Hydrogenation of Trans,Trans-Muconic Acid to Bio-Adipic Acid: Mechanism Identification and Kinetic Modelling
Abstract
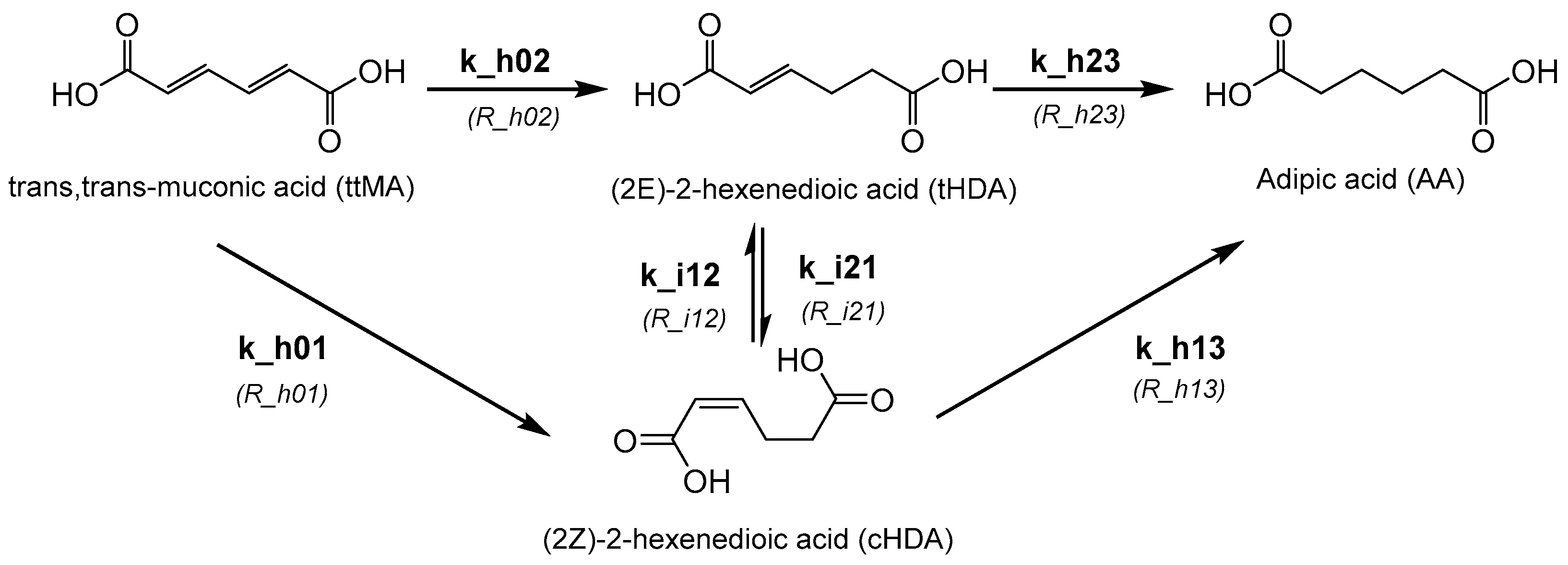
:1. Introduction
2. Materials and Methods
2.1. Experimental
2.2. Kinetic Modelling
3. Results and Discussion
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| ttMA | trans,trans-muconic acid |
| tHDA | (2E)-2-hexenedioic acid |
| cHDA | (2Z)-2-hexenedioic acid |
| AA | adipic acid |
| LHHW | Langmuir–Hinshelwood–Hougen–Watson model |
| SSE | sum of squared errors |
| R2 | coefficient of determination |
References
- Bart, J.C.J.; Cavallaro, S. Transiting from adipic acid to bioadipic acid. 1, Petroleum-based processes. Ind. Eng. Chem. Res. 2015, 54, 1–46. [Google Scholar] [CrossRef]
- Becker, J.; Lange, A.; Fabarius, C.; Wittmann, C. Top value platform chemicals: Bio-based production of organic acids. Curr. Opin. Biotechnol. 2015, 36, 168–175. [Google Scholar] [CrossRef]
- Collias, D.I.; Harris, A.M.; Nagpal, V.; Cottrell, L.W.; Schultheis, M.W. Biobased terephthalic acid technologies: A literature review. Ind. Biotechnol. 2014, 10, 91–105. [Google Scholar] [CrossRef]
- Xie, N.-Z.; Liang, H.; Huang, R.-B.; Xu, P. Biotechnological production of muconic acid: Current status and future prospects. Biotechnol. Adv. 2014, 32, 615–622. [Google Scholar] [CrossRef] [PubMed]
- Kruyer, N.S.; Peralta-Yahya, P. Metabolic engineering strategies to bio-adipic acid production. Curr. Opin. Biotechnol. 2017, 45, 136–143. [Google Scholar] [CrossRef] [PubMed]
- Polen, T.; Spelberg, M.; Bott, M. Toward biotechnological production of adipic acid and precursors from biorenewables. J. Biotechnol. 2013, 167, 75–84. [Google Scholar] [CrossRef] [PubMed]
- Bui, V.; Lau, M.K.; Macrare, D.; Schweitzer, D. Methods for Producing Isomers of Muconic Acid and Muconate Salts. U.S. Patent 20130030215 A1, 1 January 2013. [Google Scholar]
- Niu, W.; Draths, K.M.M.; Frost, J.W.W. Benzene-free synthesis of adipic acid. Biotechnol. Prog. 2002, 1, 201–211. [Google Scholar] [CrossRef] [PubMed]
- Johnson, C.W.; Salvachúa, D.; Khanna, P.; Smith, H.; Peterson, D.J.; Beckham, G.T.; Salvachua, D.; Khanna, P.; Smith, H.; Peterson, D.J.; et al. Enhancing muconic acid production from glucose and lignin-derived aromatic compounds via increased protocatechuate decarboxylase activity. Metab. Eng. Commun. 2016, 3, 111–119. [Google Scholar] [CrossRef] [PubMed]
- Vardon, D.R.; Franden, M.A.; Johnson, C.W.; Karp, E.M.; Guarnieri, M.T.; Linger, J.G.; Salm, M.J.; Strathmann, T.J.; Beckham, G.T. Adipic acid production from lignin. Energy Environ. Sci. 2015, 8, 617–628. [Google Scholar] [CrossRef]
