combinations of repurposed drugs for AD treatment should be given high priority.
AI and ML are uniquely well suited to address the challenge of identifying potentially effective combinations of repurposed drugs for AD treatment. They could be used to leverage the knowledge contained in AD databases to make the best attainable predictions as to which combinations of repurposed drugs could be used effectively to treat AD. Despite the availability of large databases on aging and dementia, no effort has yet been made to apply ML to the data they contain in order to predict effective combinations of repurposed drugs. This initial study is the first of its kind.
The goal of this project is to use ML to extract the knowledge contained in two leading AD databases, and use the extracted knowledge to identify drug combinations that could be effective in treating AD-associated neurodegeneration. The two AD datasets were obtained from the Rush Alzheimer’s Disease Center (RADC) and the National Alzheimer’s Coordinating Center (NACC). Both databases are highly regarded and used worldwide. Readers interested in epidemiological details and other database specifics are referred to the RADC and NACC websites (the relevant URLs are
https://www.radc.rush.edu/ for RADC and
https://www.alz.washington.edu/ for NACC).
NACC encompasses the data from the 29 Alzheimer’s disease centers (ADCs) that are funded by the National Institute on Aging of the National Institutes of Health of the United States [
18,
19]. RADC is an ADC but most of its dataset comes from the participants in the Religious Orders Study and the Rush Memory and Aging Project (ROSMAP) [
20,
21]. A small amount of legacy RADC data are included in NACC (less than 1.4% of total NACC data). All RADC data were removed from the NACC dataset for this study. The two datasets used for ML in this study are completely independent of each other (
Supplementary Note S1).
Owing to vast increases in high-performance computing resources, artificial neural networks (ANNs) have become the state of the art in AI for generalizing from available data and making predictions. ANNs can be configured and trained in many different ways. This initial study employed an ANN as its generalizer/predictor, and its approach had two distinguishing features.
First, rather than simply choose one from among the many possible ANNs, a global optimization procedure was used to evaluate a large number of possible configurations to find which one would provide the best predictions (see Methods section). This process showed that the ANNs that were optimal for the ROSMAP or NACC datasets were essentially the same. Second, rather than combine all the available data to train one ANN, two ANNs having the same configuration were trained separately on the ROSMAP or NACC datasets and their predictions concerning the potency of many drug combinations were compared (see Results section). The strong correlation between the two sets of predictions increases confidence in the jointly determined best drug combinations.
2. Methods
Artificial neural networks (ANNs) are computational devices that are composed of many interconnected processing units. The units can be arranged in many different ways. In principle, any unit could be connected to any other unit, but in most ANNs, units are structurally organized. The units in many commonly used ANNs are organized into layers with an input layer, an output layer, and one or more internal layers that progress in order from the input to the output layer. Forward connections project forward, in the direction from the input to the output, and can connect any unit in a previous layer (e.g., input or lower-order internal) to any unit in a subsequent layer (e.g., higher-order internal or output). Conversely, recurrent connections project back, in the direction from the output to the input, and can connect any unit in a subsequent layer to any unit in a previous layer. Recurrent connections can also occur between units in the same layer.
The variables of an ANN are the weights of the connections between the units. The values of these connection weights are usually adjusted using some form of ML. The ML technique used to train ANNs in this study is a type of supervised learning, which requires a dataset composed of many input/desired-output examples. In supervised learning, an input is processed to form an output, and the actual output is compared to the desired output to form an error. The error is used to modify the weights of the connections between the units in the network, in such a way that the error between the actual and desired output for that input is reduced. Over many training cycles (i.e., many input/desired-output presentations), the error can be reduced substantially and the match between the actual and desired outputs can become quite close.
