Abstract
Phthalic acid esters (PAEs) are mainly used as plasticizers and result in serious environmental contamination worldwide. Microbial biodegradation becomes an efficient strategy for PAE elimination. In the current study, the PAE-degrading strain Gordonia sp. LUNF6 was isolated from contaminated soil. Strain LUNF6 can efficiently degrade DBP in a wide range of temperatures, pH values, and salinity levels. This strain is also capable of degrading 11 types of PAEs and displays remediation potential in wastewater. The complete genome of strain LUNF6 was sequenced to determine its efficient degradation performance. Its genome comprises a chromosome (3,971,257 bp) and a plasmid (78,813 bp). After gene function annotation, the complete PAE degradation pathway was proposed. The gene of monoalkyl PAE hydrolase MphGd2 was cloned and heterologously expressed. The protein of MphGd2 was purified by infinity chromatography, and we hydrolyzed MBP to produce PA. These results reveal the molecular mechanism of PAE degradation by strain LUNF6, which will contribute to the application of strain LUNF6 and hydrolase MphGd2 in bioremediation.
1. Introduction
Phthalic acid esters (PAEs), the common plasticizer, can strengthen the elasticity and flexibility of plastic products. PAEs are attached to plastic polymers in the form of van der Waals forces or hydrogen bonds, and are easily released into the environment [1,2]. PAE pollution exerts an adverse influence on ecosystems and human health. A large number of studies have shown that long-term exposure to PAEs increases the risk of pregnancy disorders, cancer, deformity, and endocrine system disorders [3,4,5].
In recent years, researchers worldwide have isolated many PAE-degrading bacteria from all kinds of environmental matrices, which belong to the genus of Rhodococcus [6], Pseudomonas [7], Bacillus [8], Gordonia [9], Microbacterium [10], Burkholderia [11], and Cupriavidus [12]. The PAE degradation characteristics of these strains have also been studied. Gordonia sp. YC-JHl, isolated from oil-contaminated soil, could degrade 13 kinds of PAEs and rapidly remove di(2-ethylhexyl) phthalate (DEHP) under a wide range of temperatures and pH values [9,13]. The efficient PAE-degrading strain Pseudomonas sp. YJB6 was isolated from polluted soil [7], and its degradation ability could be significantly improved through immobilization. Rhodococcus sp. DNHP-S2 also has a wide range of substrates such as DEHP, dimethyl phthalate (DMP), di-n-butyl phthalate (DBP), di-n-octyl phthalate (DOP) and butylbenzyl phthalate (BBP) [6]. This strain could degrade 52.47% and 99.75% of DEHP within 72 h at 10 °C and 35 °C, respectively. Some strains only transform PAEs into intermediates such as phthalic acid (PA). These strains harbor enzymes involved in the hydrolysis of dialkyl or monoalkyl phthalates, but lack pathways responsible for PA metabolism [14]. However, these strains accelerate the detoxification of PAEs, and some strains also cooperate to degrade PAEs. Gordonia sp. JDC-2 could degrade DOP to generate PA, which was not further degraded [15]. However, Arthrobacter sp. JDC-32 could not degrade DOP, but could utilize PA. Therefore, DOP could be completely degraded by co-culture of Gordonia sp. JDC-2 and Arthrobacter sp. JDC-32. PAE degradation by consortium was also reported. The consortium CM9 was isolated from farmland soil contaminated by PAEs and contained strains of Rhodococcus, Xenophilus, Niabella, Tahibacter, Achromobacter, and Sphingopyxis [16]. It could degrade 94.85% and 100% of DEHP (1000 mg/L) at 24 h and 72 h, respectively. The consortium Bl, composed of Pandoraea sp. and Microbacterium sp., was capable of degrading DBP [17].
Most bacteria use dialkyl phthalate hydrolases to hydrolyze PAEs to produce monoalkyl phthalate and alcohol, and the monoalkyl phthalate is transformed to PA and alcohol under the action of monoalkyl phthalate hydrolases. This is a key step and an important way for bacteria to degrade PAEs. The PAE hydrolase genes have been cloned via construction and screening of genome libraries, as well as genome or transcriptome sequencing analysis [8,18]. Several phthalate hydrolase genes have been reported. Wu et al. cloned a hydrolase gene from Acinetobacter sp. M673 using a genome library constructed by the shotgun method [19]. This hydrolase could hydrolyze PAEs with short or medium side chains such as DMP, diethyl phthalate (DEP), dipropyl phthalate (DPrP), DBP, di-n-pentyl phthalate (DPeP), and dihexyl phthalate (DHP). Zhang et al. found that thermostable esterase EstS1 from Sulfobacillus acidophilus DSM10332 could hydrolyze DEP, DPrP, DBP, DPeP, DHP, and BBP [20]. The PAE hydrolase gene dphB was cloned from active biofilm of a sewage treatment plant [21]. DphB had the highest degradation activity at low temperature (10 °C) and could hydrolyze DPrP, DBP, and DPeP, but it could not degrade PAEs with short and long side chains. The genome of PAE-degrading strain Bacillus subrilis BJO0005 was sequenced [22]. The hydrolase GTW2817760 was identified to hydrolyze dialkyl phthalates, and the hydrolases GTW28_09400 and GTW28_13725 could hydrolyze monoalkyl phthalates. Fan et al. conducted whole-genome sequencing of Gordonia sp. YC-JH1 and detected dialkyl phthalate hydrolase EstG1 and monoalkyl phthalate hydrolase EstG2 [9].
At present, the research about PAE biodegradation is mainly focused on the isolation of functional strains and the characterization of degradation. There are few studies on the simulation application of microorganisms in the remediation of environments contaminated by PAEs. Although many PAE-degrading bacteria have been reported, the genome sequence and genetic information of the strains are still limited, which hinders the research on the molecular mechanism of PAE degradation by strains. In the present work, PAE-degrading bacterium Gordonia sp. LUNF6 was isolated. The degradation performance of strain LUNF6 under different temperatures, pH levels, and NaCl concentrations was characterized. Its complete genome was sequenced and the putative proteins were annotated. The gene of monoalkyl phthalate hydrolase MphG2 was predicted by homologous blast. MphG2 was heterologously expressed and purified by affinity chromatography, and could hydrolyze monoalkyl phthalate. The results provide more genetic information of PAE-degrading bacteria and will support the establishment of molecular degradation mechanisms and remediation applications.
2. Materials and Methods
2.1. Chemicals and Strains
The PAE chemicals (Shanghai Aladdin BioChemical Technology Co., Ltd., Shanghai, China) were dissolved in methanol. Ni-NTA Resin and the competent strain BL21 (DE3) were purchased from TransGen Biotech (Beijing, China). The expression vector pET-32a (+) was purchased from Novagen (Madison, WI, USA). The primers were synthesized for expression vector construction (Table S1). Trace element medium (TEM), binding buffer, and elution buffer were prepared according to our reports [8,9,13].
2.2. Isolation of DBP-Degrading Bacteria
In this study, 100 mL of LB medium and 5 g of soil contaminated by industrial wastewater were added to a conical flask. The mixture was cultured at 180 rpm and 30 °C for 5 days. Then, 1 mL of the mixture was transferred to TEM containing 0.5 mM DBP (≈139.17 mg/L DBP), which was incubated at the same condition. Sequential transfer to TEM was repeated five times. The TEM agar plates supplied with 0.5 mM DBP were streaked with the final culture and incubated at 30 °C. A transparent zone was observed around a colony named LUNF6. This strain was cultivated in LB liquid medium to OD600 = 1.0. The cells were collected by centrifugation (4 °C, 6000 rpm for 3 min), washed twice by liquid TEM, and resuspended in an equal volume of liquid TEM. The suspension was inoculated in TEM with 0.5 mM DBP and incubated at 30 °C and 180 rpm for 5 days. DBP and metabolites were extracted and subjected to HPLC to verify the degradation capability of strain LUNF6. The experiments were conducted in triplicate. The 16S rRNA gene of strain LUNF6 was amplified using primers 27F and 1492R [23]. The PCR products were sequenced by Sangon Biotech Co., Ltd. (Shanghai, China). The 16S rRNA gene sequence was submitted to NCBI BLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi?PROGRAM=blastx&PAGE_TYPE=BlastSearch&LINK_LOC=blasthome accessed on 23 January 2025, blastx: search protein databases using a translated nucleotide query) to search for similar sequences, which were used to construct a phylogenetic tree by MEGA 6.0.
2.3. Characterization of PAE Degradation by Strain LUNF6
The suspension of strain LUNF6 was transferred into liquid TEM (pH 8.0, 0.5 mM DBP) with 1% inoculation and was incubated at certain temperatures (20 °C, 30 °C, 40 °C and 50 °C) and 180 rpm for 8 h. To test DBP degradation under different environmental pH values, the strain suspension was cultivated in TEM within a pH range of 4.0–11.0. The samples were shaken at 30 °C and 180 rpm for 8 h. The metabolites of DBP degradation under pH 8.0 were detected by HPLC-MS [8]. To test the effect of salinity on DBP degradation, the NaCl concentration of TEM was adjusted to 0%, 2%, 4%, 6%, 8%, 10%, and 12%, respectively. The strain suspension was cultivated in TEM as above with 0.5 mM DBP at 30 °C and 180 rpm for 8 h. To investigate the substrates of strain LUNF6, this strain suspension was inoculated in liquid TEM (pH 8.0) which was supplemented with 0.5 mM of PAEs (DMP, DEP, DPrP, DBP, DPeP, DHP, diheptyl phthalate (DHeP), DOP, DEHP, BBP and dicyclohexyl phthalate (DCHP), respectively) (Table S2). The mixture was cultivated at 180 rpm and 30 °C for 2 days. The concentration of residual PAEs was determined by HPLC. The samples without strain LUNF6 were adopted as control.
