Polyphenolic Profiling of Forestry Waste by UPLC-HDMSE
Abstract
1. Introduction
2. Materials and Methods
2.1. Forestry Waste Samples
2.2. Sample Preparation
2.3. Analytical Instrumentation
2.4. UPLC Conditions
2.5. Synapt G2-Si Conditions
2.6. Data Processing
3. Results and Discussion
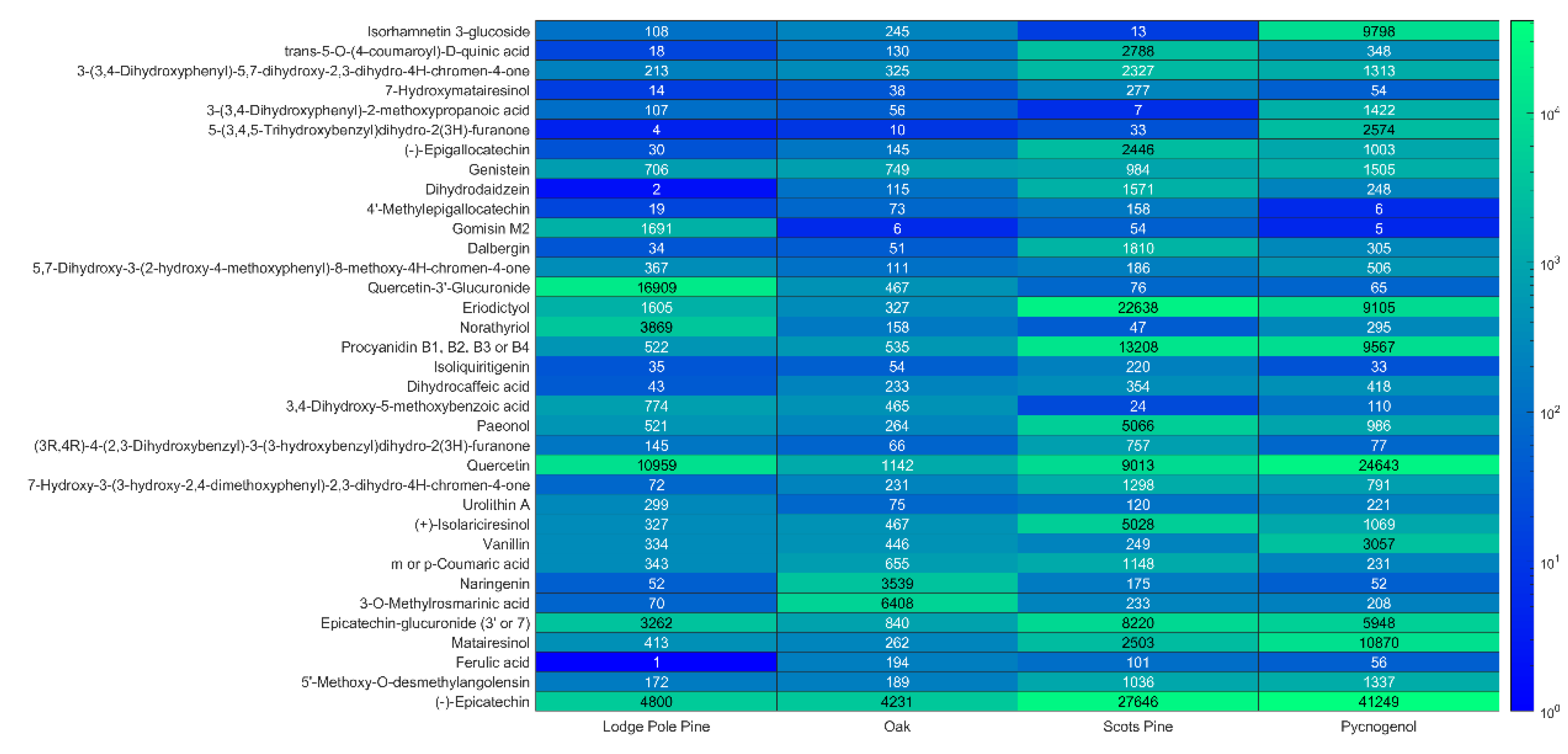
3.1. Bioactive Phenolic Compounds in Tree Bark Extracts
3.2. Tentative Identification of Polyphenols
3.3. Potential Use of Bark Waste for Nutraceutical Production
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- El Gharras, H. Polyphenols: Food sources, properties and applications—A review. Int. J. Food Sci. Technol. 2009, 44, 2512–2518. [Google Scholar] [CrossRef]
- De La Iglesia, R.; Milagro, F.I.; Campión, J.; Boqué, N.; Martínez, J.A. Healthy properties of proanthocyanidins. BioFactors 2010, 36, 159–168. [Google Scholar] [CrossRef] [PubMed]
- Krikorian, R.; Kalt, W.; Mcdonald, J.E.; Shidler, M.D.; Summer, S.S.; Stein, A.L. Cognitive performance in relation to urinary anthocyanins and their fl avonoid-based products following blueberry supplementation in older adults at risk for dementia. J. Funct. Foods 2019, 103667. [Google Scholar] [CrossRef]
- Watson, R.R.; Preedy, S.Z. Polyphenols in Human Health and Disease; Academic Press: Cambridge, MA, USA, 2014; Volume 1. [Google Scholar]
- Lattanzio, V.; Kroon, P.A.; Ralph, J.; Harris, P.; Dixon, R.A.; Dangles, O.; Lamotte, O. Recent Advances in Polyphenol Research; John Wiley & Sons: Hoboken, NJ, USA, 2008; Volume 1. [Google Scholar]
- Liu, Y.; Jia, G.; Gou, L.; Sun, L.; Fu, X.; Lan, N.; Li, S.; Yin, X. Antidepressant-like effects of tea polyphenols on mouse model of chronic unpredictable mild stress. Pharmacol. Biochem. Behav. 2013, 104, 27–32. [Google Scholar] [CrossRef] [PubMed]
- Panickar, K.S. Effects of dietary polyphenols on neuroregulatory factors and pathways that mediate food intake and energy regulation in obesity. Mol. Nutr. Food Res. 2013, 57, 34–47. [Google Scholar] [CrossRef] [PubMed]
- McDougall, G.J.; Stewart, D. The inhibitory effects of berry polyphenols on digestive enzymes. BioFactors 2005, 23, 189–195. [Google Scholar] [CrossRef]
- Zhang, W.L.; Zhu, L.; Jiang, J.G. Active ingredients from natural botanicals in the treatment of obesity. Obes. Rev. 2014, 15, 957–967. [Google Scholar] [CrossRef]
- Wang, S.; Moustaid-Moussa, N.; Chen, L.X.; Mo, H.B.; Shastri, A.; Su, R.; Bapat, P.; Kwun, I.; Shen, C.L. Novel insights of dietary polyphenols and obesity. J. Nutr. Biochem. 2014, 25, 1–18. [Google Scholar] [CrossRef]