Most units in ANNs compute the sum of the weighted connections they receive from other units. The activation level of a linear unit is simply this weighted input sum. The activation level of a nonlinear unit is the weighted input sum after it is operated on by a nonlinear function (usually a sigmoidal function that bounds the weighted input sum in the range (0, 1) or (−1, +1)). Such simple units are the most common. ANNs can sometimes contain more complex units, which themselves are composed of a set of interconnected simple units (see [
22] for further background on ANNs).
The ANNs considered here had three layers: an input, an output, and one internal layer. Forward connections connected input to internal units and internal to output units. Linear units composed the input layer. Each input unit represented the value of an input variable, scaled into the range (0, 1). The output layer was composed of nonlinear units that computed the sum of their weighted connections from internal units and imposed a sigmoidal activation function on it, squashing it into the range (0, 1). The numbers of units in the input and output layers are fixed by the dataset on which the ANN is trained (see below and
Figure 1). There is no limit on the number of internal layers, or on the number of internal units in each internal layer, of an ANN.
Compound units known as long short-term memory units (LSTMs) composed the internal layer. Several nonlinear units compose each LSTM, where some units act as gates that modulate the activations of other units [
23,
24] (
Supplementary Note S2). The LSTMs in the ANNs considered here received forward connections from input units and recurrent connections from each other. LSTMs are so called because they have their own inner memory, mediated by an inner recurrent connection. The outer recurrent connections between LSTMs can enhance this memory. The recurrent connections (inner and/or outer) of a recurrently connected layer of LSTMs endows an ANN with considerable power to process information in time. If changes over time are useful for reproducing desired outputs given temporal sequences of inputs, then a three-layered ANN having a recurrently connected internal layer of LSTMs will exploit them.
The power and versatility of an ANN having a recurrently connected layer of LSTMs can be further appreciated by noting that each time step of processing in a recurrent network is equivalent to an additional internal layer in a feedforward network [
22]. Thus, an ANN having a recurrently connected layer of LSTMs can subsume the processing capability of many types of simpler ANNs, including feedforward ANNs having multiple internal layers, and feedforward or recurrent networks of simple units or of compound units with various types of inner gating. However, a given processing task may not require the full power of an ANN having a recurrently connected layer of LSTMs. The best configuration for a given application may be a simplification of this network type. A determination of the best configuration was a distinctive feature of this study (see below).
Recurrent LSTM networks can be trained using a supervised learning algorithm known as backpropagation through time (BPTT) [
22,
25]. BPTT draws temporal sequences of input/desired-output examples from the dataset at random (with replacement), and continues training the ANN over many iterations, thereby reducing the error over the dataset. The version of BPTT used here has three parameters: starting and ending learning rate and fixed momentum. The learning rate decreases from starting to ending as the training iterations proceed from first to last, and the momentum determines how much of the previous iteration’s learning should be applied on the current iteration. These learning parameters will likely vary between ANNs having different configurations.
Once trained, the ANN not only can reproduce the desired outputs for the inputs in the dataset but can also generalize the knowledge it has extracted, and can predict the outputs for inputs on which it has not been trained. To assess generalizability, the dataset is divided into a training set (usually 75% of the dataset) and a testing set (the other 25%). Then the ANN is trained on the training set and tested on the testing set. The generalization error is the total error between the actual (predicted) and desired outputs over the testing set.
BPTT entails two sources of randomness: random initial connection weights and random presentation of input/desired-output sequences. Consequently, ANNs trained on the same dataset can vary in their outputs, and the best predictions are derived from the averaged outputs of many trained ANNs (around ten trained ANNs are usually considered sufficient) [
26]. Generalizability, which is essentially the ability of an ANN to predict desired outputs over the testing set, is likewise best assessed by averaging the actual outputs of many retrained ANNs.
The overall goal of this work was achieved in three steps. The first was to process the data contained in two AD databases into a form suitable for training ANNs using ML. The second step was to choose the best ANN configuration for this application. The third step was to use the best ANN to perform the computational drug-combination screens that provide the results of this study.