2.4. DBP Degradation by Gordonia sp. LUNF6 in Wastewater
DBP was added to industrial wastewater to final concentration of 0.5 mM. Then, the suspension of strain LUNF6 (1% inoculation amount) was inoculated into the wastewater. The control had no inoculation. The wastewater samples were shaken at 30 °C and 180 rpm for 0, 8, and 18 h, respectively. Detection of residual DBP was carried out using HPLC (Agilent 1200, Agilent Technology Co., Ltd., Santa Clara, CA, USA).
2.5. Genome Sequencing of Gordonia sp. LUNF6
The genomic DNA of strain LUNF6 was extracted using a FastPure® Bacteria DNA Isolation Mini Kit (Vazyme Biotech Co., Ltd., Nanjing, China). The concentration and integrity of extracted DNA were detected by electrophoresis. The sequencing library was constructed for second- and third-generation high-throughput sequencing by Illumina NovaSeq 600 (Illumina, San Diego, CA, USA) and Oxford Nanopore ONT PromrthION48 (Oxford Nanopore Technologies Co., Ltd., Oxford, UK), respectively. After assembly and correction of the genome sequence, average nucleotide identity (ANI) analysis of the chromosomes between strain LUNF6 and other related strains was conducted using FastANI v1.33 [24]. The genome annotation was performed in public databases such as NR, eggNOG, KEGG, SwissProt2, and GO [8,25].
2.6. Gene Cloning and Purification of PAE Hydrolase
Using the reported PAE hydrolase P8219 (WP_239582419.1) as a probe, a gene with high protein sequence consistency was detected in the genome of strain LUNF6. This gene was speculated to be the candidate gene of PAE hydrolase and named mphG2. The signal peptide and transmembrane region were predicted through SignalP5.0 and TMHMM Server v. 2.0 [26,27]. The conserved sequences of MphG2 were analyzed by CLUSTALW [28]. A phylogenetic tree of MphG2 was constructed using MEGA7.0 [29]. The gene mphG2 was amplificated using primers mphG2F and mphG2R (Table S1). The PCR products and plasmid pET-32a (+) were digested by EcoRI and HindIII. The digested products of pET-32a (+) and mphG2 were ligated to construct recombinant vector pET-mphG2. The expression vector pET-mphG2 was introduced into competent BL21 (DE3). The positive clone was cultured in LB medium (50 μg/mL ampicillin) at 37 °C and 180 rpm to OD600 = 0.8 and induced by 0.1 mM IPTG for 4 h at 30 °C. Cell disruption and protein purification were conducted following the method reported in [8]. To verify the function of MphG1, it was incubated in 0.9 mL of Tris-HCl (50 mM, pH 8.0), 0.5 mM mono-butyl phthalate (MBP) and 25 μg MphG1 at 30 °C and 180 rpm for 30 min. There was no hydrolase MphG1 in the control. The reaction products were extracted by ethyl acetate and detected by HPLC.
2.7. Analytical Method
To extract PAEs or reaction products in the mixture of strain LUNF6 or hydrolase MphG1, an equal volume of ethyl acetate was added to the samples, which were shaken vigorously. The upper organic phase was collected, and the ethyl acetate was removed by nitrogen purge. The sample was re-dissolved with an equal volume of methanol and filtered by a 0.22 μm filter membrane. The residual PAEs in the samples were detected by HPLC (Agilent 1200, Agilent Technology Co., Ltd., Santa Clara, CA, USA) with a DAD detector. HPLC detection conditions were as follows: the mobile phase was methanol/water (0.1% acetic acid) = 20%:80%, the column temperature was 25 °C, the flow rate was 0.5–0.8 mL/min, and 5 μL of the sample was injected. The column was a ZORBAX Eclipse XBD C18 column (4.6 mm × 250 mm, 5 μm). When DBP metabolites were identified by HPLC-MS, a triple quadrupole mass spectrometer (Agilent 6420, Agilent Technology Co., Ltd., Santa Clara, CA, USA) was used [13]. The column, mobile phase, and elution conditions were the same as those of HPLC mentioned above. The HPLC-MS system was equipped with an electrospray ionization (ESI) source. The capillary voltage was 4000 V. The data were collected in full-scan mode from 100 to 500 m/z.
3. Results and Discussion
3.1. Isolation and Screening of DBP-Degrading Strain Gordonia sp. LUNF6
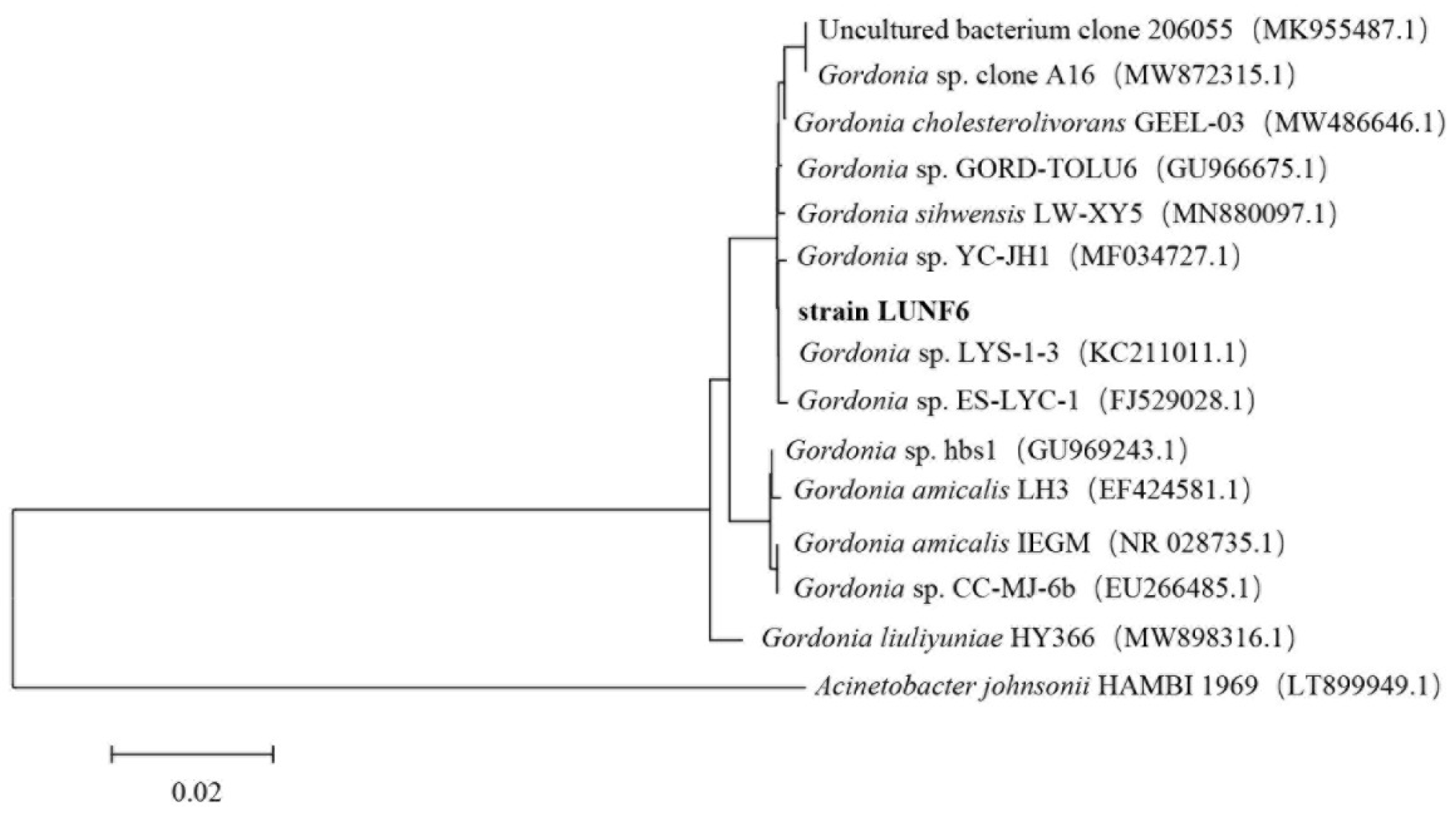
After six rounds of enrichment using DBP as a carbon source, a transparent zone around strain LUNF6 on the plate was observed, suggesting the DBP-degrading ability of this strain. After cultivation of strain LUNF6 in TEM supplied with DBP for 5 days, HPLC results showed that DBP was completely removed (Figure S1), indicating an efficient DBP degradation capability of strain LUNF6. Therefore, the DBP-degrading strain was successfully isolated by a domestication-based method. In previous reports, various pollutant-degrading strains have been obtained using this method [30,31,32,33]. The colony of strain LUNF6 was light yellow in color and had a smooth surface and neat edges (Figure S2). Based on sequence BLAST of the 16S rRNA gene in the NCBI database and phylogenetic analysis, strain LUNF6 belongs to the genus Gordonia (Figure 1). The sequence of 16S rRNA gene of Gordonia sp. LUNF6 was deposited in GenBank database under accession numbers PQ796727.1. The members of Gordonia are metabolically versatile [34,35] and capable of degrading many xenobiotics or pollutants such as β-cypermethrin [36], 3-methylpyridine [37], phenanthrene [38], carbamazepine [39], dibenzothiophene [40], and so on. Therefore, the strains of Gordonia are potential microorganism materials for environmental remediation. The isolate Gordonia sp. LUNF6 offers another option for practical remediation of PAE-contaminated environments.
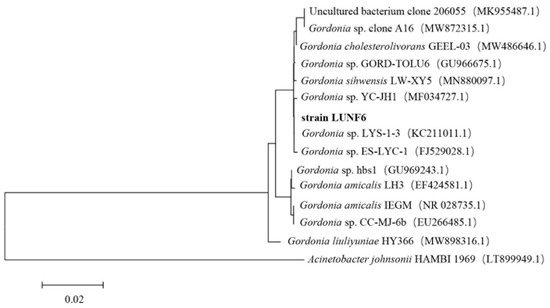
Figure 1.
The phylogenetic analysis of strain LUNF6 based on 16S rRNA gene sequence. Bar, 0.02 nucleotide substitutions per nucleotide position.