- Alonso, V.R.; Guarner, F. Linking the gut microbiota to human health. Br. J. Nutr. 2013, 109 (Suppl. 2), S21–S26. [Google Scholar] [CrossRef]
- Espley, R.V.; Bovy, A.; Bava, C.; Jaeger, S.R.; Tomes, S.; Norling, C.; Crawford, J.; Rowan, D.; McGhie, T.K.; Brendolise, C.; et al. Analysis of genetically modified red-fleshed apples reveals effects on growth and consumer attributes. Plant Biotechnol. J. 2013, 11, 408–419. [Google Scholar] [CrossRef]
- Martinez-Dominguez, E.; de la Puerta, R.; Ruiz-Gutierrez, V. Protective effects upon experimental inflammation models of a polyphenol-supplemented virgin olive oil diet. Inflamm. Res. 2001, 50, 102–106. [Google Scholar]
- Cory, H.; Passarelli, S.; Szeto, J.; Tamez, M.; Mattei, J. The role of polyphenols in human health and food systems: A mini-review. Front. Nutr. 2018, 5, 87. [Google Scholar] [CrossRef] [PubMed]
- Vejdovszky, K.; Schmidt, V.; Warth, B.; Marko, D. Combinatory estrogenic effects between the isoflavone genistein and the mycotoxins zearalenone and alternariol in vitro. Mol. Nutr. Food Res. 2017, 61, 1600526. [Google Scholar] [CrossRef]
- Jain, A.; Manghani, C.; Kohli, S.; Nigam, D.; Rani, V. Tea and human health: The dark shadows. Toxicol. Lett. 2013, 220, 82–87. [Google Scholar] [CrossRef]
- Kumar, P.S.; Kumar, N.A.; Sivakumar, R.; Kaushik, C. Experimentation on solvent extraction of polyphenols from natural waste. J. Mater. Sci. 2009, 44, 5894–5899. [Google Scholar] [CrossRef]
- Bolling, B.W.; Chen, C.-Y.O.; McKay, D.L.; Blumberg, J.B. Tree nut phytochemicals: Composition, antioxidant capacity, bioactivity, impact factors. Nutr. Res. Rev. 2011, 24, 244–275. [Google Scholar] [CrossRef] [PubMed]
- Withouck, H.; Boeykens, A.; Broucke, M.V.; Moreira, M.M.; Delerue-Matos, C.; De Cooman, L. Evaluation of the impact of pre-treatment and extraction conditions on the polyphenolic profile and antioxidant activity of Belgium apple wood. Eur. Food Res. Technol. 2019, 245, 2565–2578. [Google Scholar] [CrossRef]
- Feldman, E.B. The scientific evidence for a beneficial health relationship between walnuts and coronary heart disease. J. Nutr. 2002, 132, 1062S–1101S. [Google Scholar] [CrossRef] [PubMed]
- Debeljak, J.; Ferk, P.; Čokolič, M.; Zavratnik, A.; Tavč Benković, E.; Kreft, S.; Štrukelj, B. Randomised, double blind, cross-over, placebo and active controlled human pharmacodynamic study on the influence of silver fir wood extract (Belinal) on post-prandial glycemic response. Pharmazie 2016, 71, 566–569. [Google Scholar] [CrossRef]
