Some Eccentricity-Based Topological Indices and Polynomials of Poly(EThyleneAmidoAmine) (PETAA) Dendrimers
Abstract
:1. Introduction
2. Materials and Methods
3. Eccentricity-Based Indices and Polynomials for the PETAA Dendrimer
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Gozalbes, R.; Doucet, J.P.; Derouin, F. Application of topological descriptors in QSAR and drug design: history and new trends. Curr. Drug Targets Infect. Disord. 2002, 2, 93–102. [Google Scholar] [CrossRef] [PubMed]
- Amic, D.; Beaa, D.; Lucic, B.; Nikolic, S.; Trinajstic, N. The vertexconnectivity index revisited. J. Chem. Inf. Comput. 1998, 38, 819–822. [Google Scholar] [CrossRef]
- Ivanciuc, O. Topological Indices. In Handbook of Chemoinformatics; Gasteiger, J., Ed.; Wiley–VCH: Weinheim, Germany, 2003; Volume 3, pp. 981–1003. [Google Scholar]
- Estrada, E.; Patlewicz, G.; Uriarte, E. From molecular graphs to drugs. A review on the use of topological indices in drug design and discovery. Indian J. Chem. A 2003, 42, 1315–1329. [Google Scholar]
- Buhleier, E.; Wehner, W.; Vogtle, F. Cascade and Nonskid Chain like Syntheses of Molecular Cavity Topologies. Synthesis 1978, 2, 155–158. [Google Scholar] [CrossRef]
- Tomalia, D.A.; Baker, H.; Dewald, J.R.; Hall, M.; Kallos, G.; Martin, S.; Roeck, J.; Ryder, J.; Smith, P. Dendritic macromolecules: Synthesis of starburst dendrimers. Macromolecules 1986, 19, 2466–2468. [Google Scholar] [CrossRef]
- Newkome, G.R.; Yao, Z.; Baker, G.R.; Gupta, V.K. Micelles. Part 1. Cascade molecules: A new approach to micelles. A [27]-arborol. J. Org. Chem. 1985, 50, 2003–2004. [Google Scholar] [CrossRef]
- Abbasi, F.; Aval, S.F.; Akbarzadeh, A.; Milani, M.; Nasrabadi, H.T.; Joo, S.W.; Hanifehour, Y.; Koshki, K.N.; Pashei-Asl, R. Dendrimers: Synthesis, applications, and properties. Nano Scale Res. Lett. 2014, 247, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Huang, B.; Tang, S.; Desai, A.; Lee, K.; Leroueil, P.R.; Baker, R., Jr. Noevl Poly EThyleneAmidoAmine PETAA dendrimers produced through a unique and highly efficient synthesis. Polymer 2011, 52, 5975–5984. [Google Scholar] [CrossRef]
- Trinajstic, N. Chemical Graph Theory, 2nd ed.; CRC Press: Boca Raton, FL, USA, 1992. [Google Scholar]
- Khalifeh, M.H.; Azari, H.Y.; Ashrafi, A.R. Vertex and edge PI indices of Cartesian product graphs. Discret. Appl. Math. 2008, 156, 1780–1789. [Google Scholar] [CrossRef] [Green Version]