2.1. Processing the Data from Two AD Databases
The ROSMAP and NACC databases have similar fields which included, for each participant, age at visit, demographics (sex, race, etc.), tobacco and alcohol use, comorbidities (cancer, stroke, etc.), drugs taken, and the results over a battery of cognitive tests (e.g., number of different animal types recalled in one minute, or number of facts remembered from a text excerpt). The data in these fields were arranged into input/desired-output pairs, where the desired outputs were the cognitive test results and the inputs were the values for all the other fields for each visit. The number of desired outputs (cognitive scores) and inputs (all fields except cognitive scores) were 25 and 113 for ROSMAP, and 57 and 101 for NACC, respectively (
Figure 1).
AD is a disease process that proceeds in time. Therefore, age is a key variable, and changes with age of other input variables could also affect cognitive scores. The temporal sequences of age and other inputs could contain information useful for reproducing cognitive scores from input data. The form of ANN chosen for this study, a three-layered ANN with a recurrently connected internal layer of LSTMs, exploits information contained in temporal sequences of inputs if doing so contributes to the reproduction of desired outputs. For this reason, all of the input/desired-output pairs were arranged into temporal sequences for each participant. Specifically, the input/desired-output pairs for each participant were ordered by their age at each visit into an age-advancing sequence. On each training iteration, the ANN was presented with the age-advancing sequence for a randomly chosen participant. The input/desired-output pairs in that participant’s sequence were presented one at a time, but in order of age at visit so that temporal information was preserved. In this way, any information contained in the temporal sequences themselves would be exploited by the ANN. The ANN was trained only on age-advancing sequences of at least ten ages (
Supplementary Note S1).
Both the ROSMAP and NACC databases had missing data. Overall, 3% and 39% of values were missing from the ROSMAP and NACC databases, respectively. Missing values were spread over the dataset such that 87% and 100% of input/desired-output examples (i.e., pairs) had at least one missing value in the ROSMAP and NACC datasets, respectively. In order to ensure that an adequate number of input/desired-output pairs were available for training, workarounds of missing data were implemented.
Missing input data were replaced by the average of the data for that field. This is consistent with the maximum-entropy approach to missing data that is standard in ML applications [
27]. Missing desired-output data were left blank, but during training all errors associated with missing desired-output data were set to zero. Because connection weight modifications in BPTT are based on error, no weight modifications were made due to missing desired-output data. While the weights onto output units representing absent desired-output values were not trained, all other network connection weights were trained on the basis of the desired-output values that were present in any given input/desired-output example.
Redundant database fields were removed. Ideally, the values assigned to an input or desired output should increase monotonically with the variable it represents, but database field codes can deviate widely from that simple relationship. For example, the scoring for frontotemporal dementia in the NACC database is 0, 1, or 8 for dementia that is not frontotemporal, dementia that is frontotemporal, or no dementia at all. To make it monotonic, the frontotemporal dementia score was rearranged to 0, 1, or 2 for no dementia, dementia that is not frontotemporal, and dementia that is frontotemporal, respectively. Specific input or desired-output values were rearranged or reassigned, when necessary, to bring them all into a monotonically increasing regime. Finally, the input and desired-output data (except for missing desired-output data, which simply remained missing) were normalized to (0, 1).
2.2. Choosing the Best ANN Configuration
The combination of ANN features and ML parameters providing the best generalizability was determined using a genetic algorithm (GA) [
28,
29] (
Supplementary Note S3). Because of the randomness inherent in GAs, the best solution represents the consensus over multiple GA runs. Ten GA runs were performed separately for the ROSMAP and NACC datasets, producing twenty runs altogether (the computational intensity of GAs precluded larger numbers of GA runs). The GA results were similar between the two datasets and the consensus was obvious (
Supplementary Table S1).