- Tuck, C.O.; Perez, E.; Horváth, I.T.; Sheldon, R.A.; Poliakoff, M. Valorization of biomass: Deriving more value from waste. Science 2012, 337, 695–699. [Google Scholar] [CrossRef]
- Averesch, N.J.H.; Krömer, J.O. Tailoring strain construction strategies for muconic acid production in S. Cerevisiae and E. Coli. Metab. Eng. Commun. 2014, 1, 19–28. [Google Scholar] [CrossRef]
- Suástegui, M.; Yu Ng, C.; Chowdhury, A.; Sun, W.; Cao, M.; House, E.; Maranas, C.D.; Shao, Z. Multilevel engineering of the upstream module of aromatic amino acid biosynthesis in saccharomyces cerevisiae for high production of polymer and drug precursors. Metab. Eng. 2017, 42, 134–144. [Google Scholar] [CrossRef] [PubMed]
- Rorrer, N.A.; Dorgan, J.R.; Vardon, D.R.; Martinez, C.R.; Yang, Y.; Beckham, G.T. Renewable unsaturated polyesters from muconic acid. ACS Sustain. Chem. Eng. 2016, 4, 6867–6876. [Google Scholar] [CrossRef]
- Rorrer, N.A.; Vardon, D.R.; Dorgan, J.R.; Gjersing, E.J.; Beckham, G.T. Biomass-derived monomers for performance-differentiated fiber reinforced polymer composites. Green Chem. 2017, 19, 2812–2825. [Google Scholar] [CrossRef]
- Matthiesen, J.E.; Suàstegui, M.; Wu, Y.; Viswanathan, M.; Qu, Y.; Cao, M.; Rodriguez-Quiroz, N.; Okerlund, A.; Kraus, G.; Raman, D.R.; et al. Electrochemical conversion of biologically produced muconic acid: Key considerations for scale-up and corresponding technoeconomic analysis. ACS Sustain. Chem. Eng. 2016, 4, 7098–7109. [Google Scholar] [CrossRef] [Green Version]
- Lu, R.; Lu, F.; Chen, J.; Yu, W.; Huang, Q.; Zhang, J.; Xu, J.; Rui, L.; Lu, F.; Chen, J.; et al. Production of diethyl terephthalate from biomass-derived muconic acid. Angew. Chemie Int. Ed. 2015, 55, 249–253. [Google Scholar] [CrossRef]
- Frost, J.W.; Miermont, A.; Schweitzer, D.; Bui, V.; Wicks, D.A. Terephthalic and Trimellitic Based Acids and Carboxylate Derivatives Thereof. U.S. Patent US8367858 B2, 5 February 2013. [Google Scholar]
- Capelli, S.; Motta, D.; Evangelisti, C.; Dimitratos, N.; Prati, L.; Pirola, C.; Villa, A. Bio Adipic Acid Production from Sodium Muconate and Muconic Acid: A Comparison of Two Systems. ChemCatChem 2019, 11, 3075–3084. [Google Scholar] [CrossRef]
- Bart, J.C.J.; Cavallaro, S. Transiting from adipic acid to bioadipic acid. part II. Biosynthetic pathways. Ind. Eng. Chem. Res. 2015, 54, 567–576. [Google Scholar] [CrossRef]
- Vardon, D.R.; Rorrer, N.A.; Salvachua, D.; Settle, A.E.; Johnson, C.W.; Menart, M.J.; Cleveland, N.S.; Ciesielski, P.N.; Steirer, K.X.; Dorgan, J.R.; et al. Cis,cis-muconic acid: Separation and catalysis to bio-adipic acid for nylon-6,6 polymerization. Green Chem. 2016, 18, 3397–3413. [Google Scholar] [CrossRef]