- Sharma, V.; Goswami, R.; Madan, A.K. Eccentric-connectivity index: A novel highly discriminating topological descriptor for structure property and structure activity studies. J. Chem. Inf. Comput. Sci. 1997, 37, 273–282. [Google Scholar] [CrossRef]
- Kumar, V.; Madan, A.K. Application of graph theory: Prediction of glycogen synthase kinase-3 beta inhibitory activity of thiadiazolidinones as potential drugs for the treatment of Alzheimer’s disease. Eur. J. Pharm. Sci. 2005, 24, 213–218. [Google Scholar] [CrossRef] [PubMed]
- Sardana, S.; Madan, A.K. Application of graph theory: Relationship of molecular connectivity index, Wiener index and eccentric connectivity index with diuretic activity. MATCH Commun. Math. Comput. Chem. 2001, 43, 85–98. [Google Scholar]
- Gupta, S.; Singh, M.; Madan, A.K. Application of graph theory: relationship of eccentric connectivity index and Wiener’s index with anti-inflammatory activity. J. Math. Anal. Appl. 2002, 266, 259–268. [Google Scholar] [CrossRef]
- Lather, V.; Madan, A.K. Topological models for the prediction of HIV-protease inhibitory activity of tetrahydropyrimidin-2-ones. J. Mol. Graph. Model. 2005, 23, 339–345. [Google Scholar] [CrossRef] [PubMed]
- Sardana, S.; Madan, A.K. Topological models for prediction of antihypertensive activity of substituted benzylimidazoles. J. Mol. Struct. (Theochem.) 2003, 638, 41–49. [Google Scholar] [CrossRef]
- Dureja, H.; Madan, A.K. Topochemical models for prediction of cyclin-dependent kinase 2 inhibitory activity of indole-2-ones. J. Mol. Model. 2005, 11, 525–531. [Google Scholar] [CrossRef]
- Ilic, A.; Gutman, I. Eccentric-connectivity index of chemical trees. MATCH Commun. Math. Comput. Chem. 2011, 65, 731–744. [Google Scholar]
- Kumar, V.; Madan, A.K. Application of graph theory: Prediction of cytosolic phospholipase A2 inhibitory activity of propan-2-ones. J. Math. Chem. 2006, 39, 511–521. [Google Scholar] [CrossRef]
- Zhou, B. On eccentric-connectivity index. MATCH Commun. Math. Comput. Chem. 2010, 63, 181–198. [Google Scholar]
- Ashrafi, A.R.; Ghorbani, M.; Hossein-Zadeh, M.A. The eccentric-connectivity polynomial of some graph operations. Serdica J. Comput. 2011, 5, 101–116. [Google Scholar]