Using GA optimization, the best (i.e., consensus) ANN for this application lacked recurrent connections both inside and between LSTMs. This indicates that memory capability, and its attendant ability to process sequences temporally, did not improve ANN generalizability. The reason, probably, is because age, which provides a direct and accurate measure of time, was included as an input so the ANN did not need to infer time through temporal processing. Furthermore, the best ANN for this application had LSTMs simplified to a single gating interaction. Because the internal units in the best network were such drastic simplifications of LSTMs, they are simply referred to as compound units (CUs). The best (i.e., consensus) ANN is diagrammed in
Figure 1.
The use of gating is common in ANN applications in which different internal units learn to specialize for different subsets of the dataset [
30]. The fact that the consensus ANN employed gated units suggests that such specialization improved generalizability in this application. The consensus ANN had about 80 compound, gated units in its internal layer, and its optimal ML parameters were 0.0600, 0.0006, and 0.0002 for starting and ending learning rate and fixed momentum, respectively.
2.3. Performing the Computational Drug-Combination Screens
Consensus ANNs were trained on the ROSMAP or NACC datasets separately because their cohorts differ markedly (community versus clinical), and because their corresponding databases have many structural differences (e.g., different numbers and kinds of cognitive tests). Therefore, the datasets cannot be merged without making many unsupportable assumptions. The trained ANNs were used to screen all possible combinations of the drugs in common between the two databases.
The drugs in both databases were listed in drug categories rather than as specific drugs. The categories in common were: nonsteroidal anti-inflammatory drugs (NSAIDs, mainly cyclooxygenase-2 (COX2) inhibitors and aspirin); angiotensin converting enzyme (ACE) inhibitors; angiotensin II receptor blockers (ARBs); calcium channel blockers; beta blockers; diuretics; vasodilators; anticoagulants; lipid-lowering drugs; antidiabetics; estrogen or progestin; anti-adrenergics; antidepressants; antipsychotics; anxiolytics; anti-Parkinson’s drugs; and anti-Alzheimer’s drugs (mainly acetylcholinesterase inhibitors and N-methyl-D-aspartate receptor blockers). There are 131,072 combinations of these 17 drug categories, and consensus ANNs, trained on either the ROSMAP or NACC datasets, were tested on every combination. This constituted two, separate computational screens (ROSMAP or NACC) over all 100,000+ drug combinations.
