Abstract
Castor bean (Ricinus Communis) oil has been reported as one of the most important bio-based fuels; however, high amounts of toxic solid residue are generated in the production. This toxicity is due to several molecules, ricin protein being the most studied compound. The inhibition of the ricin protein is essential for eliminating its toxicity. The objective of this study is to predict the possible inhibition process via the interactions between the ricin protein and the flavonoids quercetin (Q) and epigallocatechin gallate (EGCG). The molecular structures of the complexes formed between the ricin protein and flavonoids were studied using quantum-chemical and molecular docking calculations to analyze the type of interaction, active site of the protein, binding energies, and different conformations in the inhibition process. Different methodologies were applied for the molecular structure determination; the best approximation was obtained with B3LYP/6-31G (d,p) theoretical methodology. Mappings of electrostatic potential (MEP) and frontier molecular orbitals were used for the identification of the probable sites of interaction, which were confirmed by molecular docking. The adjustment and alignment of flavonoid groups before and after the interaction, and charge transfer parameters, showed that Q and EGCG act as electron donors inside of the active site in ricin.
1. Introduction
Recently, castor bean seeds (Ricinus Communis) have been used as an alternative source of biofuels due to the high demand that currently exists for biomass fuels. However, during the extraction of castor oil, significant amounts of solid residue are generated; this residue presents a problem because of the high toxicity [1,2,3]. The solid residue is composed of an agglutinin, and a protein called ricin; ricin is a toxic lectin produced by the castor seed (Ricinus communis), which is found in nature as a wild plant [4]. This protein is the most studied of the ribosome inactivating proteins (RIPs), and it is composed of two polypeptide chains linked by a disulfide bridge [5]. Previous studies have suggested that the detoxification of this protein can be done with ammonia, caustic soda, lime, and heat [6,7,8,9,10]. Recently, some in vitro evaluations have confirmed the detoxification of the solid residue when combined with polyphenolic compounds [11].
Phenols are abundant phytochemical compounds in vegetables and fruits; there are 8000 phenolic structures [12]. The phenolic compounds are considered important antioxidants for human health [13]. These structures are classified in subcategories, depending on the structural skeleton that is classified according to the number of carbon atoms present in each conformation [14]. Flavonoids are constituents of the phenolic compounds, and they are a part of the human diet. Flavonoids are present in vegetables, seeds, fruits, and beverages such as wine and beer; they also provide color and flavor to food. Flavonoids are divided into several classes according to their oxidation degree in the heterocycle [13]. See Scheme 1. The structure of a flavonoid consists of a diphenylpropane (c6-c3-c6), formed by two aromatic rings linked through three carbon atoms that constitute a heterocycle oxygenated [15], and they are divided among six families, flavones, flavanones, isoflavones, flavonols, flavanols, and anthocyanidins [16]. Flavonols are colorless molecules that accumulate mainly on external and aerial tissues, such as fruit and vegetable leaves, since their biosynthesis is stimulated by light. Chemically, these molecules differ from many other flavonoids because they have a double bond between 2,3 positions and oxygen (ketone group) at the fourth position of the C ring; among them are quercetin, kaempferol, isorhamnetin, glycosides, and myricetin [17]. Quercetin is the most representative molecule of flavonols since, in addition to being a powerful antioxidant, it can chelate metals, eliminates oxygen free radicals and prevents oxidation of low-density lipoproteins, and it is also a water-soluble natural pigment that is usually present as O-glycosides in high concentrations, especially in onions, apples, and grapes [18]. Other important flavonoids are the flavanols family; these are represented by catechins such as epigallocatechin gallate (EGCG), a flavanol mainly found in green tea; it is considered as one of the most active molecules with antioxidant properties [19]. The principal reason for the study of this flavanol (epigallocatechin gallate, EGCG), is because this compound can inhibit the actions of a protein called Dyrk 1A, which is produced in excess in Down syndrome, causing damage to brain function [20]. The EGCG presents helpful effects in the prevention of pathological disorders such as obesity, insulin resistance, dyslipidemia, and hypertension [21]. In the EGCG skeleton, the saturation of the heterocyclic ring prevents the delocalization of electrons between rings A and B [22], allowing it to slow down lipid oxidation [20,23]. Furthermore, the EGCG has eight hydroxyl groups distributed in its A and B rings; has a gallate group that gives it its excellent antioxidant power [24]; and does not have a C2=C3 double bond, allowing it to generate cis–trans isomers, epimerizers, and racemic mixtures, and naturally, polymerize tannin [25]. See Scheme 1.
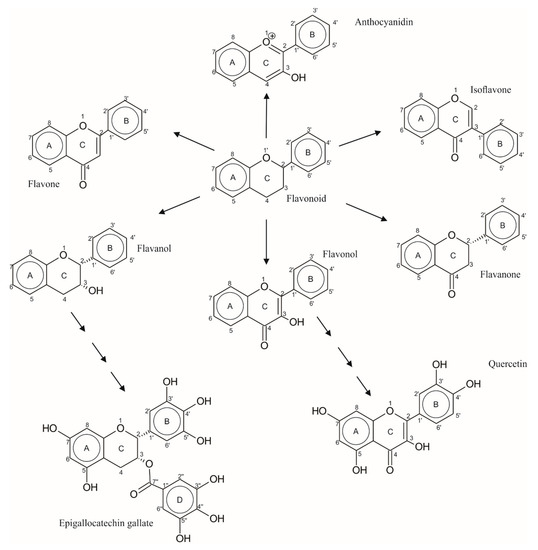
Scheme 1.
Chemical structures of flavonoids. The six flavonoid families include flavones, flavonones, isoflavones, flavonols, flavanols, and anthocyanidins. Quercetin belongs to flavonols and epigallocathechin gallate belongs to flavanols.
According to reported analysis, the most representative molecules are quercetin and epigallocatechin gallate, because these compounds are some of the most studied flavonoids in vivo for interactions with different proteins [26,27,28], principally for their inhibitory potential [29]. In the case of the EGCG, Dyer et al. found in experimental trials that this flavonoid inhibits the ricin toxin, suggesting that this molecule may reduce the lethality of the ricin [11]; however, the types of interactions present between this protein and groups of polyphenolic compounds have not been analyzed at a molecular level.