- Ogawa, S.; Matsuo, Y.; Tanaka, T.; Yazaki, Y. Utilization of flavonoid compounds from bark and wood. III. Application in health foods. Molecules 2018, 23, 1860. [Google Scholar] [CrossRef]
- Liu, J.K.; Henkel, T. Traditional Chinese medicine (TCM): Are polyphenols and saponins the key ingredients triggering biological activities? Curr. Med. Chem. 2002, 9, 1483–1485. [Google Scholar] [CrossRef]
- Gironi, F.; Piemonte, V. Temperature and solvent effects on polyphenol extraction process from chestnut tree wood. Chem. Eng. Res. Des. 2011, 89, 857–862. [Google Scholar] [CrossRef]
- Comandini, P.; Lerma-García, M.J.; Simó-Alfonso, E.F.; Toschi, T.G. Tannin analysis of chestnut bark samples (Castanea sativa Mill.) by HPLC-DAD-MS. Food Chem. 2014, 157, 290–295. [Google Scholar] [CrossRef]
- Câmara, C.R.S.; Schlegel, V. A review on the potential human health benefits of the black walnut: A comparison with the english walnuts and other tree nuts. Int. J. Food Prop. 2016, 19, 2175–2189. [Google Scholar] [CrossRef]
- Donno, D.; Boggia, R.; Zunin, P.; Cerutti, A.K.; Guido, M.; Mellano, M.G.; Prgomet, Z.; Beccaro, G.L. Phytochemical fingerprint and chemometrics for natural food preparation pattern recognition: An innovative technique in food supplement quality control. J. Food Sci. Technol. Mysore 2016, 53, 1071–1083. [Google Scholar] [CrossRef]
- Zhang, X.X.; Shi, Q.Q.; Ji, D.; Niu, L.X.; Zhang, Y.L. Determination of the phenolic content, profile, and antioxidant activity of seeds from nine tree peony (Paeonia section Moutan DC.) species native to China. Food Res. Int. 2017, 97, 141–148. [Google Scholar] [CrossRef]
- Mailer, R.; Ayton, J. Effect of irrigation and water stress on olive oil quality and yield based on a four year study. Acta Hortic. 2011, 888, 63–72. [Google Scholar] [CrossRef]
- Souza, R.T.D.A.; Silva, D.K.D.A.; Santos, M.V.F.D.; Naumann, H.D.; Magalhães, A.L.R.; Andrade, A.P.D. Association of edaphoclimatic characteristics and variability of condensed tannin content in species from Caatinga. Rev. Cienc. Agron. 2020, 51, e20196611. [Google Scholar] [CrossRef]
- Fraser, K.; Harrison, S.J.; Lane, G.A.; Otter, D.E.; Hemar, Y.; Quek, S.Y.; Rasmussen, S. Analysis of low molecular weight metabolites in tea using mass spectrometry-based analytical methods. Crit. Rev. Food Sci. Nutr. 2014, 54, 924–937. [Google Scholar] [CrossRef] [PubMed]
- Kang, K.B.; Woo, S.; Ernst, M.; van der Hooft, J.J.; Nothias, L.F.; da Silva, R.R.; Dorrestein, P.C.; Sung, S.H.; Lee, M. Assessing specialized metabolite diversity of Alnus species by a digitized LC-MS/MS data analysis workflow. Phytochemistry 2020, 173, 11229. [Google Scholar] [CrossRef]