The inputs to the ANNs were all data fields other than cognitive scores. They included demographics, tobacco and alcohol use, and comorbidities as well as drugs taken. Valid comparison of the predicted potencies of different drug combinations required testing each drug combination using a standard input, in which all inputs besides drugs were the same. The standard input was based on an age-advancing sequence of 100 (non-integer, real number) ages from 50 to 110 years, which encompasses the age range of the participants in the two databases. All of the other inputs, separately for either database, were resampled to conform to this sequence by computing a value that is representative of each input field at each age in the standard sequence (
Supplementary Note S4).
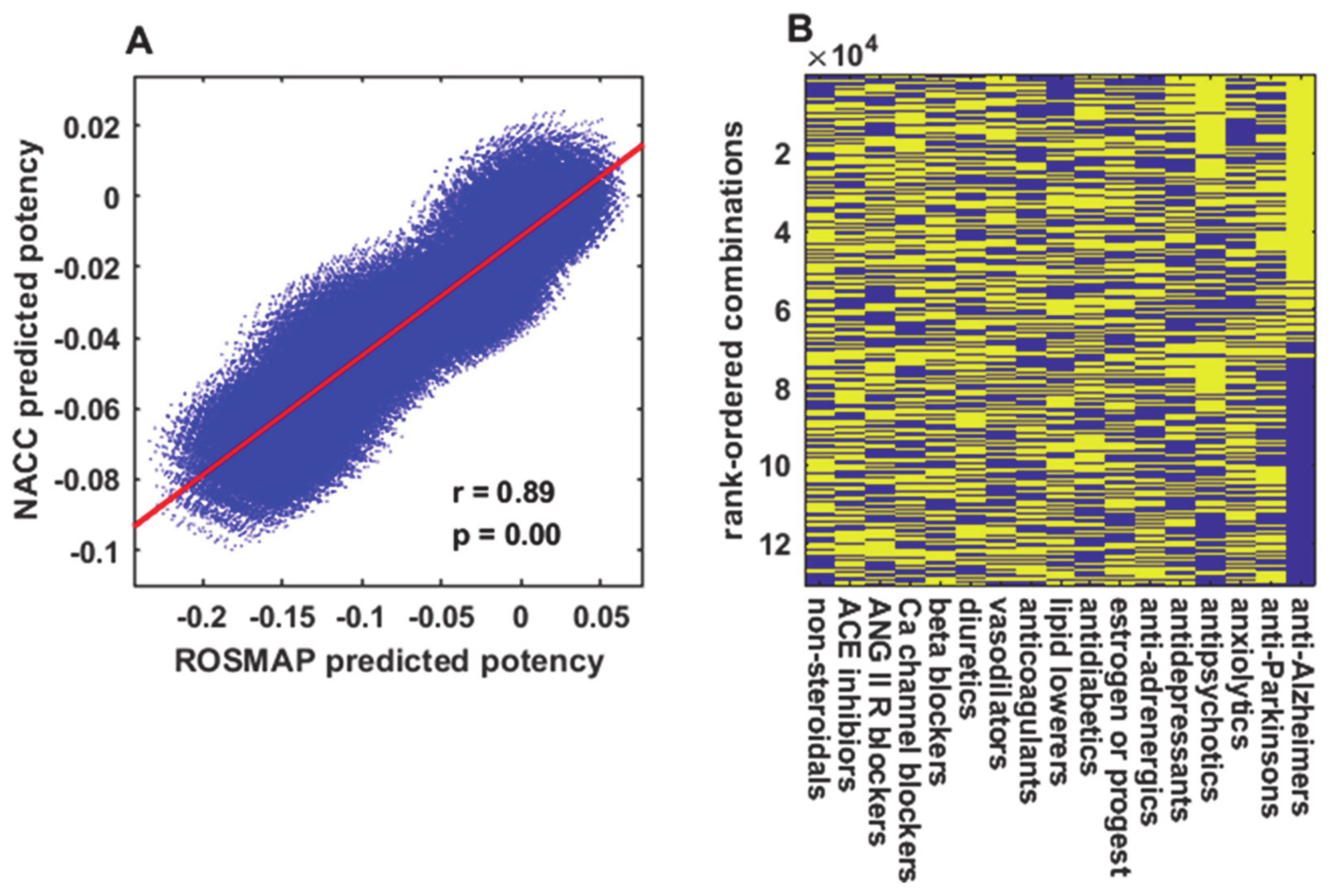
Then a consensus ANN had its weights randomized and was trained on a full dataset (either ROSMAP or NACC), and the cognitive scores predicted by the trained ANN for each drug combination at each age in the standard age-advancing sequence were found. Note that each output unit (25 for ROSMAP or 57 for NACC) represents the ANN’s prediction for one cognitive test. A combined cognitive score for each age in the standard sequence (a single number for each age) can be computed by averaging over the scores of the different cognitive tests, each represented by an individual output unit (an average over 25 output units for ROSMAP or 57 output units for NACC). For illustrative purposes, the combined cognitive scores as predicted by one ANN for many representative (not all 100,000+) drug combinations versus age are shown in
Figure 2.
To improve the accuracy of the prediction (see Methods section), the processes of randomization, retraining, and predicting were repeated 100 times for either dataset, and the predicted scores for each cognitive test, each represented by an individual output unit, were averaged over the 100 ANNs. Then the combined cognitive score was computed by averaging over the averaged predicted outputs. Thus, the combined cognitive scores actually used to compute drug-combination predicted potencies were based not on one, but on the average of 100 retrained consensus ANNs.