It is well known that theoretical analysis and quantum-chemical calculations can provide quantitative predictions of the behavior of organic compounds, such as chemical reactivity or physicochemical properties [30]. Currently, there is a great interest in the theoretical studies in polyphenolic groups [31]. However, the electronic structure methods, such as ab initio and density functional theory (DFT), are the most used due to the accurate descriptions, geometrical structures, and electronic properties obtained in these analyses [32]. Additionally, those methods are very helpful in getting the correct descriptions of the molecular interactions. For example, several functionals have been used for the prediction of many properties, the M06-2X hybrid meta-Generalized Gradient Approximation (GGA) functional being the most widely used method for biological and pharmaceutical theoretical studies, showing a higher correlation with experimental data [33,34,35]. On the other hand, the B3LYP hybrid functional is considered the best method to analyze organic molecules due to the precision of its results [36,37]. Alternatively, derivatives of flavonoid groups have been analyzed by the B3LYP and M06-2X functional, determining the molecular factors of their structural and energetic properties [38,39]. These theoretical calculations can be incredibly helpful in the prediction of chemical reactivity parameters and molecular structure; the last method mentioned is used for alignment and adjustment of a ligand, which is necessary because the initial pose of a ligand in a free solution is different from the pose of the protein-bound ligand [40,41,42]. The conformation of the ligand not only depends on the low energy in a coupling, but also on its conformational selection and the accessibility of the protein during the interactions [43,44].
Another computational procedure is molecular docking; this is a technique that attempts to predict noncovalent binding between macromolecules and ligands. Several reported studies have analyzed the molecular interactions between two or more constituent molecules, especially protein–ligand docking, thereby predicting the structures of the formed intermolecular complexes, showing how these molecules could potentially bind in vivo [45].
The objective of this research was to develop molecular docking calculations between ricin protein and specific flavonoid compounds such as quercetin and epigallocatechin gallate, in order to obtain significant and useful information about the molecular interactions present in these couplings, to predict whether these interactions are the possible cause of ricin detoxification. The DFT electronic structure method was used for geometric optimization and chemical reactivity calculations. Once the molecular interactions were defined, the electronic properties such as ionization potential, electron affinity, chemical hardness, chemical potential, and electronegativity were predicted by quantum-chemical calculations. Additionally, in order to obtain valid and reliable models, molecular alignment was employed for the quercetin and epigallocatechin gallate geometries, before and after the interaction with the ricin protein. Additionally, an exploration of the oxidation process inside of the active site was performed by a charge transfer flow direction analysis.
2. Computational Details
In this theoretical study, the ricin protein and its molecular interactions with quercetin (Q) and epigallocatechin gallate (EGCG) flavonoids were analyzed. The molecular structures of ricin (PDB ID:2AAi) and of the amino acids involved in the active site were obtained from the Protein Data Bank (http://www.rcb.org) based on X-ray diffraction data.
2.1. Quantum-Chemical Calculations
The computational strategy was carried out as follows. The ground state geometry optimization and frequency calculations of flavonoids were done using the electronic structure method DFT, with the hybrid density functionals B3LYP [46,47,48] and PBE1PBE [49], and the meta-GGA functionals M06 [50] and M06-2X [50]. All the functionals were combined with the 6–31G (d,p) basis set. The optimized geometries were used for the analytical frequency calculations, which were carried out for each stationary structure to verify whether it is the minimum of the potential energy hypersurfaces. The solubility in water is an important requirement for those compounds that can be good candidates for biological applications. For this reason all the calculations were carried out using water as a solvent, this being an analysis of great importance not only in chemistry but also in other topics of science, such as biology, medicine, and the food industry, among others [51]. All the theoretical properties have been determined in the solvent phase using water as a solvent. The solvent phase calculations were made with the polarizable continuum model (PCM) using the integral formalism variant (IEFPCM) [52,53].
The calculated geometry of EGCG molecule was compared with X-ray diffraction experimental data reported for the structure. Statistical analysis for each methodology versus experimental values was applied. This test allowed the selection of the best method for flavonoids’ geometry optimizations. Subsequently, the energies of the molecules in their neutral (EN), anionic (EN+1), and cationic (EN-1) forms were calculated with B3LYP/6-31G (d,p) methodology. The obtained energies were used for the calculation of global reactivity parameters such as ionization potential (I), electronic affinity (EA), electronegativity (χ), chemical hardness (η), and chemical potential (μ). See Equations (1)–(5). These reactivity parameters were applied for the determination of the charge transfer (ΔN), which describes the fractional number of transferred electrons from system A (flavonoids) to system B (any amino acids from the active site). See Equation (6). From the neutral molecule, the frontier molecular orbitals HOMO (highest occupied molecular orbital) and LUMO (lowest unoccupied molecular orbital) were mapped. The molecular electrostatic potential (MEP) was calculated using B3LYP/6-31G (d,p) methodology. All the quantum-chemical calculations were performed in Gaussian 09 [54] program.
2.2. Preparation of Protein for Molecular Docking Calculations
The crystal structure of ricin (PDB ID:2AAi) with a resolution of 2.5 Å was taken from the Protein Data Bank. This protein is composed of two polypeptide chains (A and B) linked by a disulfide bridge. Before the docking calculations, the ricin protein was prepared using PyMOL program [55], where water molecules were dismissed. Docking studies were carried out separately for the polypeptide chains A and B.
2.3. Molecular Docking Calculations
In the second stage, the optimized structures of flavonoids were used as ligands in the molecular docking analysis using the Autodock 4.2 software [56]. In this study, the molecular docking between each ligand (Q and EGCG) and the macromolecule (ricin protein) was performed, showing all the possible molecular interactions between both systems. The macromolecule was placed rigidly; in contrast, the ligands were flexible. The water molecules in the macromolecule were eliminated and we only kept the polar hydrogen atoms. Three boxes were designed for the Docking, one in A chain and two in B chain (B1 and B2) of the ricin protein; these boxes are located in the active sites of the chains. The conditions were: in the A chain a grid box size of 124 × 122 × 126 Å was used, centered at the coordinates x = 22.932, y = 64.561, z = 53.505, with the spacing grid point of 0.169 Å; in the B chain, the B1 the grid box was 34 × 34 × 22 Å centered at the coordinates x = 32.643, y = 42.605, z = 10.995, with a spacing grid of 0.506 Å; and in B2 the grid box was 22 × 32 × 28 Å centered at the coordinates x = −0.433, y = 34.935, z = 54.086, with a spacing grid of 0.528 Å. The coupling conformations were obtained through a genetic algorithm in combination with the Lamarckian algorithm, by selecting the lowest binding energy of the given ten conformations.