- Scelfo, S.; Pirone, R.; Russo, N. Highly efficient catalysts for the synthesis of adipic acid from cis,cis-muconic acid. Catal. Commun. 2016, 84, 98–102. [Google Scholar] [CrossRef]
- Capelli, S.; Rosengart, A.; Villa, A.; Citterio, A.; Di Michele, A.; Bianchi, C.L.L.; Prati, L.; Pirola, C. Bio-adipic acid production by catalysed hydrogenation of muconic acid in mild operating conditions. Appl. Catal. B Environ. 2017, 218, 220–229. [Google Scholar] [CrossRef]
- Matthiesen, J.E.; Carraher, J.M.; Vasiliu, M.; Dixon, D.A.; Tessonnier, J.P. Electrochemical conversion of muconic acid to biobased diacid monomers. ACS Sustain. Chem. Eng. 2016, 4, 3575–3585. [Google Scholar] [CrossRef]
- Draths, K.M.; Frost, J.W. Environmentally compatible synthesis of adipic acid from D-glucose. J. Am. Chem. Soc. 1994, 116, 399–400. [Google Scholar] [CrossRef]
- Thomas, J.M.; Raja, R.; Johnson, B.F.G.; O’Connell, T.J.; Sankar, G.; Khimyak, T. Bimetallic nanocatalysts for the conversion of muconic acid to adipic acid. Chem. Commun. 2003, 1126–1127. [Google Scholar] [CrossRef]
- Li, X.; Wu, D.; Lu, T.; Yi, G.; Su, H.; Zhang, Y. Highly efficient chemical process to convert mucic acid into adipic acid and DFT studies of the mechanism of the rhenium-catalyzed deoxydehydration. Angew. Chemie -Int. Ed. 2014, 53, 4200–4204. [Google Scholar] [CrossRef]
- She, X.; Brown, H.M.; Zhang, X.; Ahring, B.K.; Wang, Y. Selective hydrogenation of trans,trans-muconic acid to adipic acid over a titania-supported rhenium catalyst. ChemSusChem 2011, 4, 1071–1073. [Google Scholar] [CrossRef]
- Shiramizu, M.; Toste, F.D. Expanding the scope of biomass-derived chemicals through tandem reactions based on oxorhenium-catalyzed deoxydehydration. Angew. Chemie Int. Ed. 2013, 52, 12905–12909. [Google Scholar] [CrossRef]
- Sirasani, G.; Tong, L.; Balskus, E.P. A biocompatible alkene hydrogenation merges organic synthesis with microbial metabolism. Angew. Chemie-Int. Ed. 2014, 53, 7785–7788. [Google Scholar] [CrossRef] [Green Version]
- Neri, G.; Musolino, M.G.; Milone, C.; Galvagno, S. Kinetic modeling of 2,4-dinitrotoluene hydrogenation over Pd/C. Ind. Eng. Chem. Res. 1995, 34, 2226–2231. [Google Scholar] [CrossRef]
- Carraher, J.M.; Pfennig, T.; Rao, R.G.; Shanks, B.H.; Tessonnier, J.-P. Cis,cis-muconic acid isomerization and catalytic conversion to biobased Cyclic-C6-1,4-Diacid monomers. Green Chem. 2017, 21–25. [Google Scholar] [CrossRef] [Green Version]