To obtain a single number for the predicted potency of each drug combination, the differences between the combined cognitive scores predicted for that drug combination and the cognitive scores predicted for no drugs at all ages in the age-advancing sequence were averaged. Briefly summarized, the drug-combination predicted potencies are the averaged (differences) over 100 ages, of averaged predicted scores over a number of different cognitive tests (25 for ROSMAP or 57 for NACC), each represented by the averaged output unit activations of 100 retrained consensus ANNs. Thus, predicted potencies are relative measures where the reference values are the no-drug predictions. The results of this study are reported in terms of the potency of each specific drug combination, as predicted from consensus ANNs trained either on the ROSMAP or NACC datasets. For brevity, these separately trained ANN predictions will be referred to as ROSMAP predictions or NACC predictions.
4. Discussion
The ten-best combinations of the eleven drugs that excluded drugs acting mainly on the cognitive and/or psychological level included NSAID, anticoagulant, lipid-lowering, and antihypertensive drugs, as well as estrogen/progestin. Drugs from all of these categories have been shown to reduce AD risk; however they all show disappointing results when used singly in clinical trials [
31,
32,
33,
34,
35,
36,
37,
38,
39,
40,
41,
42,
43]. The use of combinations of these and other drugs remains an attractive option [
10,
11,
12,
13,
14].
However they are ranked, the ten best combinations of the ten or eleven drugs that excluded those that target cognition or mood directly, jointly determined from the ROSMAP and NACC predictions, included three to eight drugs. This finding supports the general idea that multiple drugs (including hormones) may indeed be effectively used to treat neurodegeneration, which is a multifactorial disease process [
10,
11,
12,
13,
14]. The ten best combinations are not random but tend to include drugs from the same categories: NSAID, anticoagulant, lipid-lowering, and antihypertensive drugs, as well as estrogen/progestin.
One possible interpretation of the results is that elderly individuals who take these drugs have better cognitive function than those who do not, because the conditions these drugs treat are associated with better cognitive function. This interpretation is ruled out by statistical analysis of the ROSMAP and NACC datasets showing that comorbidities, singly or in combination, do not improve cognitive function (