- Rocchetti, G.; Blasi, F.; Montesano, D.; Ghisoni, S.; Marcotullio, M.C.; Sabatini, S.; Cossignani, L.; Lucini, L. Impact of conventional/non-conventional extraction methods on the untargeted phenolic profile of Moringa oleifera leaves. Food Res. Int. 2019, 115, 319–327. [Google Scholar] [CrossRef]
- Damiani, E.; Carloni, P.; Rocchetti, G.; Senizza, B.; Tiano, L.; Joubert, E.; de Beer, D.; Lucini, L. Impact of cold versus hot brewing on the phenolic profile and antioxidant capacity of rooibos (Aspalathus linearis) herbal tea. Antioxidants 2019, 8, 499. [Google Scholar] [CrossRef]
- Zhang, C.; Zuo, T.; Wang, X.; Wang, H.; Hu, Y.; Li, Z.; Li, W.; Jia, L.; Qian, Y.; Yang, W.; et al. Integration of data-dependent acquisition (DDA) and data-independent high-definition MSE (HDMSE) for the comprehensive profiling and characterization of multicomponents from panax japonicus by UHPLC/IM-QTOF-MS. Molecules 2019, 24, 2708. [Google Scholar] [CrossRef]
- Johnson, S.R.; Rikli, H.G. Aspartic acid isomerization characterized by high definition mass spectrometry significantly alters the bioactivity of a novel toxin from poecilotheria. Toxins 2020, 12, 207. [Google Scholar] [CrossRef]
- Rohdewald, P. A review of the French maritime pine bark extract (Pycnogenol (R)), a herbal medication with a diverse clinical pharmacology. Int. J. Clin. Pharmacol. Ther. 2002, 40, 158–168. [Google Scholar] [CrossRef]
- Mármol, I.; Quero, J.; Jiménez-Moreno, N.; Rodríguez-Yoldi, M.J.; Ancín-Azpilicueta, C. A systematic review of the potential uses of pine bark in food industry and health care. Trends Food Sci. Technol. 2019, 88, 558–566. [Google Scholar] [CrossRef]
- Fogacci, F.; Tocci, G.; Sahebkar, A.; Presta, V.; Banach, M.; Cicero, A.F.G. Effect of pycnogenol on blood pressure: Findings from a PRISMA compliant systematic review and meta-analysis of randomized, double-blind, placebo-controlled, clinical studies. Angiology 2020, 71, 217–225. [Google Scholar] [CrossRef]
- Mello, B.C.B.S.; Petrus, J.C.C.; Hubinger, M.D. Concentration of flavonoids and phenolic compounds in aqueous and ethanolic propolis extracts through nanofiltration. J. Food Eng. 2010, 96, 533–539. [Google Scholar] [CrossRef]
- EU. Directive 2009/32/EC of the European Parliament and of the Council of 23 April 2009 on the Approximation of the Laws of the Member States on Extraction Solvents Used in the Production of Foodstuffs and Food Ingredients. 2009. Available online: https://eur-lex.europa.eu/legal-content/EN/ALL/?uri=CELEX%3A32009L0032 (accessed on 3 November 2020).