3.2. PAE Degradation Performance of Gordonia sp. LUNF6
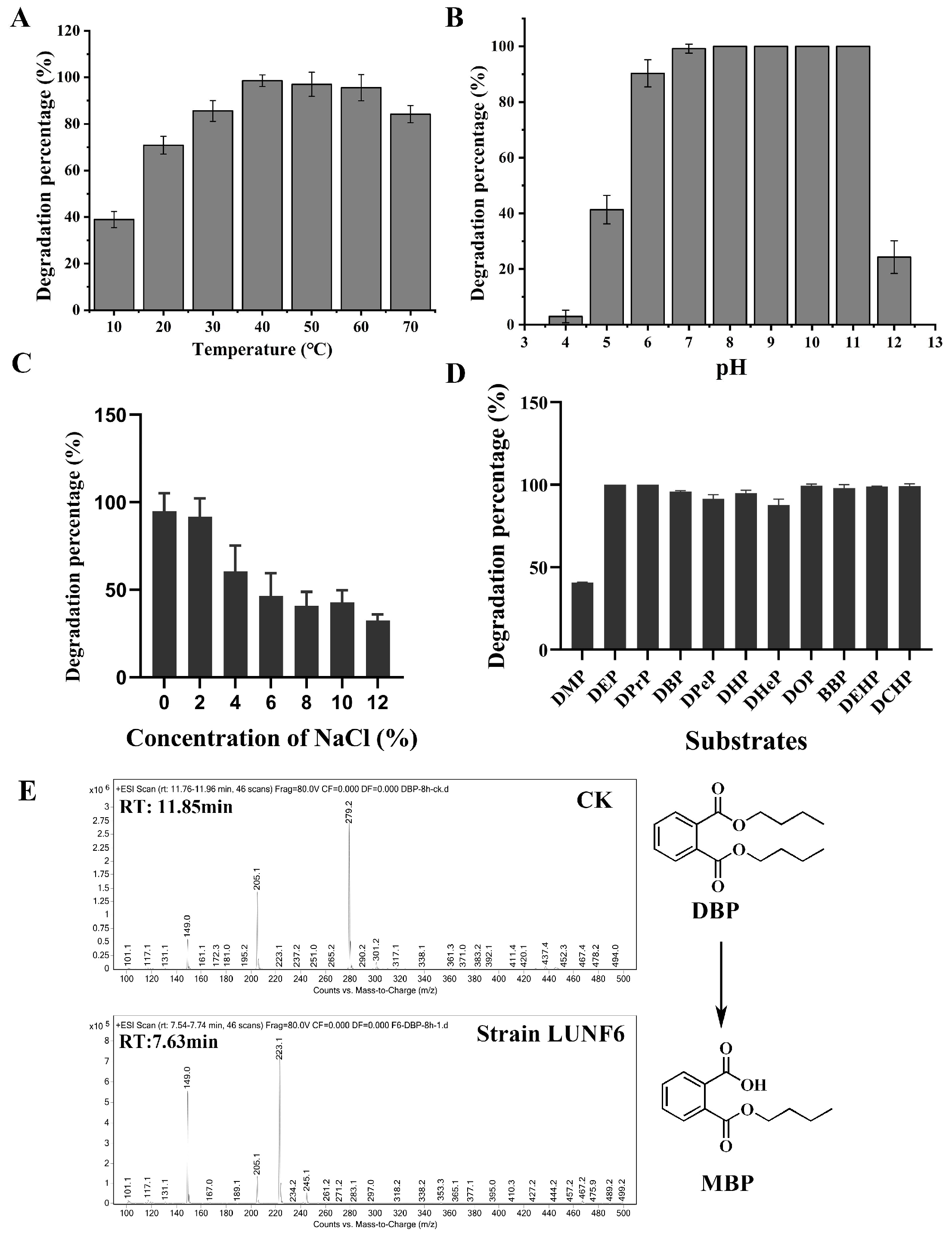
PAE degradation by Gordonia sp. LUNF6 under different conditions was assessed. After 8 h of cultivation, the degradation percentage of DBP gradually increased as the temperature rose from 10 to 40 °C and reached the maximum value of 98.57% at 40 °C (Figure 2A). Overall, 97.04% and 95.57% of DBP was removed at 50 °C and 60 °C, respectively. Even more, 78.68% of DBP was also degraded at temperatures as high as 70 °C. Therefore, strain LUNF6 possesses efficient degradation performance in a wide range of temperatures. The initial pH also plays an important role in pollutant degradation [37,41]. The degradation percentage of DBP gradually increased from 2.94% to 99.18% with initial pH from 4.0 to 7.0 (Figure 2B). Strain LUNF6 was able to completely degrade DBP at pH 8.0–11.0, while 24.27% of DBP was removed at pH 12.0. On the whole, the degradation percentage of DBP reached more than 90% at pH 6.0–11.0. This result indicates that strain LUNF6 tends prefer to neutral and alkaline environments, and this strain especially has potential practical applications in strong alkaline environments. The microorganism metabolizes pollutants mainly through a series of enzymatic reactions [42,43,44]. Environmental factors such as temperature and pH have an effect on enzymatic activity and further influence bacterial degradation. Regarding the effect of salinity on DBP degradation, the concentration of NaCl and degradation rate of DBP were negatively correlated (Figure 2C). The strain LUNF6 could degrade 60.53–94.76% of DBP at NaCl concentrations of 0–4%. The degradation percentage of 32.49% was maintained when the NaCl concentration reached as high as 12%. Thus, this strain can tolerate high NaCl concentrations and harbor high degradation efficiency, which suggests the remediation potential of strain LUNF6 in environments with high NaCl concentrations.
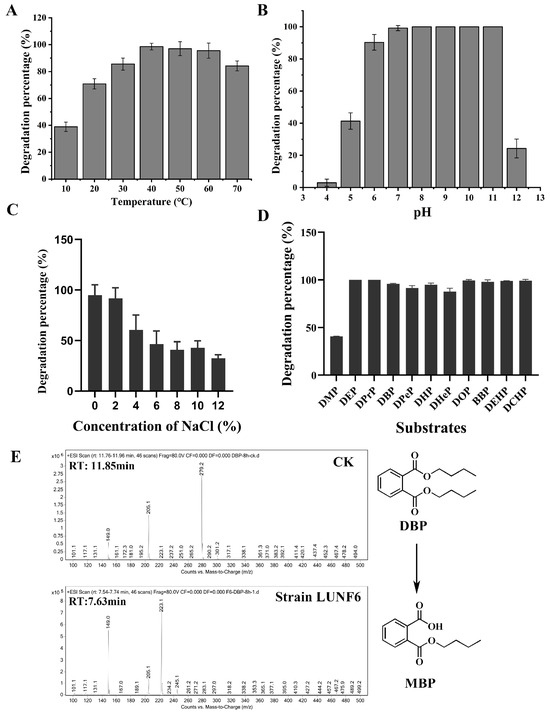
Figure 2.
PAE degradation characterization of strain LUNF6. The degradation percentage of DBP under a range of temperatures (A), pH values (B), and NaCl concentrations (C). (D) The degradation of different PAE substrates by strain LUNF6. (E) The detection of DBP metabolites by HPLC–MS.
After cultivation for 2 days, strain LUNF6 could degrade 37.74% of DMP and 85.44% of DHeP (Figure 2D). The other nine types of PAEs were almost completely removed. Compared to the control samples, obvious bacterial growth was observed in media with inoculation of strain LUNF6, indicating that these PAEs can support the growth of strain LUNF6. Strain LUNF6 could degrade PAEs with side chains of varying lengths, especially PAEs with bulky side chains such as BBP, DEHP and DCHP, indicating its high degradation capability. There are many reports on bacteria capable of degrading certain kinds of PAEs [8,41,45,46], but there are few reports on bacteria that are capable of degrading more than ten types of PAEs, except for Gordonia sp. YC-JH1 [9]. Furthermore, some kinds of PAEs usually coexist in the environment [47,48]. Strain LUNF6 is a microbial resource with a versatile degradation capability for actual applications. The metabolites of DBP were detected by HPLC-MS (Figure 2E and Figure S3). In the control group, a molecule with m/z of 279.2 was found, which was identified as an authentic DBP with a molecular weight of 278.34. In the treatment group, a molecule with m/z of 223.1 was detected, which was consistent with the molecular weight of protonated MBP (222.24). Therefore, strain LUNF6 could degrade DBP to produce MBP. In previous reports, the PAE degradation performance of many bacterial strains was investigated under various conditions of pH, temperature, and other factors [46,49,50,51]. Some strains exhibit excellent degradation performance. These experiments were conducted under a single variable. Here, similar methods were used to explore the degradation capability of strain LUNF6. In the future, 2k factorial design, a highly effective experimental design method [52,53,54], will be adopted to better examine the effects of multiple variables on PAE degradation efficiency.