- Rosengart, A.; Capelli, S.; Pirola, C.; Citterio, A.; Bianchi, C.L.; Prati, L.; Villa, A. Renewable adipic acid from the hydrogenation of trans, trans-muconic acid: Selection of a three phases kinetic model. Chem. Eng. Trans. 2017, 57, 931–936. [Google Scholar] [CrossRef]
- Froment, G.F.; Bischoff, K.B.; De Wilde, J. Chemical Reactor Analysis and Design, 3rd ed.; Wiley: New York, NY, USA, 2011. [Google Scholar]
- Lylykangas, M.S.; Rautanen, P.A.; Krause, A.O.I. Hydrogenation and deactivation kinetics in the liquid-phase hydrogenation of isooctenes on Pt/Al2O3. Ind. Eng. Chem. Res. 2004, 43, 1641–1648. [Google Scholar] [CrossRef]
- Buzzi-Ferraris, G.; Manenti, F. Interpolation and Regression Models for the Chemical Engineer: Solving Numerical Problems; John Wiley & Sons: Hoboken, NJ, USA, 2010. [Google Scholar]
- Veldsink, J.W.; Bouma, M.J.; Schöön, N.H.; Beenackers, A.C.M. Heterogeneous hydrogenation of vegetable oils: A literature review. Catal. Rev. 1997, 39, 253–318. [Google Scholar] [CrossRef]
- Rajashekharam, M.V.; Nikalje, D.D.; Jaganathan, R.; Chaudhari, R.V. Hydrogenation of 2,4-dinitrotoluene using a Pd/Al2O3 catalyst in a slurry reactor: A molecular level approach to kinetic modeling and nonisothermal effects. Ind. Eng. Chem. Res. 1997, 36, 592–604. [Google Scholar] [CrossRef]
- Jones, W.H. Catalysis in Organic Syntheses; Science, E., Ed.; Academic Press: Cambridge, MA, USA, 2013. [Google Scholar] [CrossRef]
- Chaudhari, R.V.; Rode, C.V.; Deshpande, R.M.; Jaganathan, R.; Leib, T.M.; Mills, P.L. Kinetics of hydrogenation of maleic acid in a batch slurry reactor using a bimetallic Ru-Re/C catalyst. Chem. Eng. Sci. 2003, 58, 627–632. [Google Scholar] [CrossRef]
- Zhan, Z.; Jackson, J.E.; Miller, D.J.; Zhang, Z. Kinetics of aqueous-phase hydrogenation of lactic acid to propylene glycol. Ind. Eng. Chem. Res. 2002, 41, 691–696. [Google Scholar] [CrossRef]
- Feng, Y.; Zhou, L.; Wan, Q.; Lin, S.; Guo, H. Selective Hydrogenation of 1,3-Butadiene Catalyzed by a Single Pd Atom Anchored on Graphene: The Importance of Dynamics. Chem. Sci. 2018, 9, 5890–5896. [Google Scholar] [CrossRef] [Green Version]

| Catalyst | T (°C) | P (bar) | Time (h) | Conversion (%) | AA yield (%) | Solvent | Ref. |
|---|---|---|---|---|---|---|---|
| Pt/C 10% | 25 | 3.5 | 3 | 100 | 90 | Water | [25] |
| Pt/C 10% | 25 | 34 | 2.5 | 100 | 97 | Water | [8] |
| Ru10Pt2/SiO2 | 80 | 30 | 5 | 91 | 96 | Ethanol | [26] |
| Pt/C 5% | 160 | n.a. | 12 | n.a. | 99 | Pentanol | [27] |
| Re/TiO2 | 210 | 68 | 5 | 100 | 90 | Methanol | [28] |
| Pd/C 10% | 25 | 7 | 4 | n.a. | 62 | n-butanol | [29] |
| Royer | 37 | 25 | 18 | n.a. | 75 | [30] | |
| Pd/C 1% | 24 | 24 | 0.3 | >97 | >97 | Water | [10] |
| Ni/Al2O3 14.2% | 60 | 10 | 5 | 100 | >99 | [22] | |