- Vos, F.; Crespy, V.; Chaffaut, L.; Mennen, L.; Knox, C.; Neveu, V. Original article phenol-explorer: An online comprehensive database on polyphenol contents in foods. Database 2010, 2010, bap024. [Google Scholar] [CrossRef]
- Salehi, B.; Fokou, P.V.T.; Sharifi-Rad, M.; Zucca, P.; Pezzani, R.; Martins, N.; Sharifi-Rad, J. The therapeutic potential of naringenin: A review of clinical trials. Pharmaceuticals 2019, 12, 11. [Google Scholar] [CrossRef]
- Mastino, P.M.; Mauro, M.; Jean, C.; Juliano, C.; Marianna, U. Analysis and potential antimicrobial activity of phenolic compounds in the extracts of cistus creticus subspecies from sardinia. Nat. Prod. J. 2018, 8, 166–174. [Google Scholar] [CrossRef]
- Forney, L.; Lenard, N.; Stewart, L.; Henagan, T. Dietary quercetin attenuates adipose tissue expansion and inflammation and alters adipocyte morphology in a tissue-specific manner. Int. J. Mol. Sci. 2018, 19, 895. [Google Scholar] [CrossRef]
- Rotelli, A.E.; Guardia, T.; Juárez, A.O.; De La Rocha, N.E.; Pelzer, L.E. Comparative study of flavonoids in experimental models of inflammation. Pharmacol. Res. 2003, 48, 601–606. [Google Scholar] [CrossRef]
- Hameed, A.; Hafizur, R.M.; Hussain, N.; Raza, S.A.; Rehman, M.; Ashraf, S.; Ul-Haq, Z.; Khan, F.; Abbas, G.; Choudhary, M.I. Eriodictyol stimulates insulin secretion through cAMP/PKA signaling pathway in mice islets. Eur. J. Pharmacol. 2018, 820, 245–255. [Google Scholar] [CrossRef] [PubMed]
- de Oliveira Ferreira, E.; Fernandes, M.Y.S.D.; de Lima, N.M.R.; Neves, K.R.T.; do Carmo, M.R.S.; Lima, F.A.V.; Fonteles, A.A.; Menezes, A.P.F.; de Andrade, G.M. Neuroinflammatory response to experimental stroke is inhibited by eriodictyol. Behav. Brain Res. 2016, 312, 321–332. [Google Scholar] [CrossRef]
- Wang, Y.; Chen, Y.; Chen, Y.; Zhou, B.; Shan, X.; Yang, G. Biomedicine & pharmacotherapy eriodictyol inhibits IL-1β-induced in fl ammatory response in human osteoarthritis chondrocytes. Biomed. Pharmacother. 2018, 107, 1128–1134. [Google Scholar] [CrossRef] [PubMed]
- Prakash, M.; Basavaraj, B.V.; Chidambara Murthy, K.N. Biological functions of epicatechin: Plant cell to human cell health. J. Funct. Foods 2019, 52, 14–24. [Google Scholar] [CrossRef]
- Bernatova, I. Biological activities of (−)-epicatechin and (−)-epicatechin-containing foods: Focus on cardiovascular and neuropsychological health. Biotechnol. Adv. 2018, 36, 666–681. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Needs, P.W.; Lu, Y.; Kroon, P.A.; Ren, D.; Yang, X. Different antitumor effects of quercetin, quercetin-3′-sulfate and quercetin-3-glucuronide in human breast cancer MCF-7 cells. Food Funct. 2018, 9, 1736–1746. [Google Scholar] [CrossRef]
- Li, J.; Malakhova, M.; Mottamal, M.; Reddy, K.; Kurinov, I.; Carper, A.; Langfald, A.; Oi, N.; Kim, M.O.; Zhu, F.; et al. Norathyriol suppresses skin cancers induced by solar ultraviolet radiation by targeting ERK kinases. Cancer Res. 2012, 72, 260–271. [Google Scholar] [CrossRef]