- Hasni, R.; Arif, N.E.; Alikhani, S. Eccentric connectivity polynomials of some families of dendrimers. J. Comput. Theor. Nanosci. 2014, 11, 450–453. [Google Scholar] [CrossRef]
- Ghorbani, M.; Iranmanesh, M. Computing eccentric connectivity polynomial of fullerenes. Fuller. Nanotub. Carbon Nanostruct. 2013, 21, 134–139. [Google Scholar] [CrossRef]
- Ghorbani, M.; Hemmasi, M. Eccentric connectivity polynomial of C18n+10 fullerenes. Bulg Chem. Commun. 2013, 45, 5–8. [Google Scholar]
- Fathalikhani, K.; Faramarzi, H.; Yousefi-Azari, H. Total eccentricity of some graph operations. Electron. Notes Discret. Math. 2014, 45, 125–131. [Google Scholar] [CrossRef]
- Ghorbani, M.; Hosseinzadeh, M.A. A new version of Zagreb indices. Filomat 2012, 26, 93–100. [Google Scholar] [CrossRef] [Green Version]
- Das, K.C.; Lee, D.W.; Graovac, A. Some properties of the Zagreb eccentricity indices. ARS Math. Contemp. 2013, 6, 117–125. [Google Scholar] [CrossRef]
- Vukičević, D.; Graovac, A. Note on the comparison of the first and second normalized Zagreb eccentricity indices. Acta Chim. Slov. 2010, 57, 524–528. [Google Scholar]
- Du, Z.; Zhou, B.; Trinajstić, N. Extremal properties of the Zagreb eccentricity indices. Croat. Chem. Acta 2012, 85, 359–362. [Google Scholar] [CrossRef]
- Qi, X.; Du, Z. On Zagreb eccentricity indices of trees. MATCH Commun. Math. Comput. Chem. 2017, 78, 241–256. [Google Scholar]
- Gupta, S.; Singh, M.; Madan, A.K. Connective eccentricity index: A novel topological descriptor for predicting biological activity. J. Mol. Graph. Model. 2000, 18, 18–25. [Google Scholar] [CrossRef]
- Bajaj, S.; Sambi, S.S.; Gupta, S.; Madan, A.K. Model for prediction of anti-HIV activity of 2-pyridinone derivatives using novel topological descriptor. QSAR Comb. Sci. 2006, 25, 813–823. [Google Scholar] [CrossRef]
- De, N. Relationship between augmented eccentric connectivity index and some other graph invariants. Int. J. Adv. Math. Sci. 2013, 1, 26–32. [Google Scholar] [CrossRef]
- Doślić, T.; Saheli, M. Augmented eccentric-connectivity index. Miskolc Math. Notes 2011, 12, 149–157. [Google Scholar]
- Alaeiyan, M.; Asadpour, J.; Mojarad, R. A numerical method for MEC polynomial and MEC index of one-pentagonal carbon nanocones, Fullerenes. Nanotubes Carbon Nanostruct. 2013, 21, 825–835. [Google Scholar] [CrossRef]