| Ni electrode | 25 | 25 | 1 | 50 | <5 | Acidic solution | [24] |
| Pt/C 5% | 60 | 4 | 2.5 | 100 | 100 | Water | [23] |
| Species | Kinetic Equation | |
|---|---|---|
| ttMA | (7) | |
| cHDA | (8) | |
| tHDA | (9) | |
| AA | (10) | |
| H2 | (11) |
| LHHW_13P | Case 1 | Case 2 | Case 3 | ||||
|---|---|---|---|---|---|---|---|
| Reference Parameters | Calculated Parameters | Divergence | Calculated Parameters | Divergence | Calculated Parameters | Divergence | |
| KttMA | 9.40 × 100 | 9.40 × 100 | 0.00% | 9.41 × 100 | +0.11% | 9.85 × 100 | +4.79% |
| KcHDA | 3.44 × 104 | 3.21 × 104 | −6.69% | 3.59 × 104 | +4.36% | 4.95 × 104 | +43.90% |
| KtHDA | 2.28 × 101 | 2.27 × 101 | −0.44% | 2.28 × 101 | 0.00% | 2.52 × 101 | +10.53% |
| Ah02 | 8.86 × 10−6 | 1.00 × 10−5 | limited | 1.00 × 10−3 | limited | 1.00 × 10−1 | limited |
| Eah02 | 2.75 × 104 | 2.75 × 104 | 0.00% | 2.75 × 104 | 0.00% | 2.73 × 104 | −0.73% |
| Ah23 | 1.39 × 10−1 | 1.39 × 10−1 | 0.00% | 1.40 × 10−1 | +0.72% | 2.40 × 10−1 | +72.66% |
| Eah23 | 4.01 × 104 | 4.01 × 104 | 0.00% | 4.01 × 104 | 0.00% | 3.97 × 104 | −1.00% |
| Ai12 | 2.23 × 101 | 2.23 × 101 | 0.00% | 2.23 × 101 | 0.00% | 2.30 × 101 | +3.14% |
| Ai21 | 5.17 × 100 | 5.23 × 100 | +1.16% | 5.24 × 100 | +1.35% | 5.02 × 100 | −2.90% |
| Eai12 | 2.95 × 104 | 2.05 × 104 | −30.51% | 2.92 × 104 | −1.02% | 2.60 × 104 | −11.86% |
| Eai21 | 7.63 × 105 | 7.53 × 105 | −1.31% | 7.51 × 105 | −1.57% | 8.25 × 105 | +8.13% |
| Ah13 | 8.66 × 100 | 8.66 × 100 | 0.00% | 8.72 × 100 | +0.69% | 8.89 × 100 | −2.66% |
| Eah13 | 3.89 × 104 | 3.29 × 104 | −15.42% | 3.80 × 104 | −2.31% | 4.67 × 104 | +20.05% |
| RR | 7.76 × 10−4 | 7.69 × 10−4 | 7.69 × 10−4 | 7.71 × 10−4 | |||
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rosengart, A.; Pirola, C.; Capelli, S. Hydrogenation of Trans,Trans-Muconic Acid to Bio-Adipic Acid: Mechanism Identification and Kinetic Modelling. Processes 2020, 8, 929. https://doi.org/10.3390/pr8080929
Rosengart A, Pirola C, Capelli S. Hydrogenation of Trans,Trans-Muconic Acid to Bio-Adipic Acid: Mechanism Identification and Kinetic Modelling. Processes. 2020; 8(8):929. https://doi.org/10.3390/pr8080929
Chicago/Turabian StyleRosengart, Alessandro, Carlo Pirola, and Sofia Capelli. 2020. "Hydrogenation of Trans,Trans-Muconic Acid to Bio-Adipic Acid: Mechanism Identification and Kinetic Modelling" Processes 8, no. 8: 929. https://doi.org/10.3390/pr8080929
APA StyleRosengart, A., Pirola, C., & Capelli, S. (2020). Hydrogenation of Trans,Trans-Muconic Acid to Bio-Adipic Acid: Mechanism Identification and Kinetic Modelling. Processes, 8(8), 929. https://doi.org/10.3390/pr8080929