3.3. Remediation of Wastewater by Gordonia sp. LUNF6
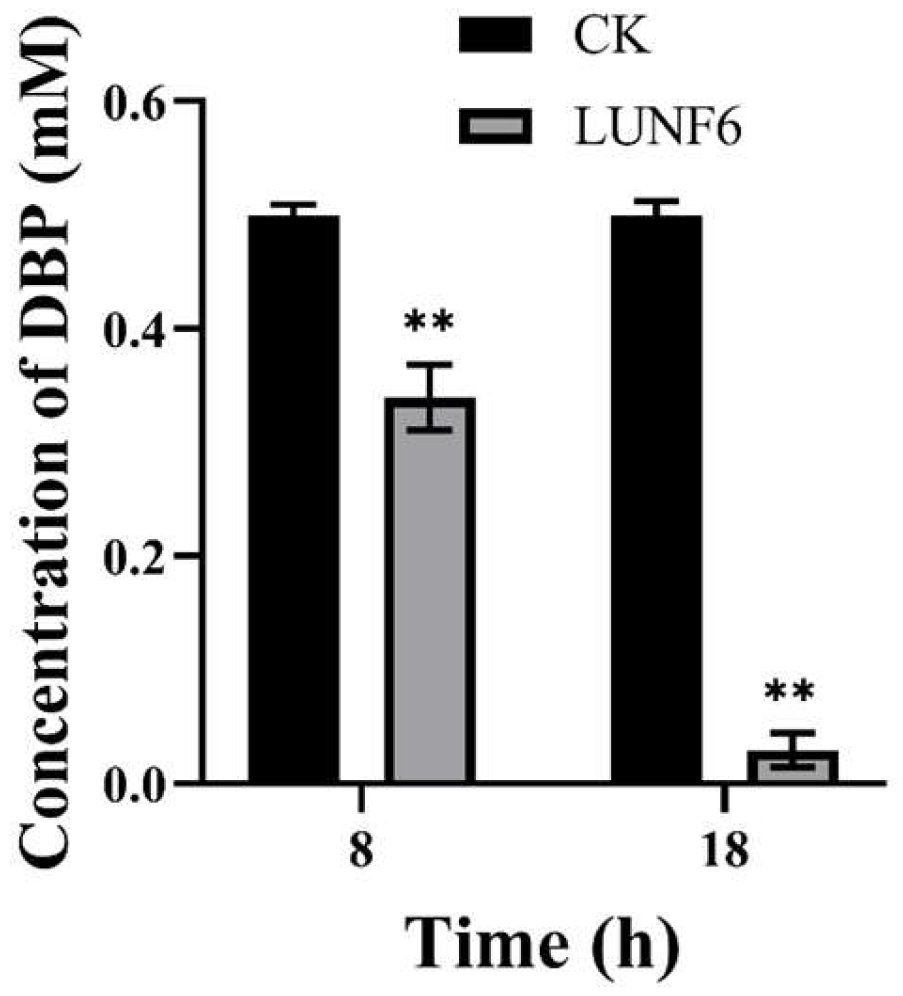
After incubation of wastewater and strain LUNF6, DBP in the wastewater was degraded rapidly; 32.11% of DBP was removed within 8 h (Figure 3) and 94.14% of DBP was degraded after 18 h, indicating that strain LUNF6 was able to remediate wastewater contaminated by DBP. Industrial and domestic discharge results in the distribution of PAEs in aqueous environments such as wastewater [47,55]. However, PAEs are of high hydrophobicity and not easily hydrolyzed, which leads to their accumulation in wastewater. In wastewater treatment plants, microbial metabolism greatly contributes to PAE removal [56,57]. In wastewater, Bacillus sp. LUNF1 degraded only 38.46% of DBP after 24 h and 57.08% and 100% of DBP after 5 d and 7 d, respectively [8]. Based on the discussion above, Gordonia sp. LUNF6 is a potential strain for wastewater remediation.
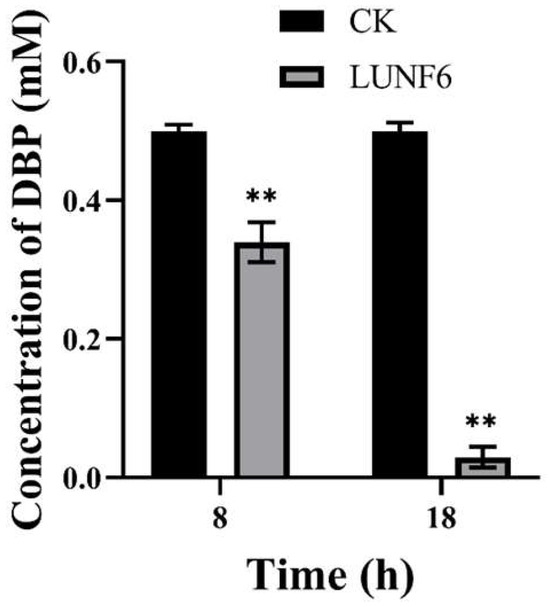
Figure 3.
The removal of DBP from wastewater by strain LUNF6 (** p < 0.01). CK, the control sample containing wastewater without inoculation of strain LUNF6.
3.4. Genomic Analysis of Gordonia sp. LUNF6
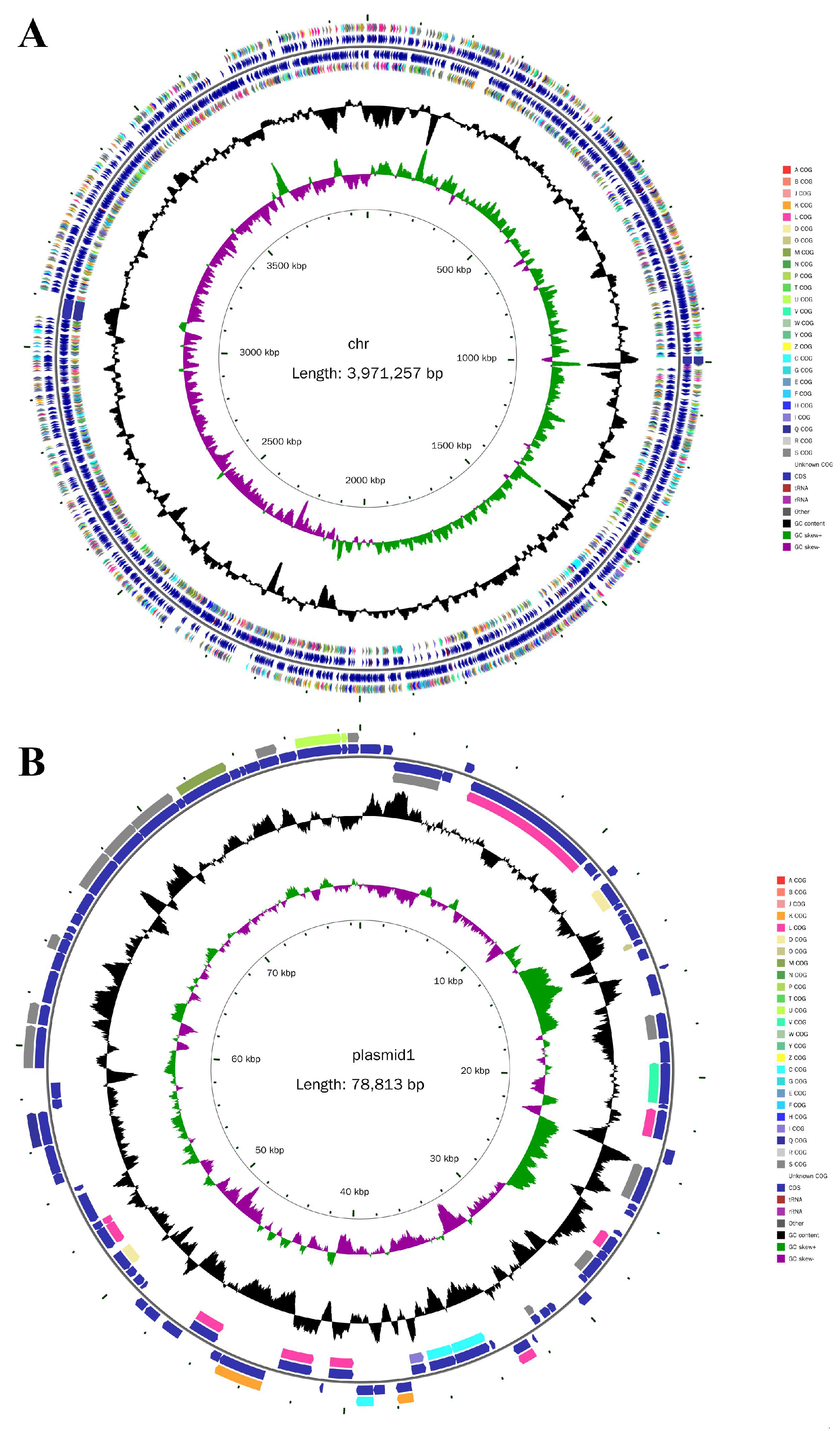
To further investigate the molecular basis of strain LUNF6 degrading DBP, its genome was sequenced by technology of Illumina NovaSeq and PacBio Sequel. After sequencing and quality control, high-quality sequencing data of 1,635,742,091 bp and 1,382,692,007 bp were obtained from Illumina and PacBio platforms, respectively (Table S3). After genome assembly and correction, one chromosome and one plasmid were obtained (Figure 4A,B). The genome sequence has been submitted to the GenBank database under accession numbers CP178181 and CP178182 for the chromosome and plasmid, respectively. After ANI analysis, strain LUNF6 might be a member of Gordonia sihwensis based on its ANI value of 99.1977% (Table S4).
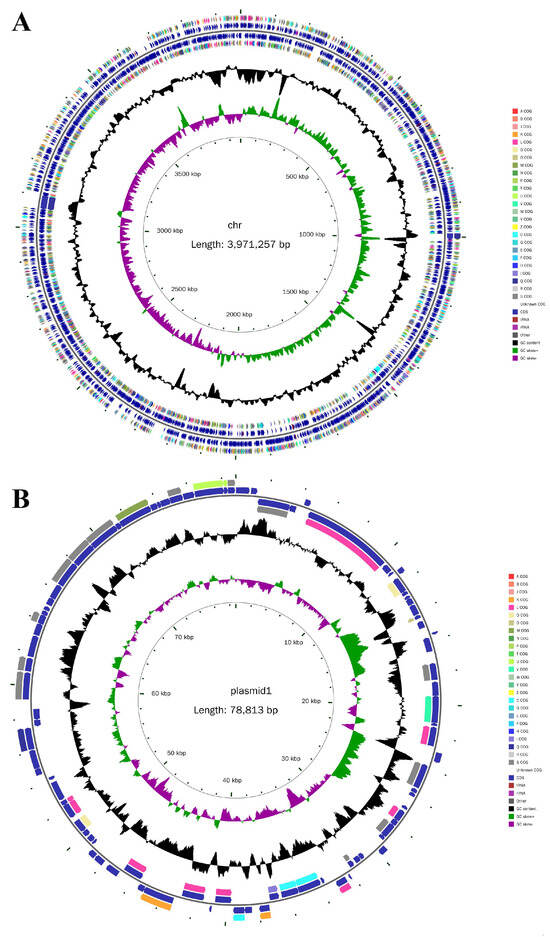
Figure 4.