- De, N.; Nayeem, S.M.A.; Pal, A. Modified eccentric-connectivity of Generalized Thorn Graphs. Int. J. Comput. Math. 2014, 2014, 436140. [Google Scholar] [CrossRef]
- Xu, X.; Guo, Y. The edge version of eccentric connectivity index. Int. Math. Forum 2012, 7, 273–280. [Google Scholar]
- Odaba, Z.N. The edge eccentric-connectivity index of dendrimers. J. Comput. Theor. Nanosci. 2013, 10, 783–784. [Google Scholar] [CrossRef]
- Gao, W.; Iqbal, Z.; Ishaq, M.; Sarfraz, R.; Aamir, M.; Aslam, A. On Eccentricity-Based Topological Indices Study of a Class of Porphyrin-Cored Dendrimers. Biomolecules 2018, 8, 71. [Google Scholar] [CrossRef]
- Iqbal, Z.; Ishaq, M.; Aslam, A.; Gao, W. On eccentricity-based topological descriptors of water-soluble dendrimers. Z. Naturforsch. 2018, 74, 25–33. [Google Scholar] [CrossRef]
- Gao, W.; Iqbal, Z.; Ishaq, M.; Aslam, A.; Sarfraz, R. Topological aspects of dendrimers via distance-based descriptors. IEEE Access 2019, 7, 35619–35630. [Google Scholar] [CrossRef]
- Devillers, J.; Balaban, A.T. Topological Indices and Related Descriptors in QSAR and QSPR; Gordon and Breach Science Publishers: Amsterdam, The Netherlands, 1999. [Google Scholar]
- Randić, M. Quantitative structure-property relationship: Boiling points and planar benzenoids. New J. Chem. 1996, 20, 1001–1009. [Google Scholar]
- Randić, M. Comparative regression analysis. Regressions based on a single descriptor. Croatica Chem. Acta 1993, 66, 289–312. [Google Scholar]
- Randić, M.; Pompe, M. On characterization of the CC double bond in alkenes. SAR QSAR Environ. Res. 1999, 10, 451–471. [Google Scholar]
- Karelson, M.; Lobanov, V.S. Quantum-Chemical descriptors in QSAR/QSPR studies. Chem. Rev. 1996, 96, 1027–1043. [Google Scholar] [CrossRef] [PubMed]
- Dureja, H.; Gupta, S.; Madan, A.K. Predicting anti-HIV-1 activity of 6-arylbenzonitriles: Computational Approach using superaugmented eccentric connectivity topochemical indices. J. Mol. Graph. Model. 2008, 26, 1020–1029. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.; Singh, M.; Madan, A.K. Eccentric distance sum: A novel graph invariant for predicting biological and physical properties. J. Math. Anal. Appl. 2002, 275, 386–401. [Google Scholar] [CrossRef] [Green Version]
- Kumar, V.; Sardana, S.; Madan, A.K. Predicting anti-HIV activity of 2,3-diaryl-1,3 thiazolidin-4-ones: Computational approach using reformed eccentric connectivity index. J. Mol. Model. 2004, 10, 399–407. [Google Scholar] [CrossRef] [PubMed]