- Lin, H.; Tu, C.; Niu, Y.; Li, F.; Yuan, L.; Li, N.; Xu, A.; Gao, L.; Li, L. Dual actions of norathyriol as a new candidate hypouricaemic agent: Uricosuric effects and xanthine oxidase inhibition. Eur. J. Pharmacol. 2019, 853, 371–380. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Liu, M.; Yu, H.; Wang, W.; Han, L.; Chen, Q.; Ruan, J.; Wen, S.; Zhang, Y.; Wang, T. Mangiferin improves hepatic lipid metabolism mainly through its metabolite-norathyriol by modulating SIRT-1/AMPK/SREBP-1c signaling. Front. Pharmacol. 2018, 9, 1–13. [Google Scholar] [CrossRef]
- Lee, Y. Cancer chemopreventive potential of procyanidin. Toxicol. Res. 2017, 33, 273–282. [Google Scholar] [CrossRef] [PubMed]
- Lopes, N.P. Natural product reports. Nat. Prod. Rep. 2016, 33. [Google Scholar] [CrossRef]
- Pandey, R.; Chandra, P.; Arya, K.R.; Kumar, B. Development and validation of an ultra high performance liquid chromatography electrospray ionization tandem mass spectrometry method for the simultaneous determination of selected flavonoids in Ginkgo biloba. J. Sep. Sci. 2014, 37, 3610–3618. [Google Scholar] [CrossRef]
- Hsieh, C.L.; Cheng, C.Y.; Tsai, T.H.; Lin, I.H.; Liu, C.H.; Chiang, S.Y.; Lin, J.G.; Lao, C.J.; Tang, N.Y. Paeonol reduced cerebral infarction involving the superoxide anion and microglia activation in ischemia-reperfusion injured rats. J. Ethnopharmacol. 2006, 106, 208–215. [Google Scholar] [CrossRef]
- Kim, S.H.; Kim, S.A.; Park, M.K.; Kim, S.H.; Park, Y.D.; Na, H.J.; Kim, H.M.; Shin, M.K.; Ahn, K.S. Paeonol inhibits anaphylactic reaction by regulating histamine and TNF-α. Int. Immunopharmacol. 2004, 4, 279–287. [Google Scholar] [CrossRef]
- Wu, J.B.; Song, N.N.; Wei, X.B.; Guan, H.S.; Zhang, X.M. Protective effects of paeonol on cultured rat hippocampal neurons against oxygen-glucose deprivation-induced injury. J. Neurol. Sci. 2008, 264, 50–55. [Google Scholar] [CrossRef]
- Ye, S.; Liu, X.; Mao, B.; Yang, L.; Liu, N. Paeonol enhances thrombus recanalization by inducing vascular endothelial growth factor 165 via ERK1/2 MAPK signaling pathway. Mol. Med. Rep. 2016, 13, 4853–4858. [Google Scholar] [CrossRef]
- Zhang, B.; Cai, J.; Duan, C.-Q.; Reeves, M.J.; He, F. A review of polyphenolics in oak woods. Int. J. Mol. Sci. 2015, 16, 6978–7014. [Google Scholar] [CrossRef]
- Mill, O.L.; Antunes-ricardo, M.; García-cayuela, T.; Ibañez, E. Enzyme-assisted in situ supercritical fluid extraction of isorhamnetin conjugates from Opuntia ficus-indica (L.) Mill Marilena Antunes-Ricardo. Inst. Food Sci. Res. 2019, 141, 71–72. [Google Scholar] [CrossRef]
- Arshadi, M.; Gref, R.; Geladi, P.; Dahlqvist, S.A.; Lestander, T. The influence of raw material characteristics on the industrial pelletizing process and pellet quality. Fuel Process. Technol. 2008, 89, 1442–1447. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Potter, C.M.; Jones, D.L. Polyphenolic Profiling of Forestry Waste by UPLC-HDMSE. Processes 2020, 8, 1411. https://doi.org/10.3390/pr8111411
Potter CM, Jones DL. Polyphenolic Profiling of Forestry Waste by UPLC-HDMSE. Processes. 2020; 8(11):1411. https://doi.org/10.3390/pr8111411
Chicago/Turabian StylePotter, Colin M., and David L. Jones. 2020. "Polyphenolic Profiling of Forestry Waste by UPLC-HDMSE" Processes 8, no. 11: 1411. https://doi.org/10.3390/pr8111411
APA StylePotter, C. M., & Jones, D. L. (2020). Polyphenolic Profiling of Forestry Waste by UPLC-HDMSE. Processes, 8(11), 1411. https://doi.org/10.3390/pr8111411