Circle maps of the chromosome (A) and plasmid (B) of strain LUNF6. From inside to outside, circle 1 represents the scale; circle 2 represents GC Skew; circle 3 represents GC content; circle 4 and 7 represent the COG to which each CDS belongs; circle 5 and 6 represent the positions of CDS, tRNA, and rRNA on the genome.
The length of the chromosome and plasmid was 3,971,257 bp and 78,813 bp, respectively. The GC content of the chromosome and plasmid was 68.31% and 63.80%, respectively. The genome size and GC content of strain LUNF6 approximately correspond to those of other strains of the Gordonia genus [9,36]. There are 3718 and 89 protein-coding genes (CDSs) predicted in the chromosome and plasmid, respectively (Table S3). In addition, the chromosome harbors 12 rRNA genes, 51 tRNA genes, and 7 CRISPR structures.
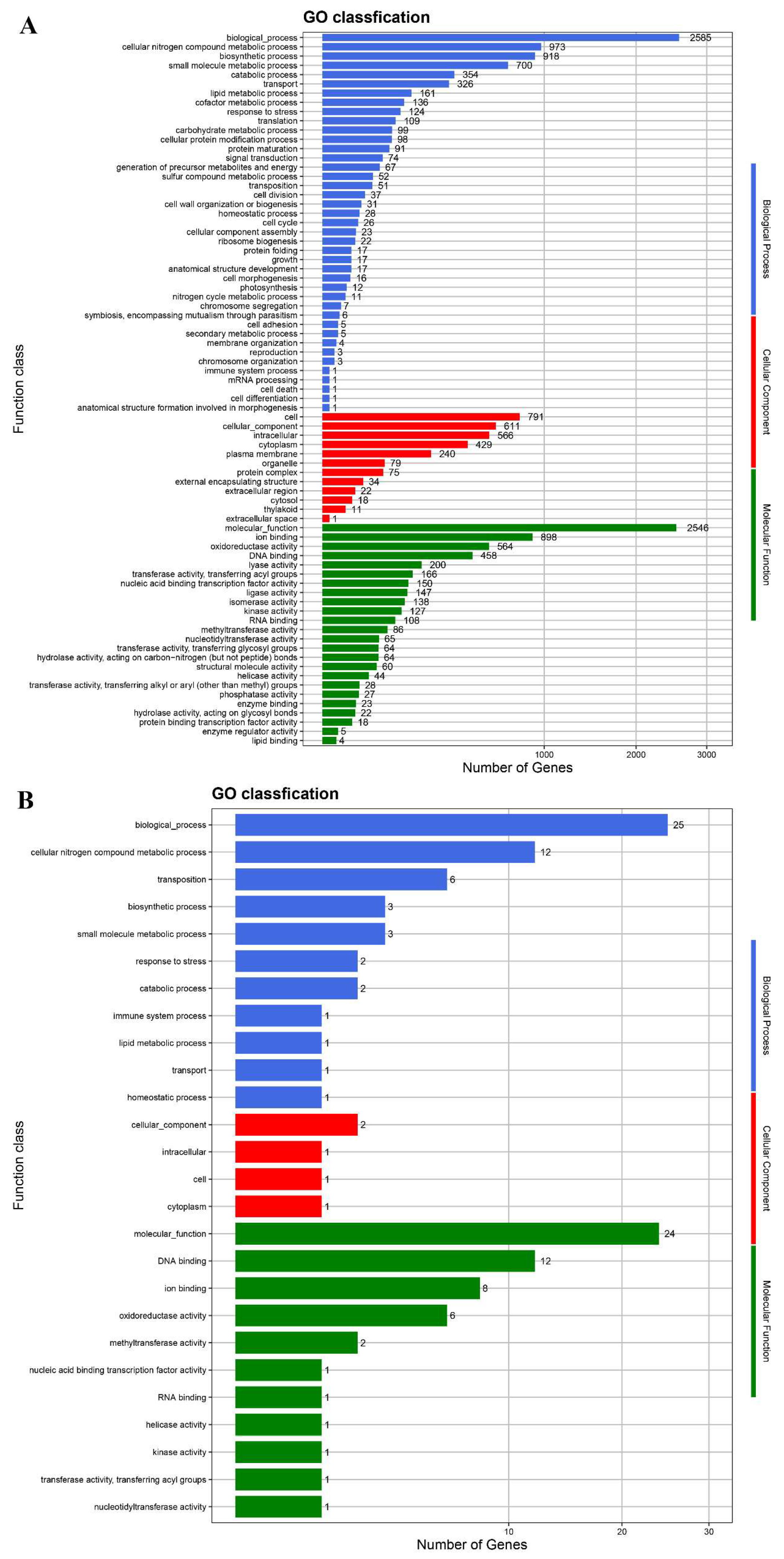
The protein-coding genes were annotated in the databases GO, NR, COG, KEGG, and Swiss-Prot (Table S3). In the GO database, 2585 and 2546 genes from the chromosome were annotated under biological process and molecular function, respectively (Figure 5). After annotation in the COG database, 3319 genes from the chromosome belong to 21 COG function classifications (Figure S4). According to KEGG annotation, 154 genes of strain LUNF6 are responsible for xenobiotic biodegradation and metabolism (Figure S5). Among them, most genes function in the degradation of aromatic compounds, such as benzoate, polycyclic aromatic hydrocarbon, xylene, toluene, and chlorobenzene. In addition, some genes are annotated under the degradation process of other pollutants such as atrazine, methane, dioxin, and chloroalkane. These results suggest that strain LUNF6 has potential degradation ability for other pollutants, which needs to be verified by further investigation.
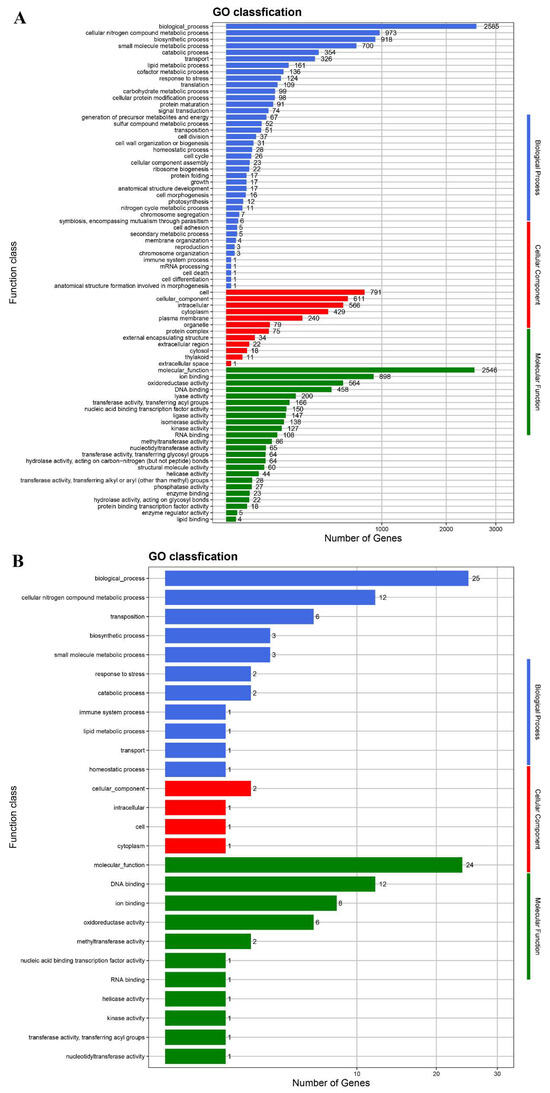
Figure 5.
The genes from the chromosome (A) and plasmid (B) of strain LUNF6 annotated in the GO database.
3.5. PAE Degradation Pathway of Gordonia sp. LUNF6
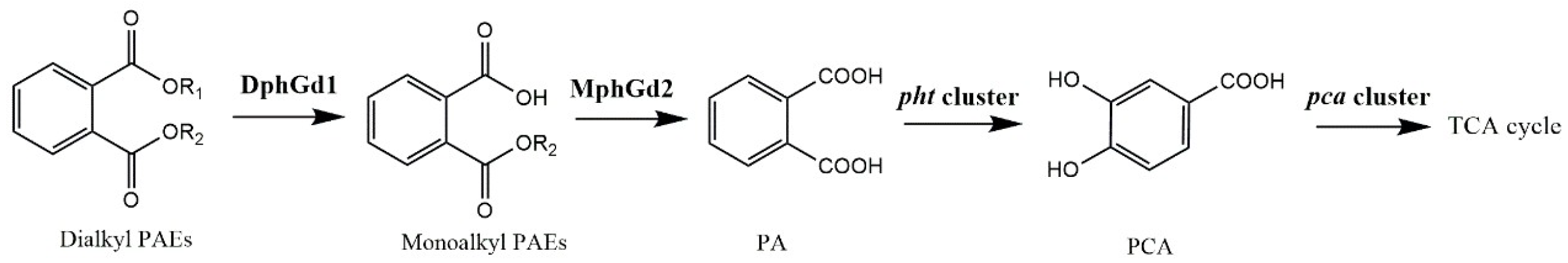
Microorganisms can degrade PAEs rapidly under aerobic conditions. PAE degradation by bacteria is usually initiated by hydrolysis of ester bonds, where hydrolases play important roles [22,25,58]. The genes of putative dialkyl PAE hydrolase DphGd1 and monoalkyl PAE hydrolase MphGd2 were detected from the genome of strain LUNF6. These hydrolases catalyzed the hydrolysis of ester bonds of PAEs to produce PA. In Gram-positive bacteria, PA was metabolized by a pht cluster (phtR-phtAaAbAcAd-phtB-phtC) [9,59,60], and the product protocatechuic acid (PCA) was degraded into the tricarboxylic acid cycle under the function of a pca cluster (pcaR-pcaGH-pcaB-pcaL-pcaIJ-phtF) [9,61]. A pht cluster and pca cluster were also predicted in Gordonia sp. LUNF6 (Table S5). All of these genes are located in the chromosome of strain LUNF6. Overall, a PAE metabolic pathway in strain LUNF6 has been proposed (Figure 6).