Supplementary Figure S6 and Figure S7).
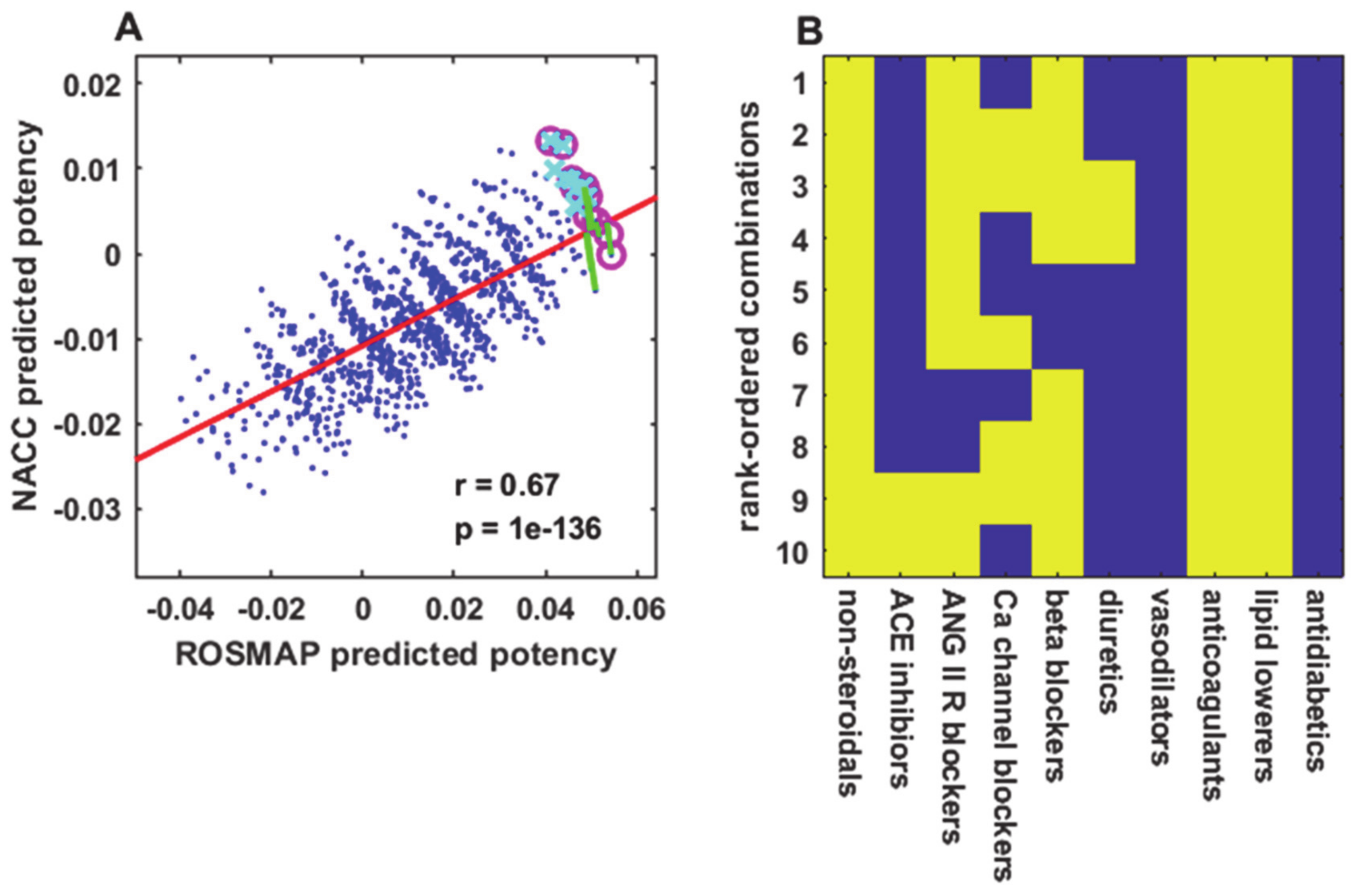
It is interesting that the predicted cognitive scores for many drug combinations are higher than those for the no-drug case. This can be seen in
Figure 2 with reference to the no-drug prediction (red line), and in
Figure 3A,
Figure 4A, and
Figure 5A for all points with ROSMAP and NACC predictions higher than zero (the no-drug prediction). This result implies that certain drug combinations may improve cognitive function beyond what would be expected if each drug merely normalized its primary treatment target (e.g., lipid-lowering and antihypertensive drugs that normalize lipid levels and blood pressure, respectively). Such improvements beyond the no-drug condition could result from synergies in primary and/or secondary drug effects. For example, certain NSAIDs (mainly COX2 inhibitors) not only reduce inflammation but also reduce amyloid-β production [
44,
45,
46], while certain antihypertensive drugs also have anti-inflammatory properties [
47,
48,
49].
Drugs that act mainly on the cognitive or psychological level were excluded from most analyses because they could mask the effects of other drugs, and combinations thereof, that might improve cognitive function by slowing down an underlying process of neurodegeneration. The leading cases in point were the anti-Alzheimer’s drugs (mainly acetylcholinesterase inhibitors and N-methyl-D-aspartate receptor blockers), which practically dichotomized the entire set of 100,000+ drug combination predictions into those with and those without anti-Alzheimer’s drugs (
Figure 3B). Other drugs that act on the cognitive or psychological level, mainly antipsychotics and antidepressants, may similarly mask the potential effects of other drugs on neurodegeneration (
Supplementary Figure S3B). Anti-Alzheimer’s drug use was 6% and 27% in the ROSMAP and NACC datasets, respectively, and antidepressant and antipsychotic drugs were used more often when participants also used anti-Alzheimer’s drugs. Usage of other drugs was about the same whether or not participants also used anti-Alzheimer’s drugs (
Supplementary Figure S8).