- Iqbal, Z.; Ishaq, M.; Farooq, R. Computing different versions of atom-bond connectivity index of dendrimers. J. Inform. Math. Sci. 2017, 9, 217–229. [Google Scholar]
- Kang, S.M.; Iqbal, Z.; Ishaq, M.; Sarfraz, R.; Aslam, A.; Nazeer, W. On Eccentricity-Based Topological Indices and Polynomials of Phosphorus-Containing Dendrimers. Symmetry 2018, 10, 237. [Google Scholar] [CrossRef]
- Iqbal, Z.; Ishaq, M.; Aamir, M. On Eccentricity-Based Topological Descriptors of Dendrimers. Iran. J. Sci. Technol. Trans. Sci. 2018. [Google Scholar] [CrossRef]
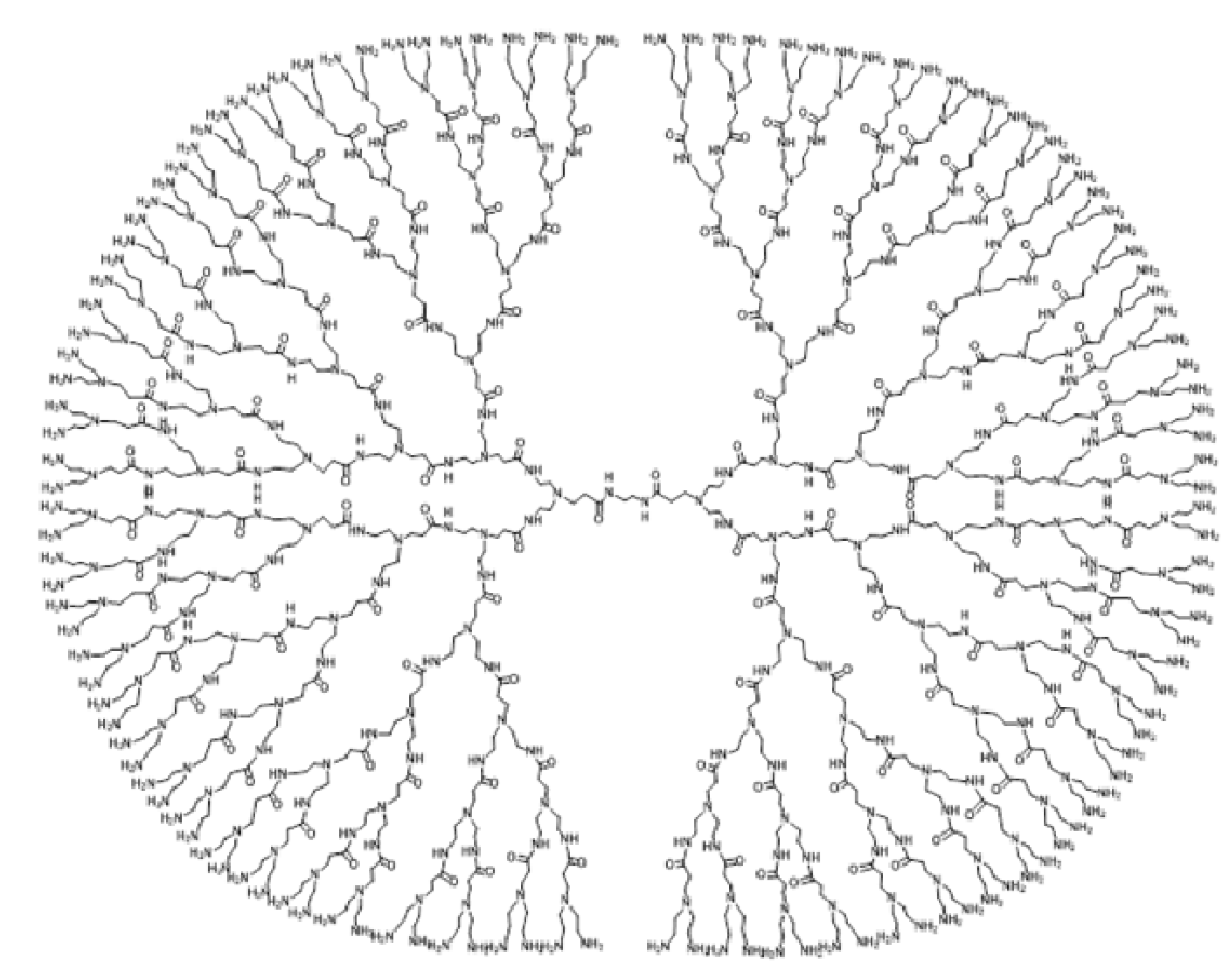
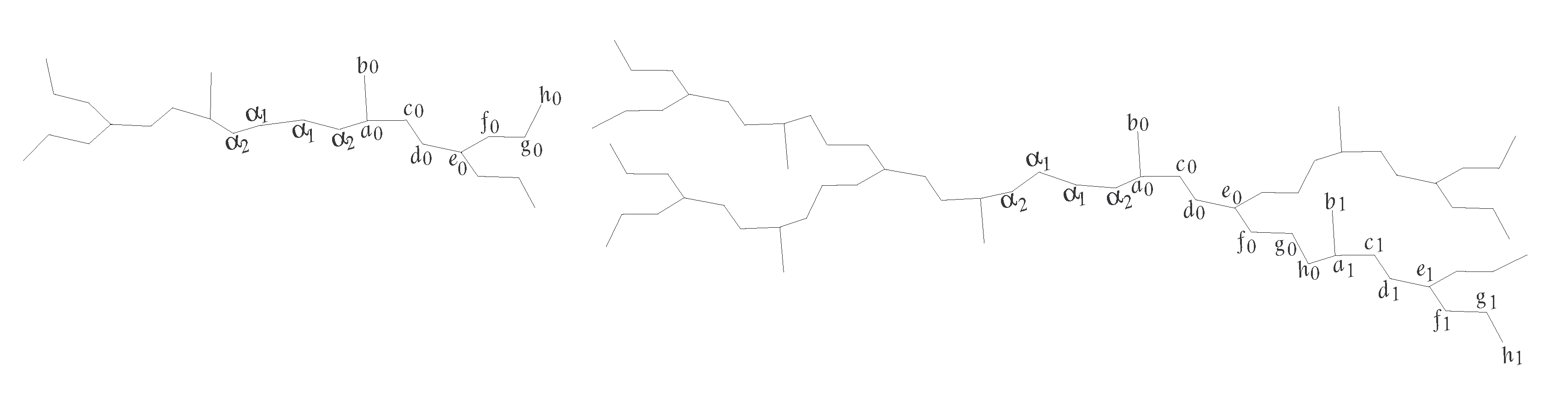
| Representative | Degree | Eccentricity | Frequency | ||
|---|---|---|---|---|---|
| 2 | 4 | 4 | 2 | ||
| 2 | 5 | 6 | 2 | ||
| 3 | 5 | 4 | |||
| 1 | 3 | 3 | |||
| 2 | 5 | 6 | |||
| 2 | 5 | 6 | |||
| 3 | 6 | 8 | |||
| 2 | 5 | 6 | |||
| 2 | 4 | 4 | |||
| when | 2 | 5 | 6 | ||
| 1 | 2 | 2 |
| Representative | Eccentricity | Frequency | Eccentricity of an Edge | Degree of an Edge |
|---|---|---|---|---|
| 1 | 2 | |||
| 2 | 2 | |||
| 2 | 3 | |||
| 2 | ||||
| 3 | ||||
| 2 | ||||
| 3 | ||||
| 3 | ||||
| 2 | ||||
| when | 2 | |||
| 1 | ||||
| when | 3 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zheng, J.; Iqbal, Z.; Fahad, A.; Zafar, A.; Aslam, A.; Qureshi, M.I.; Irfan, R. Some Eccentricity-Based Topological Indices and Polynomials of Poly(EThyleneAmidoAmine) (PETAA) Dendrimers. Processes 2019, 7, 433. https://doi.org/10.3390/pr7070433
Zheng J, Iqbal Z, Fahad A, Zafar A, Aslam A, Qureshi MI, Irfan R. Some Eccentricity-Based Topological Indices and Polynomials of Poly(EThyleneAmidoAmine) (PETAA) Dendrimers. Processes. 2019; 7(7):433. https://doi.org/10.3390/pr7070433
Chicago/Turabian StyleZheng, Jialin, Zahid Iqbal, Asfand Fahad, Asim Zafar, Adnan Aslam, Muhammad Imran Qureshi, and Rida Irfan. 2019. "Some Eccentricity-Based Topological Indices and Polynomials of Poly(EThyleneAmidoAmine) (PETAA) Dendrimers" Processes 7, no. 7: 433. https://doi.org/10.3390/pr7070433
APA StyleZheng, J., Iqbal, Z., Fahad, A., Zafar, A., Aslam, A., Qureshi, M. I., & Irfan, R. (2019). Some Eccentricity-Based Topological Indices and Polynomials of Poly(EThyleneAmidoAmine) (PETAA) Dendrimers. Processes, 7(7), 433. https://doi.org/10.3390/pr7070433