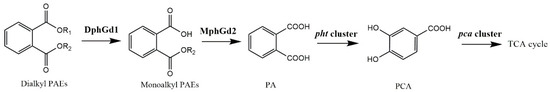
Figure 6.
PAE degradation pathway in strain LUNF6.
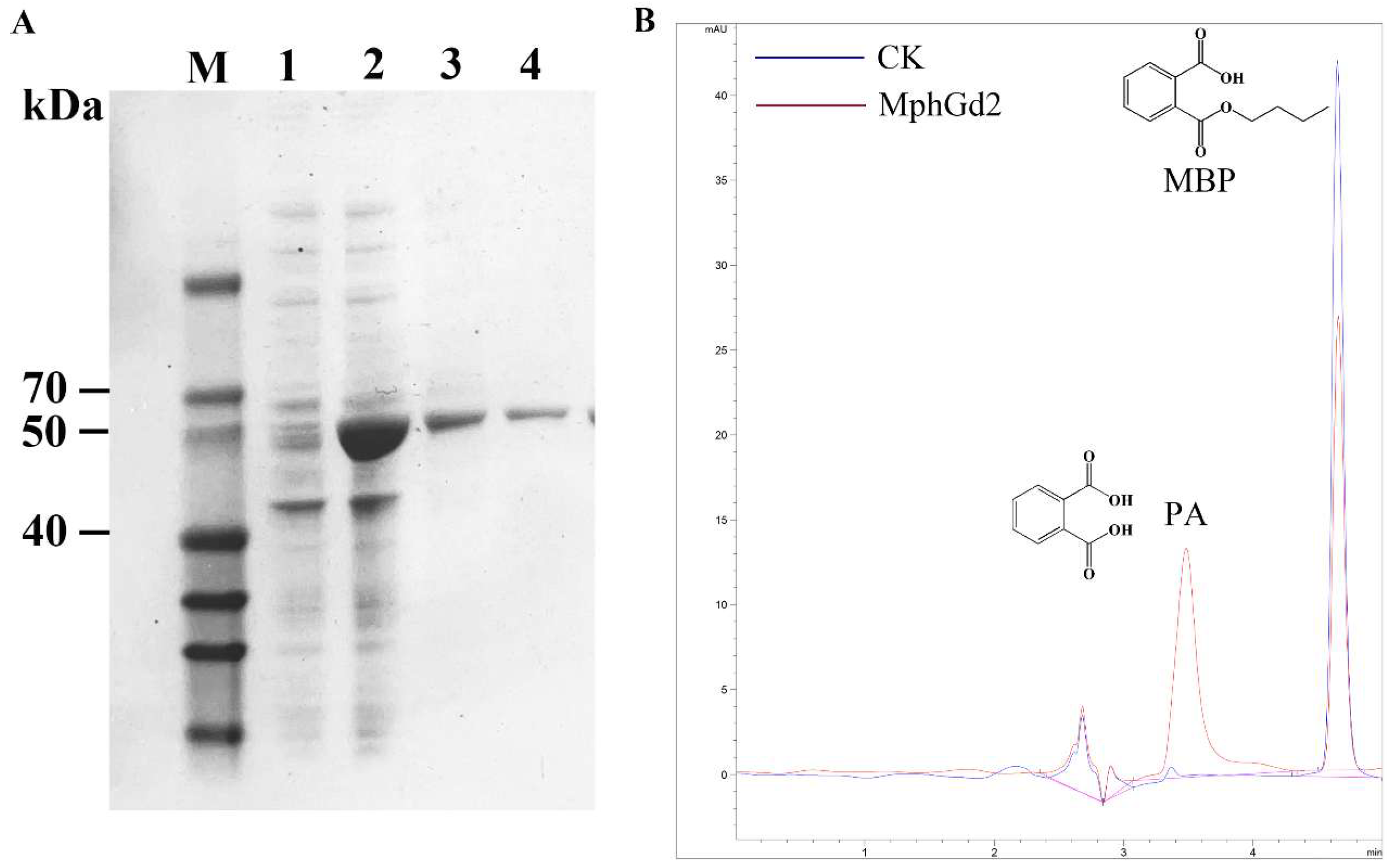
The hydrolase MphGd2 contains 297 amino acids with a molecular weight of about 32.3 kDa and an isoelectric point of 5.10. There is no signal peptide in the MphG2 sequence after analysis by SignalP 5.0, indicating that MphG2 is an intracellular enzyme. The conserved motif of MphGd2 was analyzed by multiple sequence comparison, and the catalytic triad of MphG2 was inferred to be Ser111-Asp245-His277 (Figure S6). The expression of protein MphGd2 was induced by IPTG. According to SDS-PAGE analysis, the whole-cell sample contains obvious bands near 50 kDa after induction (Figure 7A), which is similar to the theoretical molecular weight of MphGd2 recombinant protein (52 kDa). This result indicates that IPTG successfully induced the expression of the mphGd2 gene. After affinity purification, an obvious band of about 50 kDa was also detected, indicating successful purification of MphG2. The enzymatic activity of MphGd2 towards MBP was also detected. The HPLC results showed that a peak appeared in the control group samples at 4.652 min (Figure 7B), which was consistent with the retention time of the MBP standard substance. Therefore, this peak represented MBP. In the MphGd2 treatment sample, this peak at 4.652 min was significantly smaller than that of the control group. However, a new peak appeared at 3.483 min, which was consistent with the retention time of PA. These results indicate that the hydrolase MphGd2 can hydrolyze the ester bond of MBP to produce PA.
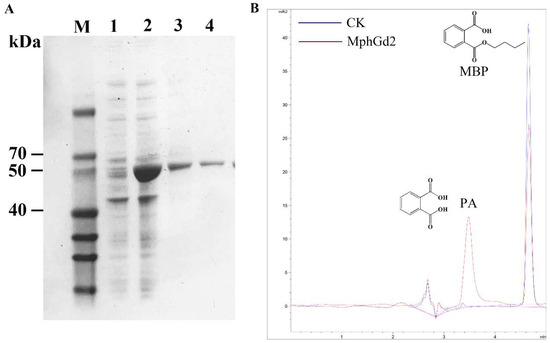
Figure 7.
Purification and identification of monoalkyl PAE hydrolase MphGd2. (A) SDS-PAGE detection of recombinant protein MphGd2. Lane M, protein marker; lane 1, whole-cell extracts of expression strains without induction; lane 2, whole-cell extracts of expression strains after IPTG induction; lane 3, supernatant of sample lane 2; lane 4, purified MphGd2. (B) HPLC analysis of MBP hydrolyzed by MphGd2.
4. Conclusions
PAEs are a common type of environmental pollutant with the effect of environmental estrogen. The isolated strain Gordonia sp. LUNF6 can efficiently degrade PAEs, especially under conditions of 30 °C, neutral to alkaline pH, and less than 6% salinity. Strain LUNF6 has broad a substrate spectrum of PAEs and has the best degradation effect on DBP and BBP. It also can remediate wastewater contaminated by DBP. The PAE hydrolase gene mphGd2 was cloned through genome sequencing and genomic analysis. After induction expression and purification, the hydrolase MphGd2 was obtained, which can hydrolyze MBP, revealing the molecular mechanism of strain LUNF6 in degrading PAEs. This study provides bacteria and enzyme resources capable of efficiently degrading PAEs. The application of PAE-degrading strains and hydrolytic enzymes in environmental remediation will be systematically investigated in the future.
Supplementary Materials
The following supporting information can be downloaded at www.mdpi.com/article/10.3390/pr13030731/s1: Figure S1: The HPLC profile of DBP degraded by strain LUNF6 compared with control; Figure S2: The strain LUNF6 on the LB agar plate containing DBP; Figure S3: The HPLC profile of DBP degraded by strain LUNF6. CK, the control sample without inoculation of strain LUNF6; Figure S4: The functional classification of genes from chromosome (A) and plasmid (B) based on COG database; Figure S5: The distribution of genes from chromosome (A) and plasmid (B) of strain LUNF6 in metabolic pathways based on KEGG database; Figure S6: Sequence alignment of MphGd2 and other hydrolases. The catalytic triad (Ser111-Asp245-His277) was marked by pentastar; Table S1: The primer sequences for PCR of gene mphGd2; Table S2: The conversion of PAEs concentrations described in this study; Table S3: The genome characteristics of Gordinia sp. LUNF6; Table S4: The ANI analysis based on chromosome sequence of strain LUNF6 and its relatives; Table S5: PAEs degradation related genes in Gordonia sp. LUNF6.
Author Contributions
Conceptualization, S.F.; methodology, X.H.; software, Z.F.; validation, S.F., Z.F. and M.X.; formal analysis, S.F.; investigation, S.F.; resources, S.F.; data curation, Z.S.; writing—original draft preparation, S.F.; writing—review and editing, X.H.; visualization, Y.Z. and P.Z.; supervision, S.F. and X.H.; project administration, S.F.; funding acquisition, S.F., X.H., Z.S., Y.Z. and P.Z. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Natural Science Foundation of China (32101364), the Project of Central Government Guidance on Local Science and Technology Development of Hebei Province (236Z3807G and 246Z2907G), the Hebei Natural Science Foundation (B2023408001, H2020408003 and C2020408016), the Science Research Project of Hebei Education Department (ZD2020194), the Doctoral (Postdoctoral) Research Initiation Fund Project (XBQ202031), the Innovation and entrepreneurship training program for college students of Langfang Normal University (S202410100008), and the Special Project of Hebei Province Key Research and Development Plan Project on Rural Revitalization Technology Innovation (22327103D).