The exclusion of drugs that act mainly on the cognitive or psychological level in no way implies that those drugs would not also slow down the neurodegenerative process, or would not synergize with other drugs that do. Though predictions were derived for combinations of subsets of the 17 drug categories in common between the ROSMAP and NACC databases (i.e., subsets that excluded drugs that target cognition or mood), the ANNs were trained on input patterns that included all 17 categories. Thus, any synergies present in the datasets between the drugs in the subsets (i.e., NSAID, anticoagulant, lipid-lowering, and antihypertensive drugs, as well as estrogen/progestin) and drugs that act mainly on the cognitive or psychological level (i.e., anti-Alzheimer’s and anti-Parkinson’s drugs as well as anti-adrenergics, antidepressants, antipsychotics, and anxiolytics) would have been learned by the ANNs and factored into all predictions.
The drugs appearing in the ten best combinations in this study overlap with those in a previous study in which ML was used to train a computational model of brain inflammation [
50]. Model predictions of drug-combination efficacy were significantly correlated with the cognitive benefits of the same drug combinations as determined from the ROSMAP dataset. The ten best combinations from the previous study also included NSAIDs (COX2 inhibitors and aspirin) as well as antihypertensive drugs (ACE inhibitors, angiotensin II receptor blockers, calcium channel blockers, and beta blockers). Earlier computational studies of potential anti-Alzheimer’s drug interactions were based on process-driven models, rather than on data-driven models as trained by ML. Those models suggested that NSAIDs could be effectively combined with antidiabetic drugs or estrogen as anti-Alzheimer’s therapies [
51,
52]. The drug categories in the previous studies were not the same as in this study. That discrepancy limits the value of directly comparing their results.
All drugs in the computational combination screens in this study are approved. Many elderly people already take some of the drugs included in the ten best combinations. Where not contraindicated, other drugs could be added to their regimens to complete some of the predicted combinations, for the purpose of conducting controlled clinical trials to test them. The most promising candidates for testing include combinations of NSAID (COX2 inhibitors and aspirin), anticoagulant, and lipid-lowering drugs along with antihypertensive drugs such as angiotensin II receptor, calcium channel, or beta blockers, or ACE inhibitors. Combinations of NSAID, anticoagulant, and lipid-lowering drugs that also include more than one antihypertensive drug and, where not contraindicated, estrogen/progestin would be of particular interest.
For simplicity in this initial effort, the trained ANNs were used to predict the efficacy of drug combinations given a representative or “standard” patient (
Supplementary Note S4), but they could be used alternatively to predict the efficacy of drug combinations given patients with specific demographic, comorbid, and other characteristics (there are tens of thousands of such combinations given the various ROSMAP or NACC input fields). For example, the ANNs could be used to predict the efficacy of all combinations of any set of drugs in common between the ROSMAP and NACC databases, including certain antihypertensive drugs, in elderly Hispanic women who neither smoke tobacco nor drink alcohol, and who are diabetic and take diabetes medication as well as a statin, but who are not hypertensive. No retraining would be required to make predictions concerning patients with any set of existing database characteristics. Retraining ANNs on augmented datasets that include additional input fields as they become available, such as genetic variants, would allow even finer tuning of predictions for specific subclasses of patients.
The current study has two major limitations. First, the drugs in the databases are not identified individually but by category. Many categories include drugs of different classes, which may affect neurodegeneration in different ways. Second, the ideal desired endpoints for the clinical trials would be reductions in biomarkers of neurodegeneration rather than improvements in cognitive ability, but the databases so far exclude biomarkers. AIs trained via ML on large datasets from databases that identify drugs individually, and that include biomarkers of neurodegeneration, could be used to make better predictions concerning combinations of repurposed drugs that would be effective in treating AD specifically and neurodegeneration more generally.