3. Results and Discussion
3.1. Quantum-Chemical Calculations
The obtained geometrical parameters are shown in Table 1. These parameters were calculated using different methodologies: PBE1PBE/6-31G (d,p), M06/6-31G (d,p), M06-2X/6-31G (d,p), and B3LYP/6-31G (d,p). The bond lengths and bond angles of the Q and EGCG molecules (calculated in the solvent phase) were compared with the reported X-Ray diffraction values [57,58]. The correlation analysis showed that the best methodology employed to analyze the molecular structure of Q and EGCG was B3LYP/6-31G (d,p), with a correlation of 0.94 for bond lengths and 0.82 for bond angles for Q, and correlations of 0.95 and 0.93 for bond lengths and bond angles in EGCG. The absence of imaginary frequencies confirmed the global minima on the potential energy surface. These optimized structures of Q and EGCG calculated with B3LYP/6-31G (d,p) in the solved phase were used in the molecular docking calculations (See Figure 1).

Table 1.
Q and EGCG geometrical parameters calculated in the solvent phase (water) using IEFPCM solvation model, and the X-ray diffraction data reported for the same compounds.
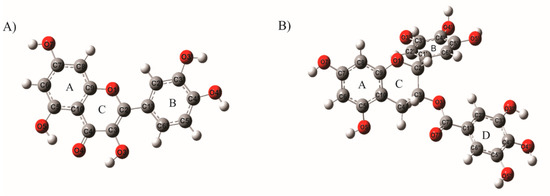
Figure 1.
Optimized molecular structures of (A) quercetin (Q) and (B) epigallocatechin gallate (EGCG) calculated with the best methodology B3LYP/6-31G (d,p) in the solvent phase.
3.2. Global Chemical Reactivity
The optimized geometries obtained with B3LYP/6-31G (d,p), were used for global chemical reactivity calculations. The reactivity parameters were calculated according to the approximations of energies in their neutral (EN), anionic (EN+1), and cationic (EN-1) states, based on the optimization of the fundamental state geometry. The calculated parameters were: ionization potential (I), electronic affinity (EA), chemical potential (μ), electronegativity (χ), and chemical hardness (η).
The obtained results are shown in Table 2, in which is observed that Q presents a higher capacity to form anions due to the electronegativity value (χ), which represents a greater tendency to attract electrons. Regarding chemical hardness (η), Q shows a lower value compared to EGCG; this parameter is indicative of the good interaction of this molecule with the surrounding environment. The chemical harness and chemical potential values were applied in the calculation of the fractional number of transferred electrons (ΔN).

Table 2.
Chemical reactivity parameters and total electronic energy of quercetin (Q) and epigallocatechin gallate (EGCG) calculated with B3LYP/6-31G (d,p) in solvent phase. For the reactivity calculation parameters see Equations (1)–(5). All the values are in eV.
3.3. Molecular Electrostatic Potential (MEP)
The molecular electrostatic potential (MEP) can predict the reactive sites of electrophilic and nucleophilic attack in a molecule. These MEPs were realized to show the electron density within the Q and EGCG on surfaces that surround the molecules, and the locations of sites for nucleophilic and electrophilic attacks. Red indicates negative regions promoting electrophilic attack and blue indicates the positive parts showing favorable sites for nucleophilic attack. Molecular electrostatic potentials of Q and EGCG were calculated using B3LYP/6-31G (d,p) methodology. The surface map is shown in Figure 2 and Figure 3. Figure 2 shows MEP map of quercetin with three possible electrophilic attack sites localized on O4 (−0.655 e), O5 (−0.590 e), and O13 (−0.586 e) atoms, while the possible nucleophilic attack sites are localized on hydrogens, mainly H7 (0.353 e) and H14 (0.357 e). Figure 3 shows the MEP map of EGCG with five possible electrophilic attack sites localized on O7′′(−0.531 e), O3′′(−0.585 e), O7 (−0.560 e), O3′(0.582 e), and O1 (−0.570 e) atoms, and the possible nucleophilic attack sites are localized on hydrogens of the three-ring, mainly H3′′ (0.356 e), H4′′ (0.360 e), H5′′ (0.346 e), H5 (0.351 e), H6 (0.342 e), H7 (0.345 e), H3′ (0.340 e), and H4′ (0.359 e). The values are shown in units of electric charge.
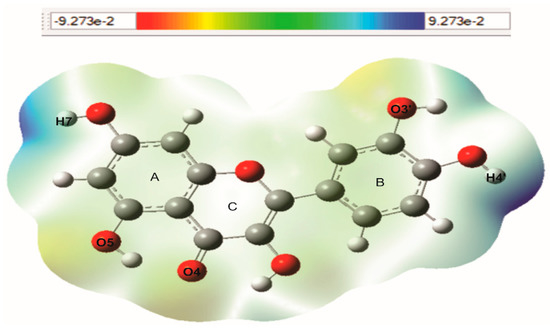
Figure 2.
Molecular electrostatic potential (MEP) map of quercetin (Q), calculated with B3LYP/6-31G (d,p) methodology.
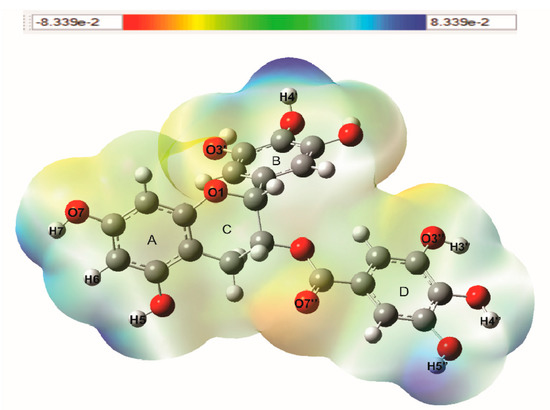
Figure 3.
Molecular electrostatic potential (MEP) map of epigallocatechin gallate (EGCG), calculated with B3LYP/6-31G (d,p) methodology.