Data Availability Statement
No new data were created or analyzed in this study.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Hahladakis, J.N.; Velis, C.A.; Weber, R.; Iacovidou, E.; Purnell, P. An overview of chemical additives present in plastics: Migration, release, fate and environmental impact during their use, disposal and recycling. J. Hazard. Mater. 2018, 344, 179–199. [Google Scholar] [CrossRef] [PubMed]
- Akhbarizadeh, R.; Dobaradaran, S.; Schmidt, T.C.; Nabipour, I.; Spitz, J. Worldwide bottled water occurrence of emerging contaminants: A review of the recent scientific literature. J. Hazard. Mater. 2020, 392, 122271. [Google Scholar] [CrossRef]
- Benjamin, S.; Masai, E.; Kamimura, N.; Takahashi, K.; Anderson, R.C.; Faisal, P.A. Phthalates impact human health: Epidemiological evidences and plausible mechanism of action. J. Hazard. Mater. 2017, 340, 360–383. [Google Scholar] [CrossRef] [PubMed]
- Katsikantami, I.; Sifakis, S.; Tzatzarakis, M.; Vakonaki, E.; Kalantzi, O.; Tsatsakis, A.; Rizos, A. A global assessment of phthalates burden and related links to health effects. Environ. Int. 2016, 97, 212–236. [Google Scholar] [CrossRef] [PubMed]
- Mariana, M.; Feiteiro, J.; Verde, I.; Cairrao, E. The effects of phthalates in the cardiovascular and reproductive systems: A review. Environ. Int. 2016, 94, 758–776. [Google Scholar] [CrossRef]
- Wang, L.; Gan, D.; Gong, L.; Zhang, Y.; Wang, J.; Guan, R.; Zeng, L.; Qu, J.; Dong, M.; Wang, L. Analysis of the performance of the efficient di-(2-ethylhexyl) phthalate-degrading bacterium rhodococcus pyridinovorans dnhp-s2 and associated catabolic pathways. Chemosphere 2022, 306, 135610. [Google Scholar] [CrossRef]
- Feng, N.; Feng, Y.; Liang, Q.; Mo, C. Complete biodegradation of di-n-butyl phthalate (dbp) by a novel Pseudomonas sp. Yjb6. Sci. Total Environ. 2021, 761, 143208. [Google Scholar] [CrossRef]
- Fan, S.; Li, C.; Guo, J.; Johansen, A.; Liu, Y.; Feng, Y.; Xue, J.; Li, Z. Biodegradation of phthalic acid esters (paes) by Bacillus sp. Lunf1 and characterization of a novel hydrolase capable of catalyzing paes. Environ. Technol. Innov. 2023, 32, 103269. [Google Scholar] [CrossRef]
- Fan, S.; Wang, J.; Li, K.; Yang, T.; Jia, Y.; Zhao, B.; Yan, Y. Complete genome sequence of Gordonia sp. Yc-jh1, a bacterium efficiently degrading a wide range of phthalic acid esters. J. Biotechnol. 2018, 279, 55–60. [Google Scholar] [CrossRef]
- Sun, Y.; Zhang, Y.; Ma, Y.; Xin, R.; Li, X.; Niu, Z. Exploring the potential of a new marine bacterium associated with plastisphere to metabolize dibutyl phthalate and bis(2-ethylhexyl) phthalate by enrichment cultures combined with multi-omics analysis. Environ. Pollut. 2024, 342, 123146. [Google Scholar] [CrossRef]
- Li, J.; Zhang, J.; Yadav, M.P.; Li, X. Biodegradability and biodegradation pathway of di-(2-ethylhexyl) phthalate by burkholderia pyrrocinia b1213. Chemosphere 2019, 225, 443–450. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; Li, X.; Dong, Y.; Li, J.; Li, Y.; Li, H.; Chen, L.; Zhou, M.; Hou, H. Biodegradation of phthalic acid esters (paes) by cupriavidus oxalaticus strain e3 isolated from sediment and characterization of monoester hydrolases. Chemosphere 2021, 266, 129061. [Google Scholar] [CrossRef] [PubMed]
- Fan, S.; Wang, J.; Yan, Y.; Wang, J.; Jia, Y. Excellent degradation performance of a versatile phthalic acid esters-degrading bacterium and catalytic mechanism of monoalkyl phthalate hydrolase. Int. J. Mol. Sci. 2018, 19, 2803. [Google Scholar] [CrossRef] [PubMed]
- Jin, D.; Kong, X.; Li, Y.; Bai, Z.; Zhuang, G.; Zhuang, X.; Deng, Y. Biodegradation of di-n-butyl phthalate by Achromobacter sp. Isolated from rural domestic wastewater. Int. J. Environ. Res. Public Health 2015, 12, 13510–13522. [Google Scholar] [CrossRef]
- Wu, X.; Liang, R.; Dai, Q.; Jin, D.; Wang, Y.; Chao, W. Complete degradation of di-n-octyl phthalate by biochemical cooperation between Gordonia sp. Strain jdc-2 and Arthrobacter sp. Strain jdc-32 isolated from activated sludge. J. Hazard. Mater. 2010, 176, 262–268. [Google Scholar] [CrossRef]
- Bai, N.; Li, S.; Zhang, J.; Zhang, H.; Zhang, H.; Zheng, X.; Lv, W. Efficient biodegradation of dehp by cm9 consortium and shifts in the bacterial community structure during bioremediation of contaminated soil. Environ. Pollut. 2020, 266, 115112. [Google Scholar] [CrossRef]
- Yang, J.; Guo, C.; Liu, S.; Liu, W.; Wang, H.; Dang, Z.; Lu, G. Characterization of a di-n-butyl phthalate-degrading bacterial consortium and its application in contaminated soil. Environ. Sci. Pollut. Res. 2018, 25, 17645–17653. [Google Scholar] [CrossRef]
- Jin, D.; Bai, Z.; Chang, D.; Hoefel, D.; Jin, B.; Wang, P.; Wei, D.; Zhuang, G. Biodegradation of di-n-butyl phthalate by an isolated Gordonia sp. Strain qh-11: Genetic identification and degradation kinetics. J. Hazard. Mater. 2012, 221–222, 80–85. [Google Scholar] [CrossRef]
- Wu, J.; Liao, X.; Yu, F.; Wei, Z.; Yang, L. Cloning of a dibutyl phthalate hydrolase gene from Acinetobacter sp. Strain m673 and functional analysis of its expression product in Escherichia coli. Appl. Microbiol. Biotechnol. 2013, 97, 2483–2491. [Google Scholar] [CrossRef]
- Zhang, X.; Fan, X.; Qiu, Y.; Li, C.; Xing, S.; Zheng, Y.; Xu, J. Newly identified thermostable esterase from sulfobacillus acidophilus: Properties and performance in phthalate ester degradation. Appl. Environ. Microbiol. 2014, 80, 6870–6878. [Google Scholar] [CrossRef]
- Jiao, Y.; Chen, X.; Wang, X.; Liao, X.; Xiao, L.; Miao, A.; Wu, J.; Yang, L. Identification and characterization of a cold-active phthalate esters hydrolase by screening a metagenomic library derived from biofilms of a wastewater treatment plant. PLoS ONE 2013, 8, e75977. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Liu, X.; Zhao, J.; Huang, H.; Wu, M.; Li, X.; Li, W.; Sun, X.; Sun, B. An efficient phthalate ester-degrading bacillus subtilis: Degradation kinetics, metabolic pathway, and catalytic mechanism of the key enzyme. Environ. Pollut. 2021, 273, 116461. [Google Scholar] [CrossRef]
- Wang, P.; Zhang, Y.; Jin, J.; Wang, T.; Wang, J.; Jiang, B. A high-efficiency phenanthrene-degrading Diaphorobacter sp. Isolated from pah-contaminated river sediment. Sci. Total Environ. 2020, 746, 140455. [Google Scholar] [CrossRef] [PubMed]
- Mispelaere, M.; De Rop, A.S.; Hermans, C.; De Maeseneire, S.L.; Soetaert, W.K.; De Mol, M.L.; Hulpiau, P. Whole genome-based comparative analysis of the genus streptomyces reveals many misclassifications. Appl. Microbiol. Biotechnol. 2024, 108, 453. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Tang, J.; Wu, M.; Zhou, X.; Wang, S.; Ye, H.; Xiang, W.; Zhang, Q.; Cai, T. Whole genome sequencing exploitation analysis of dibutyl phthalate by strain Stenotrophomonas acidaminiphila bdbp 071. Food Biosci. 2023, 51, 102185. [Google Scholar] [CrossRef]
- Almagro, A.J.; Tsirigos, K.D.; Sonderby, C.K.; Petersen, T.N.; Winther, O.; Brunak, S.; von Heijne, G.; Nielsen, H. Signalp 5.0 improves signal peptide predictions using deep neural networks. Nat. Biotechnol. 2019, 37, 420–423. [Google Scholar] [CrossRef]
- Kumar, A.; Kumar, P.; Saumya, K.U.; Giri, R. Investigating the conformational dynamics of SARS-CoV-2 nsp6 protein with emphasis on non-transmembrane 91–112 & 231–290 regions. Microb. Pathog. 2021, 161, 105236. [Google Scholar]
- Pang, J.; Wei, Z.; Wang, L.; Guo, X.; Chen, Q.; Wei, Y.; Peng, Y.; Zhang, Z.; Zhang, Y.; Liu, J.; et al. Acanthamoeba keratitis in china: Genotypic and clinical correlations. Transl. Vis. Sci. Technol. 2024, 13, 5. [Google Scholar] [CrossRef]
- Li, S.; Hou, R.; Zhang, M.F.; Shang, J.X. First report of fusarium commune causing root rot of blueberry in guizhou province, china. Plant Dis. 2023, 107, 1227. [Google Scholar] [CrossRef]
- Huang, J.; Zhang, A.; Yang, Q.; Ding, Y.; Xiao, Z. Degradation of nitrocellulose film under aerobic conditions by a newly isolated rhodococcus pyridinivorans strain. Bioresour. Technol. 2024, 413, 131464. [Google Scholar] [CrossRef]