3.4. Electronic Density of the Frontier Molecular Orbitals (HOMO and LUMO)
The electron density of the frontier molecular orbitals was obtained by performing the energy calculations of Q and EGCG molecules in neutral state. A detailed analysis of the molecular orbitals can provide useful information about the electron density distribution of a molecule. It can be observed for Q, that both the HOMO orbital (donor) and LUMO orbital (acceptor) are distributed throughout the entire structure of the molecule (see Figure 4). The distribution of the electronic density of HOMO is an indication of the susceptibility to electrophilic attacks, and the possible interactions of quercetin (Q) with the active site of ricin protein.
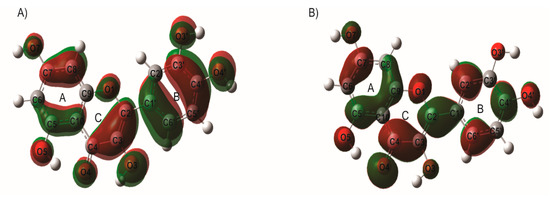
Figure 4.
Localization of (A) HOMO and (B) LUMO molecular orbitals of quercetin (Q), calculated with B3LYP/6-31G (d,p) methodology.
In the epigallocatechin gallate (EGCG), it can be seen that the HOMO orbital is distributed in the A and C rings of the structure, indicating this part of the molecule as an electron donor zone and the more susceptible atoms for an electrophilic attack. See Figure 5. On the other hand, the D-ring appears to be more susceptible to a nucleophilic attack; it was confirmed with the results of molecular docking.
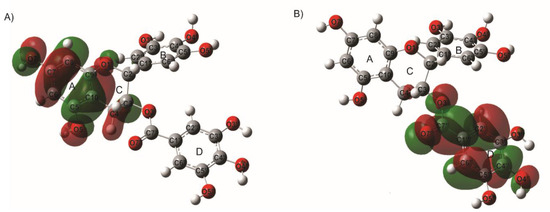
Figure 5.
Localization of (A) HOMO and (B) LUMO molecular orbitals of epigallocatechin gallate (EGCG), calculated with B3LYP/6-31G (d,p) methodology.
3.5. Docking Analysis
The molecular docking was realized between the ricin protein and the best geometries of quercetin (Q) and epigallocatechin gallate (EGCG). The ricin protein presents two chains, A and B; these chains have different active sites, one site for the A chain and two active sites for the B chain, which are distinguished as site 1 (B1) and site 2 (B2); this analysis was carried out with the Autodock 4.2 program [56].
The results show the interactions between the active site of A chain and the ligands Q and EGCG. The obtained result for Q with A chain showed four hydrogen bonds with one π–π interaction between TYR 80 and C7 in A ring, while in the results for EGCG with A chain showed six hydrogen bonds (Figure 6).
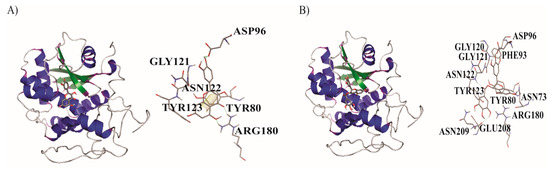
Figure 6.
Detailed molecular interactions of (A) quercetin (Q) and (B) epigallocatechin gallate (EGCG) both with the active site of the A chain of the protein ricin.
On the other hand, Figure 7 shows the interactions between the active sites of B chain (B1 y B2) and the ligands Q and EGCG. It was observed that the ligands are joined in a shallow pocket site in the B chain, where five hydrogen bonds were found in Q with B1 and B2 interactions, and only two π–cation interactions between LYS 40 with C1’ and C3’ in B1 site. In the EGCG interactions with B chain, seven (in B1) and two (in B2) hydrogen bonds were observed.
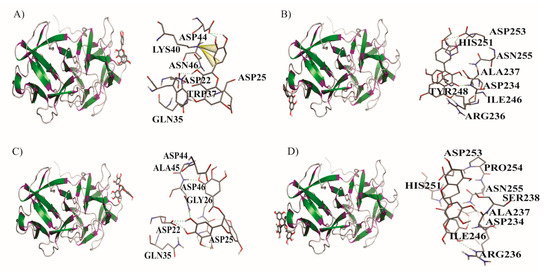
Figure 7.
Detailed molecular interactions of quercetin (Q) and epigallocatechin gallate (EGCG) with the active sites of the B chain of the protein ricin. (A) Q with B1. (B) Q with B2. (C) EGCG with B1. (D) EGCG with B2.
The results in Table 3 show that the most stable energy was obtained for A chain with both molecules: registered values of −6.30 kcal/mol for Q and −7.43 kcal/mol for EGCG, which coincide with Monzingo and Robertus, who found that the rings of Tyr80 and Tyr123 in the active site of chain A, are the most energetically favorable region for stacking of aromatic molecules and formation of hydrogen bonds [59]. In the case of B chain, the most stable energy was found in B1 site for both molecules. The amino acids of the active sites match with those reported in the literature [60,61], particularly for the A chain; the docking of EGCG with B chain (B1) shows a higher number of hydrogen bonds due to the conformation of the structure in the active site.

Table 3.
Molecular interaction analysis for the ricin protein (A and B chains) and quercetin (Q) and epigallocatechin gallate (EGCG).
The coupling between ricin and ligands had more interactions in the case of the quercetin molecule, as shown by five hydrogen bonds and two π–cation interactions in B1 and five hydrogen bonds in B2 active site; and in the A chain this molecule presented four hydrogen bonds and one π–π interaction. Besides that, the EGCG, presented seven hydrogen bonds in the B1 active site, and six hydrogen bonds in the A active site, which can be attributed to the higher number of –OH groups in the structure; this agrees with M. Sahihi et. al., who reported that the number of hydrogen bond interactions increases when the number of –OH groups in flavonoids structure increases [62].
3.6. Adjustment and Alignment after Interaction
The molecular docking results were verified with a superimposition of the ligands. This analysis was determined with the adjustment and alignment of the Q and EGCG geometries, before (in green) and after (in blue) the interactions in the active sites of A and B chains. The energy difference between both geometries allows one to know the feasibility of the interaction with the active site of the protein. The structural changes in the molecules depend on the interactions with the amino acids of the active sites. The obtained results did not show a considerable energy difference for the quercetin before and after the interaction, with values in the range of −0.036 to −0.042 kcal/mol in both chains. For EGCG, the difference is remarkable: values of −0.11 kcal/mol for A chain, −0.12 kcal/mol for B1, and −0.25 kcal/mol for B2 site; this means that EGCG requires more energy to interact with the active site B2 due to several rotational angles in the structure. Figure 8 shows the geometries of the Q and EGCG before and after the interactions with the active sites, specifying a difference of total electronic energy.
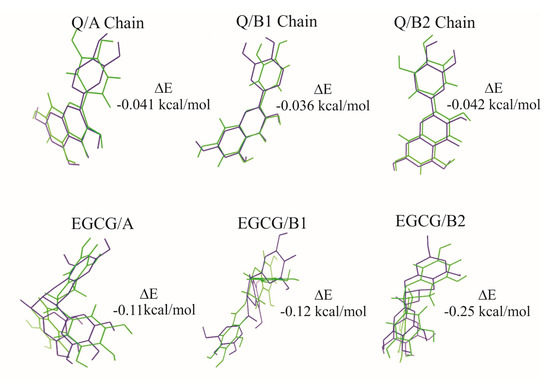
Figure 8.
An adjustment and alignment of the ligands Q and EGCG before (green) and after (blue) the interaction with A and B chains of the ricin protein.
3.7. Fractional Number of Transferred Electrons (ΔN)
Charge transfer between amino acids of the active site of A and B (B1 and B2) chains with the ligands was calculated using Equation (6). The active amino acid sites were isolated using PyMOL [38] software, without modifying their compositions or conformations. The reactivity parameters of chemical hardness (η) and chemical potential (µ) were used for the determination of the charge transfer parameter (ΔN) [63]. This parameter describes the fractional number of electrons transferred from A system to B system. In this case, the polyphenols Q and EGCG were selected as the A system, and the amino acids in the active site were selected as B system. If ΔN values are positive, it indicates that the system is an electron donor. In contrast, if ΔN values are negative, the system is an electron acceptor. For the interaction between Q and the A, B1, and B2 active sites, the obtained values of ΔN yielded only negative values, indicating that the charge transfer flows from quercetin to the amino acids of ricin, thereby meaning Q acts as an electron donor. In the case of EGCG, the ΔN parameter showed some positive values, such as the TYR80 with 0.001 and ARG180 with 0.788 in the A chain, and ASP25 with 0.196 in B1 active site, thereby it acts as an electron acceptor with those amino acids. The rest amino acids of the active site presented negative values of ΔN, so the EGCG acts as an electron donor. These results are shown in Figure 9 and Figure 10.
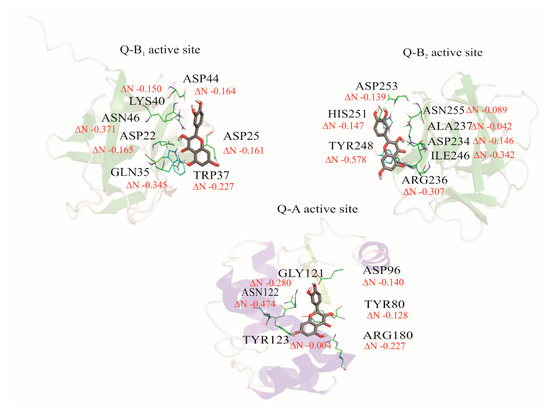
Figure 9.
The schematization of the fractional number of transferred electrons (ΔN) between quercetin and A, B1, and B2 active sites of the ricin protein.
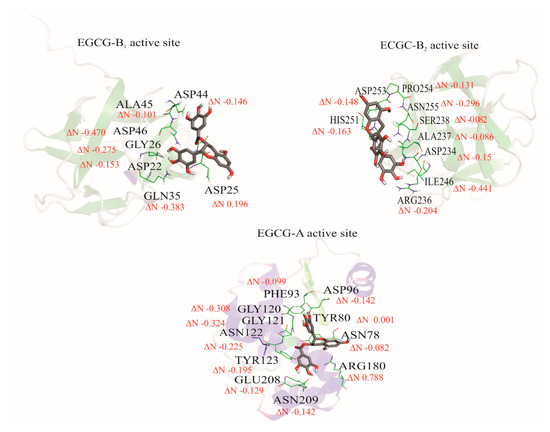
Figure 10.
The schematization of the fractional number of transferred electrons (ΔN) between epigallocatechin gallate and A, B1, and B2 active sites of the ricin protein.
4. Conclusions
This study provides essential information about quantum-chemical calculations of quercetin (Q) and epigallocatechin gallate (EGCG) molecules, and valuable information about the interactions of Q and EGCG with the ricin protein for possible inhibition.
For geometrical parameters of Q and EGCG, the methodology B3LYP/6-31G (d,p) was defined as the best approximation with X-ray diffraction experimental values.
According to molecular electrostatic potential (MEP), the site for nucleophilic attack situated in quercetin (Q) coincided with the interactions obtained in A, B1, and B2 chains. For epigallocatechin gallate (EGCG) the electrophilic attack sites coincided with both chains A and B1. In contrast, the nucleophilic attack partially coincided with the interactions obtained for the A chain and the B1 and B2 chains. This behavior in EGCG could be related to the dihedral angles in the structure, which generated a steric impediment between the molecule and amino acids of the active site.
The analysis of the frontier molecular orbitals (HOMO and LUMO) showed that the electronic density of quercetin is distributed throughout the entire structure of the molecule, confirming the molecular coupling results. For epigallocatechin gallate, the HOMO orbital is positioned in the A and B rings, which coincides with the interactions shown for A and B1 chains, whereas the LUMO orbital matches with the interactions obtained for A, B1, and B2.
The comparison of the Q geometry before and after the interaction with the ricin chains showed the lowest values of ΔE. In contrast, for EGCG, these values indicated a greater deformation of the structure. Moreover, the charge transfer parameter (ΔN) calculated for both molecules, Q and EGCG, showed that the tendency of these systems is toward electrons donation because of the negative values; it is an indication that these compounds suffer an oxidation process inside of the active site.