- Ke, Z.; Song, J.; Ma, J.; Wang, M.; Mao, H.; Xia, C.; Qi, L.; Zhou, Y.; Wang, J. Isolation and characterization of the aspartame-degrading strain Pseudarthrobacter sp. As-1. Environ. Pollut. 2024, 340, 122883. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Sung, E.J.; Seo, S.; Min, E.K.; Lee, J.Y.; Shim, I.; Kim, P.; Kim, T.Y.; Lee, S.; Kim, K.T. Integrated multi-omics analysis reveals the underlying molecular mechanism for developmental neurotoxicity of perfluorooctanesulfonic acid in zebrafish. Environ. Int. 2021, 157, 106802. [Google Scholar] [CrossRef] [PubMed]
- Kaur, G.; Lecka, J.; Krol, M.; Brar, S.K. Novel btex-degrading strains from subsurface soil: Isolation, identification and growth evaluation. Environ. Pollut. 2023, 335, 122303. [Google Scholar] [CrossRef] [PubMed]
- Arenskotter, M.; Broker, D.; Steinbuchel, A. Biology of the metabolically diverse genus gordonia. Appl. Environ. Microbiol. 2004, 70, 3195–3204. [Google Scholar] [CrossRef]
- Sowani, H.; Kulkarni, M.; Zinjarde, S. Harnessing the catabolic versatility of gordonia species for detoxifying pollutants. Biotechnol. Adv. 2019, 37, 382–402. [Google Scholar] [CrossRef]
- Zhou, X.; Tang, J.; Wang, S.; Zhang, Y.; Ye, H.; Zhang, Q.; Xiang, W.; Cai, T.; Zeng, C. Whole genome sequencing and transcriptomics-based characterization of a novel beta-cypermethrin-degrading Gordonia alkanivorans gh-1 isolated from fermented foods. Chemosphere 2023, 320, 138017. [Google Scholar] [CrossRef]
- Hu, J.; Yang, P.; Mei, K.; Chen, J.; Yang, F.; Wu, M.; Yu, J.; Chen, J.; Zheng, J. Biodegradation of 3-methylpyridine by an isolated strain, Gordonia rubripertincta zjj. Bioresour. Technol. 2024, 412, 131303. [Google Scholar] [CrossRef]
- Mai, Z.; Wang, L.; Li, Q.; Sun, Y.; Zhang, S. Biodegradation and metabolic pathway of phenanthrene by a newly isolated bacterium Gordonia sp. Scsio19801. Biochem. Biophys. Res. Commun. 2021, 585, 42–47. [Google Scholar] [CrossRef]
- Wang, Y.; Gao, J.; Zhou, S.; Lian, M. Microbial degradation of carbamazepine by a newly isolated of Gordonia polyophrenivorans. Environ. Technol. Innov. 2023, 32, 103322. [Google Scholar] [CrossRef]
- Delegan, Y.; Kocharovskaya, Y.; Frantsuzova, E.; Streletskii, R.; Vetrova, A. Characterization and genomic analysis of Gordonia alkanivorans 135, a promising dibenzothiophene-degrading strain. Biotechnol. Rep. 2021, 29, e591. [Google Scholar] [CrossRef]
- Song, X.; Zhang, Z.; Dai, Y.; Cun, D.; Cui, B.; Wang, Y.; Fan, Y.; Tang, H.; Qiu, L.; Wang, F.; et al. Biodegradation of phthalate acid esters by a versatile pae-degrading strain Rhodococcus sp. Lw-xy12 and associated genomic analysis. Int. Biodeterior. Biodegrad. 2022, 170, 105399. [Google Scholar] [CrossRef]
- Geiger, R.A.; Junghare, M.; Mergelsberg, M.; Ebenau-Jehle, C.; Jesenofsky, V.J.; Jehmlich, N.; von Bergen, M.; Schink, B.; Boll, M. Enzymes involved in phthalate degradation in sulphate-reducing bacteria. Environ. Microbiol. 2019, 21, 3601–3612. [Google Scholar] [CrossRef] [PubMed]
- Wei, S.T.; Chen, Y.L.; Wu, Y.W.; Wu, T.Y.; Lai, Y.L.; Wang, P.H.; Ismail, W.; Lee, T.H.; Chiang, Y.R. Integrated multi-omics investigations reveal the key role of synergistic microbial networks in removing plasticizer di-(2-ethylhexyl) phthalate from estuarine sediments. mSystems 2021, 6, e35821. [Google Scholar] [CrossRef] [PubMed]
- Ebenau-Jehle, C.; Soon, C.; Fuchs, J.; Geiger, R.; Boll, M. An aerobic hybrid phthalate degradation pathway via phthaloyl-coenzyme a in denitrifying bacteria. Appl. Environ. Microbiol. 2020, 86, e00498-20. [Google Scholar] [CrossRef]
- Kanaujiya, D.K.; Sivashanmugam, S.; Pakshirajan, K. Biodegradation and toxicity removal of phthalate mixture by Gordonia sp. In a continuous stirred tank bioreactor system. Environ. Technol. Innov. 2022, 26, 102324. [Google Scholar] [CrossRef]
- Xu, W.; Wan, Q.; Wenfeng, W.; Wang, Y.; Feng, F.; Cheng, J.; Yuan, J.; Yu, X. Biodegradation of dibutyl phthalate by a novel endophytic bacillus subtilis strain hb-t2 under in-vitro and in-vivo conditions. Environ. Technol. 2020, 43, 1917–1926. [Google Scholar] [CrossRef]
- Tuan, T.H.; Lin, C.; Bui, X.T.; Ky, N.M.; Dan, T.C.N.; Mukhtar, H.; Giang, H.H.; Varjani, S.; Hao, N.H.; Nghiem, L.D. Phthalates in the environment: Characteristics, fate and transport, and advanced wastewater treatment technologies. Bioresour. Technol. 2022, 344, 126249. [Google Scholar] [CrossRef]
- Zhang, Y.; Jiao, Y.; Li, Z.; Tao, Y.; Yang, Y. Hazards of phthalates (paes) exposure: A review of aquatic animal toxicology studies. Sci. Total Environ. 2021, 771, 145418. [Google Scholar] [CrossRef]
- Li, C.; Liu, Q.; Mao, L.; Zhang, W.; Zhang, J.; Niu, D.; Yin, D.; Taoli, H.; Ren, J. Characterization of modified rape straw biochar in immobilizing aspergillus sydowii w1 pellets and evaluation on its role as a novel composite for di(2-ethylhexyl) phthalate degradation. J. Hazard. Mater. 2025, 489, 137533. [Google Scholar] [CrossRef]
- Peng, C.; Tang, J.; Yu, X.; Zhou, X.; Wang, M.; Zhang, Y.; Zhou, H.; Huang, S.; Wen, Q.; Chen, S.; et al. Biodegradation of various phthalic acid esters at high concentrations by Gordonia alkanivorans gh-1 and its degradation mechanism. Environ. Technol. Innov. 2025, 38, 104066. [Google Scholar] [CrossRef]
- Xie, Y.; Guo, X.; Liang, Z.; Shim, H. Biochemical pathways and enhanced degradation of endocrine disruptor di-2-ethylhexyl phthalate by an indigenous isolate Bacillus sp. My156. Int. Biodeterior. Biodegrad. 2023, 176, 105523. [Google Scholar] [CrossRef]
- Maneechakr, P.; Karnjanakom, S. A combination of 2k factorial with box-behnken designs for fame production via methanolysis of waste cooking palm oil over low-cost catalyst. J. Environ. Chem. Eng. 2019, 7, 103389. [Google Scholar] [CrossRef]
- Noguera, J.; Jiménez-Cabas, J.; Álvarez, B.; Caicedo-Ortiz, J.; Ruiz-Ariza, J. Analysis of buñuelos growth rate using 2k factorial design. Procedia Comput. Sci. 2020, 177, 267–275. [Google Scholar] [CrossRef]
- Moyo, F.; van der Merwe, J.; Wamwangi, D. Ruthenium implantation for corrosion resistance: Using 2k factorial design to determine significant implantation parameters. Results Surf. Interfaces 2025, 18, 100465. [Google Scholar] [CrossRef]
- Prasad, B. Phthalate pollution: Environmental fate and cumulative human exposure index using the multivariate analysis approach. Environ. Sci.-Process Impacts 2021, 23, 389–399. [Google Scholar] [CrossRef]
- Marttinen, S.K.; Kettunen, R.H.; Sormunen, K.M.; Rintala, J.A. Removal of bis(2-ethylhexyl) phthalate at a sewage treatment plant. Water Res. 2003, 37, 1385–1393. [Google Scholar] [CrossRef]
- Oliver, R.; May, E.; Williams, J. The occurrence and removal of phthalates in a trickle filter stw. Water Res. 2005, 39, 4436–4444. [Google Scholar] [CrossRef]
- Xu, Y.; Minhazul, K.A.H.M.; Wang, X.; Liu, X.; Li, X.; Meng, Q.; Li, H.; Zhang, C.; Sun, X.; Sun, B. Biodegradation of phthalate esters by Paracoccus kondratievae bjq0001 isolated from jiuqu (baijiu fermentation starter) and identification of the ester bond hydrolysis enzyme. Environ. Pollut. 2020, 263, 114506. [Google Scholar] [CrossRef]
- Li, D.; Yan, J.; Wang, L.; Zhang, Y.; Liu, D.; Geng, H.; Xiong, L. Characterization of the phthalate acid catabolic gene cluster in phthalate acid esters transforming bacterium-Gordonia sp. Strain hs-nh1. Int. Biodeterior. Biodegrad. 2016, 106, 34–40. [Google Scholar] [CrossRef]
- Liu, T.; Li, J.; Qiu, L.; Zhang, F.; Linhardt, R.; Zhong, W. Combined genomic and transcriptomic analysis of dibutyl phthalate metabolic pathway in Arthrobacter sp. Zjutw. Biotechnol. Bioeng. 2020, 117, 3712–3726. [Google Scholar] [CrossRef]
- Gerischer, U.; Segura, A.; Ornston, L.N. Pcau, a transcriptional activator of genes for protocatechuate utilization in acinetobacter. J. Bacteriol. 1998, 180, 1512–1524. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).