Based on the above-mentioned results, we can say that both flavonoids present significant interactions with the ricin protein, which could be considered as a probable inactivation of the protein, triggering a possible detoxification process.
Author Contributions
Formal analysis, M.C.M.-C. and L.-L.L.-M.; investigation, M.C.M.-C.; methodology, L.M.R.-V.; writing—original draft, M.C.M.-C., L.-L.L.-M., and L.M.R.-V.; writing—review and editing, L.-L.L.-M., N.-A.S.-B., F.S.-S., H.A.P.-C., L.R.H.-O., and L.M.R.-V. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Consejo Nacional de Ciencia y Tecnología.
Acknowledgments
M.C.M-C thanks Consejo Nacional de Ciencia y Tecnología. All the researchers are grateful to Universidad Autónoma de Chihuahua for the support and infrastructure provided.
Conflicts of Interest
The authors declare no conflict of interest. The founding sponsors had no role in the design in the analyses and writing of the study.
References
- Voisin, G.; Jansen, F.; Gros, P. Cytotoxic Products Formed by Covalent Bonding of the A Chain of Ricin with an Antibody and the Process for Their Preparation and Use. U.S. Patent 4,340,535, 20 July 1982. [Google Scholar]
- Bozza, W.P.; Tolleson, W.H.; Rivera Rosado, L.A.; Zhang, B. Ricin detection: Tracking active toxin. Biotechnol. Adv. 2015, 33, 117–123. [Google Scholar] [CrossRef]
- Polito, L.; Bortolotti, M.; Battelli, M.G.; Calafato, G.; Bolognesi, A. Ricin: An ancient story for a timeless plant toxin. Toxins 2019, 11, 324. [Google Scholar] [CrossRef]
- Olsnes, S.; Kozlov, J.V. Ricin. Toxicon 2001, 39, 1723–1728. [Google Scholar] [CrossRef]
- Iglesias, R.; Citores, L.; Ferreras, J. Ribosomal RNA N-glycosylase activity assay of ribosome-inactivating proteins. Bio-Protocol 2017, 7, e2180. [Google Scholar] [CrossRef]
- Silitonga, A.S.; Masjuki, H.H.; Ong, H.C.; Yusaf, T.; Kusumo, F.; Mahlia, T.M.I. Synthesis and optimization of Hevea brasiliensis and Ricinus communis as feedstock for biodiesel production: A comparative study. Ind. Crops Prod. 2016, 85, 274–286. [Google Scholar] [CrossRef]
- Akande, T.O.; Odunsi, A.A.; Akinfala, E.O. A review of nutritional and toxicological implications of castor bean (Ricinus communis L.) meal in animal feeding systems. J. Anim. Physiol. Anim. Nutr. 2016, 100, 201–210. [Google Scholar] [CrossRef] [PubMed]
- Sousa, N.L.; Cabral, G.B.; Vieira, P.M.; Baldoni, A.B.; Aragão, F.J.L. Bio-detoxification of ricin in castor bean (Ricinus communis L.) seeds. Sci. Rep. 2017, 7, 15385. [Google Scholar] [CrossRef] [PubMed]
- Adedapo, A.M.; Babalola, T.O. Effect of Autoclaved Castor Seed Meal on Growth Performance, Body Composition and Haematology of Clarias Gariepinus Fingelings; Federal University Oye-Ekiti: Oye-Ekiti, Nigeria, 2017. [Google Scholar]
- Ashraf Ashfaq, M.; Soma Sekhar Reddy, P.; Anil Kumar, C.; Selvaraj, V.M.; Dinesh Kumar, V. Ricin and RCA—The enemies within castor (Ricinus communis L.): A perspective on their biogenesis, mechanism of action, detection methods and detoxification strategies. In The Castor Bean Genome; Springer: Cham, Switzerland, 2018; pp. 215–235. ISBN 9783319972800. [Google Scholar]
- Dyer, P.D.R.; Kotha, A.K.; Gollings, A.S.; Shorter, S.A.; Shepherd, T.R.; Pettit, M.W.; Alexander, B.D.; Getti, G.T.M.; El-Daher, S.; Baillie, L.; et al. An in vitro evaluation of epigallocatechin gallate (eGCG) as a biocompatible inhibitor of ricin toxin. Biochim. Biophys. Acta Gen. Subj. 2016, 1860, 1541–1550. [Google Scholar] [CrossRef] [PubMed]
- Bravo, L. Polyphenols: Chemistry, dietary sources, metabolism, and nutritional significance. Nutr. Rev. 2009, 56, 317–333. [Google Scholar] [CrossRef]
- Guo, W.; Kong, E.; Meydani, M. Dietary polyphenols, inflammation, and cancer. Nutr. Cancer 2009, 61, 807–810. [Google Scholar] [CrossRef]
- Wang, B.; Guo, C.; Zhang, M.; Park, B.; Xu, B. High-resolution single-molecule recognition imaging of the molecular details of Ricin–aptamer interaction. J. Phys. Chem. B 2012, 116, 5316–5322. [Google Scholar] [CrossRef] [PubMed]
- Rice-evans, C.A.; Miller, N.J.; Bolwell, P.G.; Bramley, P.M.; Pridham, J.B. The relative antioxidant activities of plant-derived polyphenolic flavonoids. Free Radic. Res. 1995, 22, 375–383. [Google Scholar] [CrossRef] [PubMed]
- Okawa, M.; Kinjo, J.; Nohara, T.; Ono, M. DPPH (1,1-Diphenyl-2-Picrylhydrazyl) radical scavenging activity of Flavonoids obtained from some medicinal plants. Biol. Pharm. Bull. 2001, 24, 1202–1205. [Google Scholar] [CrossRef] [PubMed]
- Han, X.; Shen, T.; Lou, H. Dietary polyphenols and their biological significance. Int. J. Mol. Sci. 2007, 8, 950–988. [Google Scholar] [CrossRef]
- Lesjak, M.; Beara, I.; Simin, N.; Pintać, D.; Majkić, T.; Bekvalac, K.; Orčić, D.; Mimica-Dukić, N. Antioxidant and anti-inflammatory activities of quercetin and its derivatives. J. Funct. Foods 2018, 40, 68–75. [Google Scholar] [CrossRef]
- Higdon, J.V.; Frei, B. Tea catechins and polyphenols: Health effects, metabolism, and antioxidant functions. Crit. Rev. Food Sci. Nutr. 2003, 43, 89–143. [Google Scholar] [CrossRef]
- De la Torre-Robles, A.; Rivas, A.; Lorenzo-Tovar, M.L.; Monteagudo, C.; Mariscal-Arcas, M.; Olea-Serrano, F. Estimation of the intake of phenol compounds from virgin olive oil of a population from southern Spain. Food Addit. Contam. Part A 2014, 31, 1460–1469. [Google Scholar] [CrossRef]
- Legeay, S.; Rodier, M.; Fillon, L.; Faure, S.; Clere, N. Epigallocatechin gallate: A review of its beneficial properties to prevent metabolic syndrome. Nutrients 2015, 7, 5443–5468. [Google Scholar] [CrossRef]
- Kim, D.-O.; Lee, C.Y. Comprehensive study on vitamin C equivalent antioxidant capacity (VCEAC) of various polyphenolics in scavenging a free radical and its structural relationship. Crit. Rev. Food Sci. Nutr. 2004, 44, 253–273. [Google Scholar] [CrossRef]
- Lopez del Amo, J.M.; Fink, U.; Dasari, M.; Grelle, G.; Wanker, E.E.; Bieschke, J.; Reif, B. Structural properties of EGCG-induced, nontoxic Alzheimer’s disease Aβ oligomers. J. Mol. Biol. 2012, 421, 517–524. [Google Scholar] [CrossRef]
- Soobrattee, M.A.; Neergheen, V.S.; Luximon-Ramma, A.; Aruoma, O.I.; Bahorun, T. Phenolics as potential antioxidant therapeutic agents: Mechanism and actions. Mutat. Res. Mol. Mech. Mutagen. 2005, 579, 200–213. [Google Scholar] [CrossRef] [PubMed]
- Seeram, N.P.; Nair, M.G. Inhibition of lipid peroxidation and structure−activity-related studies of the dietary constituents anthocyanins, anthocyanidins, and catechins. J. Agric. Food Chem. 2002, 50, 5308–5312. [Google Scholar] [CrossRef] [PubMed]
- Ferry, D.R.; Smith, A.; Malkhandi, J.; Fyfe, D.W.; DeTakats, P.G.; Anderson, D.; Baker, J.; Kerr, D.J. Phase I clinical trial of the flavonoid quercetin: Pharmacokinetics and evidence for in vivo tyrosine kinase inhibition. Clin. Cancer Res. 1996, 2, 659–668. [Google Scholar] [PubMed]
- Sengupta, B.; Sengupta, P.K. The interaction of quercetin with human serum albumin: A fluorescence spectroscopic study. Biochem. Biophys. Res. Commun. 2002, 299, 400–403. [Google Scholar] [CrossRef]
- Gutzeit, H.O.; Henker, Y.; Kind, B.; Franz, A. Specific interactions of quercetin and other flavonoids with target proteins are revealed by elicited fluorescence. Biochem. Biophys. Res. Commun. 2004, 318, 490–495. [Google Scholar] [CrossRef]
- Dahiya, R.; Mohammad, T.; Roy, S.; Anwar, S.; Gupta, P.; Haque, A.; Khan, P.; Kazim, S.N.; Islam, A.; Ahmad, F.; et al. Investigation of inhibitory potential of quercetin to the pyruvate dehydrogenase kinase 3: Towards implications in anticancer therapy. Int. J. Biol. Macromol. 2019, 136, 1076–1085. [Google Scholar] [CrossRef]
- Avakyan, V.G.; Nazarov, V.B.; Alfimov, M.V.; Bagatur, A.A.; Voronezheva, N.I. The role of intra-and intermolecular hydrogen bonds in the formation of β-cyclodextrin head-to-head and head-to-tail dimers. The results of ab initio and semiempirical quantum-chemical calculations. Russ. Chem. Bull. 2001, 50, 206–216. [Google Scholar] [CrossRef]
- Pithayanukul, P.; Ruenraroengsak, P.; Bavovada, R.; Pakmanee, N.; Suttisri, R.; Saen-oon, S. Inhibition of Naja kaouthia venom activities by plant polyphenols. J. Ethnopharmacol. 2005, 97, 527–533. [Google Scholar] [CrossRef]
- Parr, R.G.; Szentpály, L.V.; Liu, S. Electrophilicity index. J. Am. Chem. Soc. 1999, 121, 1922–1924. [Google Scholar] [CrossRef]
- Landeros-Martínez, L.L.; Chavez-Flores, D.; Orrantia-Borunda, E.; Flores-Holguin, N. Construction of a nanodiamond-tamoxifen complex as a breast cancer drug delivery vehicle. J. Nanomater. 2016. [Google Scholar] [CrossRef]
- Landeros-Martínez, L.L.; Glossman-Mitnik, D.; Orrantia-Borunda, E.; Flores-Holguín, N. Theoretical calculation of UV-Vis, IR spectra and reactivity properties of tamoxifen drug: A methodology comparison. MOJ Bioorganic Org. Chem. 2017, 1, 87–95. [Google Scholar] [CrossRef]
- García-Montoya, I.A.; Flores-Holguín, N.R.; Landeros-Martínez, L.-L.; Alvarado-González, M.; Rascón-Cruz, Q.; Fuentes-Montero, M.E.; Palomares-Báez, P.; Rodríguez-Valdez, L.M. Analysis of the molecular interactions between cytochromes P450 3A4 and 1A2 and aflatoxins: A docking study. Appl. Sci. 2019, 9, 2467. [Google Scholar] [CrossRef]
- Tirado-Rives, J.; Jorgensen, W.L. Performance of B3LYP density functional methods for a large set of organic molecules. J. Chem. Theory Comput. 2008, 4, 297–306. [Google Scholar] [CrossRef] [PubMed]
- Winter, N.O.C.; Graf, N.K.; Leutwyler, S.; Hättig, C. Benchmarks for 0-0 transitions of aromatic organic molecules: DFT/B3LYP, ADC(2), CC2, SOS-CC2 and SCS-CC2 compared to high-resolution gas-phase data. Phys. Chem. Chem. Phys. 2013, 15, 6623–6630. [Google Scholar] [CrossRef]
- Aparicio, S. A systematic computational study on flavonoids. Int. J. Mol. Sci. 2010, 11, 2017–2038. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Li, X.; Shang, Y.; Chen, K. Radical scavenging activity of puerarin: A theoretical study. Antioxidants 2019, 8, 590. [Google Scholar] [CrossRef]
- Vieth, M.; Hirst, J.D.; Brooks, C.L. Do active site conformations of small ligands correspond to low free-energy solution structures? J. Comput. Aided. Mol. Des. 1998, 12, 563–572. [Google Scholar] [CrossRef]
- Perola, E.; Charifson, P.S. Conformational analysis of drug-like molecules bound to proteins: An extensive study of ligand reorganization upon binding. J. Med. Chem. 2004, 47, 2499–2510. [Google Scholar] [CrossRef]
- Hao, M.H.; Haq, O.; Muegge, I. Torsion angle preference and energetics of small-molecule ligands bound to proteins. J. Chem. Inf. Model. 2007, 47, 2242–2252. [Google Scholar] [CrossRef]
- Seo, M.H.; Park, J.; Kim, E.; Hohng, S.; Kim, H.S. Protein conformational dynamics dictate the binding affinity for a ligand. Nat. Commun. 2014, 5, 1–7. [Google Scholar] [CrossRef]
- Greives, N.; Zhou, H.X. Both protein dynamics and ligand concentration can shift the binding mechanism between conformational selection and induced fit. Proc. Natl. Acad. Sci. USA 2014, 111, 10197–10202. [Google Scholar] [CrossRef] [PubMed]
- Kitchen, D.B.; Decornez, H.; Furr, J.R.; Bajorath, J. Docking and scoring in virtual screening for drug discovery: Methods and applications. Nat. Rev. Drug Discov. 2004, 3, 935–949. [Google Scholar] [CrossRef] [PubMed]
- Becke, A.D. Density-functional thermochemistry. IV. A new dynamical correlation functional and implications for exact-exchange mixing. J. Chem. Phys. 1996, 104, 1040–1046. [Google Scholar] [CrossRef]
- Becke, A.D.; Johnson, E.R. A density-functional model of the dispersion interaction. J. Chem. Phys. 2005, 123, 154101. [Google Scholar] [CrossRef]
- Stephens, P.J.; Devlin, F.J.; Chabalowski, C.F.; Frisch, M.J. Ab initio calculation of vibrational absorption and circular dichroism spectra using density functional force fields. J. Phys. Chem. 1994, 98, 11623–11627. [Google Scholar] [CrossRef]
- Perdew, J.P.; Burke, K.; Ernzerhof, M. Generalized gradient approximation made simple. Phys. Rev. Lett. 1996, 77, 3865–3868. [Google Scholar] [CrossRef]
- Zhao, Y.; Truhlar, D.G. The M06 suite of density functionals for main group thermochemistry, thermochemical kinetics, noncovalent interactions, excited states, and transition elements: Two new functionals and systematic testing of four M06-class functionals and 12 other function. Theor. Chem. Acc. 2008, 120, 215–241. [Google Scholar] [CrossRef]
- Ghalami-Choobar, B.; Ghiami-Shomami, A. Theoretical calculation of pKa values of the Nortryptiline and Amitryptiline drugs in aqueous and non-aqueous solvents. Comput. Theor. Chem. 2013, 1018, 66–70. [Google Scholar] [CrossRef]
- Amovilli, C.; Barone, V.; Cammi, R.; Cancès, E.; Cossi, M.; Mennucci, B.; Pomelli, C.S.; Tomasi, J. Recent Advances in the Description of Solvent Effects with the Polarizable Continuum Model. In Advances in Quantum Chemistry; Academic Press Inc.: Cambridge, MA, USA, 1998; Volume 32, pp. 227–261. [Google Scholar]
- Mennucci, B.; Cancès, E.; Tomasi, J. Evaluation of solvent effects in isotropic and anisotropic dielectrics and in ionic solutions with a unified integral equation method: Theoretical bases, computational implementation, and numerical applications. J. Phys. Chem. B 1997, 101, 10506–10517. [Google Scholar] [CrossRef]
- Frisch, M.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Mennucci, B.; Petersson, G. Gaussian 09, Revision d. 01; Gaussian, Inc.: Wallingford, CT, USA, 2009. [Google Scholar]
- Delano, W.L. PyMOL Molecular Graphics System; Delano Scientific, L.L.C.: San Carlos, CA, USA, 2002. [Google Scholar]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef]
- Spek, A.L.; Kojić-Prodić, B.; Labadie, R.P. Structure of (−)-epicatechin: (2R,3R)-2-(3,4-dihydroxyphenyl)-3,4-dihydro-2H-1-benzopyran-3,5,7-triol, C15H14O6. Acta Crystallogr. Sect. C Cryst. Struct. Commun. 1984, 40, 2068–2071. [Google Scholar] [CrossRef]
- Jin, G.Z.; Yamagata, Y.; Tomita, K. Structure of quercetin dihydrate. Acta Crystallogr. Sect. C Cryst. Struct. Commun. 1990, 46, 310–313. [Google Scholar] [CrossRef]
- Monzingo, A.F.; Robertus, J.D. X-ray analysis of substrate analogs in the ricin A-chain active site. J. Mol. Biol. 1992, 227, 1136–1145. [Google Scholar] [CrossRef]
- Katzin, B.J.; Collins, E.J.; Robertus, J.D. Structure of ricin A-chain at 2.5 Å. Proteins Struct. Funct. Genet. 1991, 10, 251–259. [Google Scholar] [CrossRef]
- Rutenber, E.; Robertus, J.D. Structure of ricin B-chain at 2.5 Å resolution. Proteins Struct. Funct. Genet. 1991, 10, 260–269. [Google Scholar] [CrossRef]
- Sahihi, M.; Heidari-Koholi, Z.; Bordbar, A.-K. The interaction of polyphenol flavonoids with β-lactoglobulin: molecular docking and molecular dynamics simulation studies. J. Macromol. Sci. Part. B 2012, 51, 2311–2323. [Google Scholar] [CrossRef]
- Parr, R.G.; Pearson, R.G. Absolute hardness: Companion parameter to absolute electronegativity. J. Am. Chem. Soc. 1983, 105, 7512–7516. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).