Impact of Oil on the Bacterial Community of the Sierozems of the ‘Daulet Asia’ Landfill in Southern Kazakhstan
Abstract
1. Introduction
2. Materials and Methods
2.1. Site for Collecting Soil for Subsequent Microfield Experiment
2.2. Properties of the Oil Used in the Study
2.3. Model Experiment in Soil
2.4. Sample Collection and Processing
2.5. Metagenomic Sequencing of the Soil Microbiome
2.5.1. Sample Preparation and Sequencing
2.5.2. Bioinformatic Analysis
2.6. Analysis of Cultivable Bacteria Diversity in Soil Samples and Characterization of Individual Strains
2.6.1. Culture Media and Conditions
2.6.2. Determination of the Number of Cultivable Microorganisms
2.6.3. Isolation of Oil-Degrading Strains from Soil Samples
2.6.4. Determination of the Spectrum of Substrates Utilized by Degrading Microorganisms
2.6.5. Determination of the Temperature Range of the Studied Strains
2.6.6. Determination of Halotolerance of the Studied Strains When Grown on Diesel Fuel
2.6.7. Determination of the pH Range of the Studied Strains During Growth on Diesel Fuel
2.6.8. Determination of Degradation Efficiency
2.6.9. Identification of Strains
3. Results and Discussion
3.1. Physicochemical Analysis of Soil at the Daulet Asia Test Site
3.2. Physicochemical Analysis of Crude Oil (Fractions and Individual Hydrocarbons)
3.3. Estimation of Oil Loss in Field Conditions
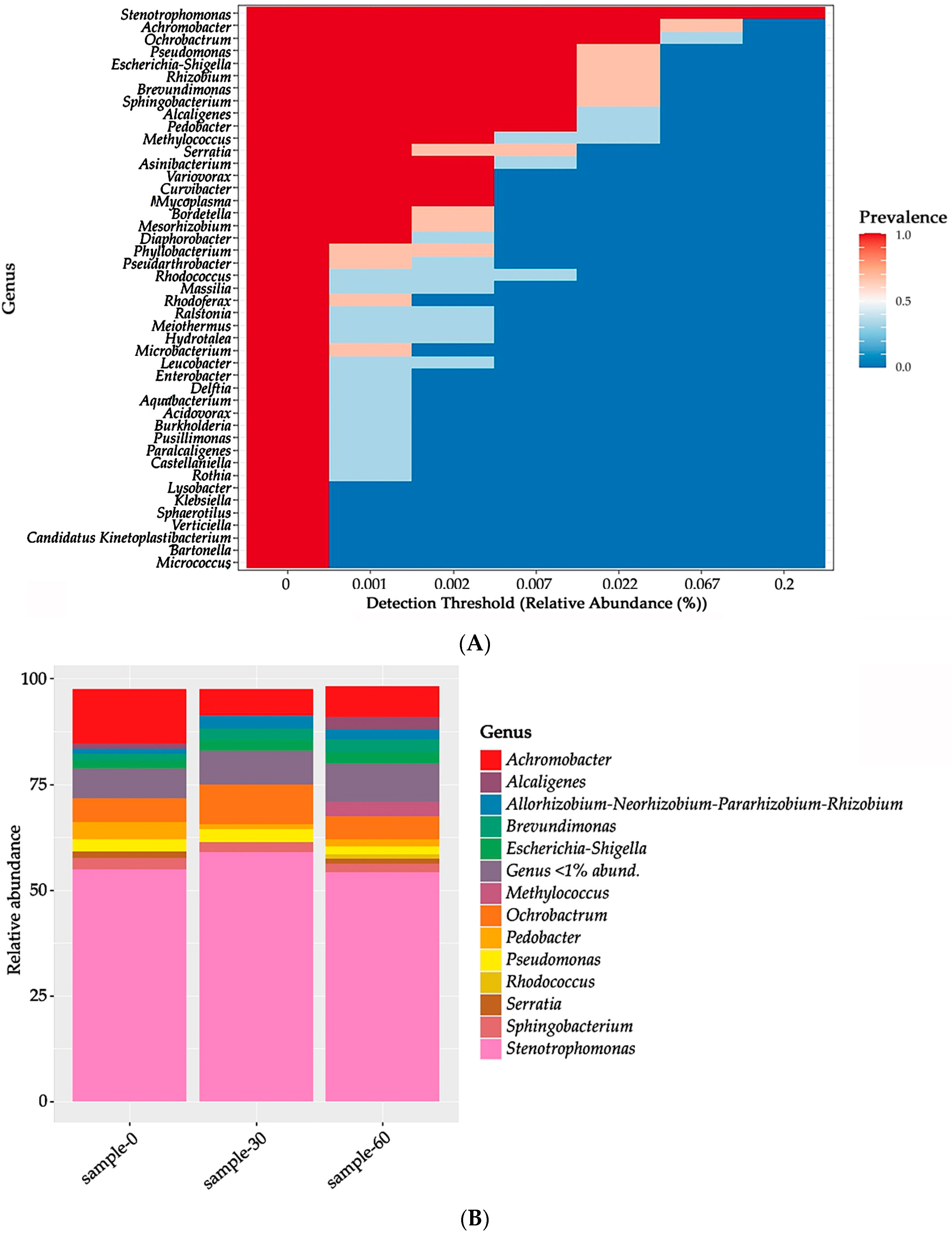
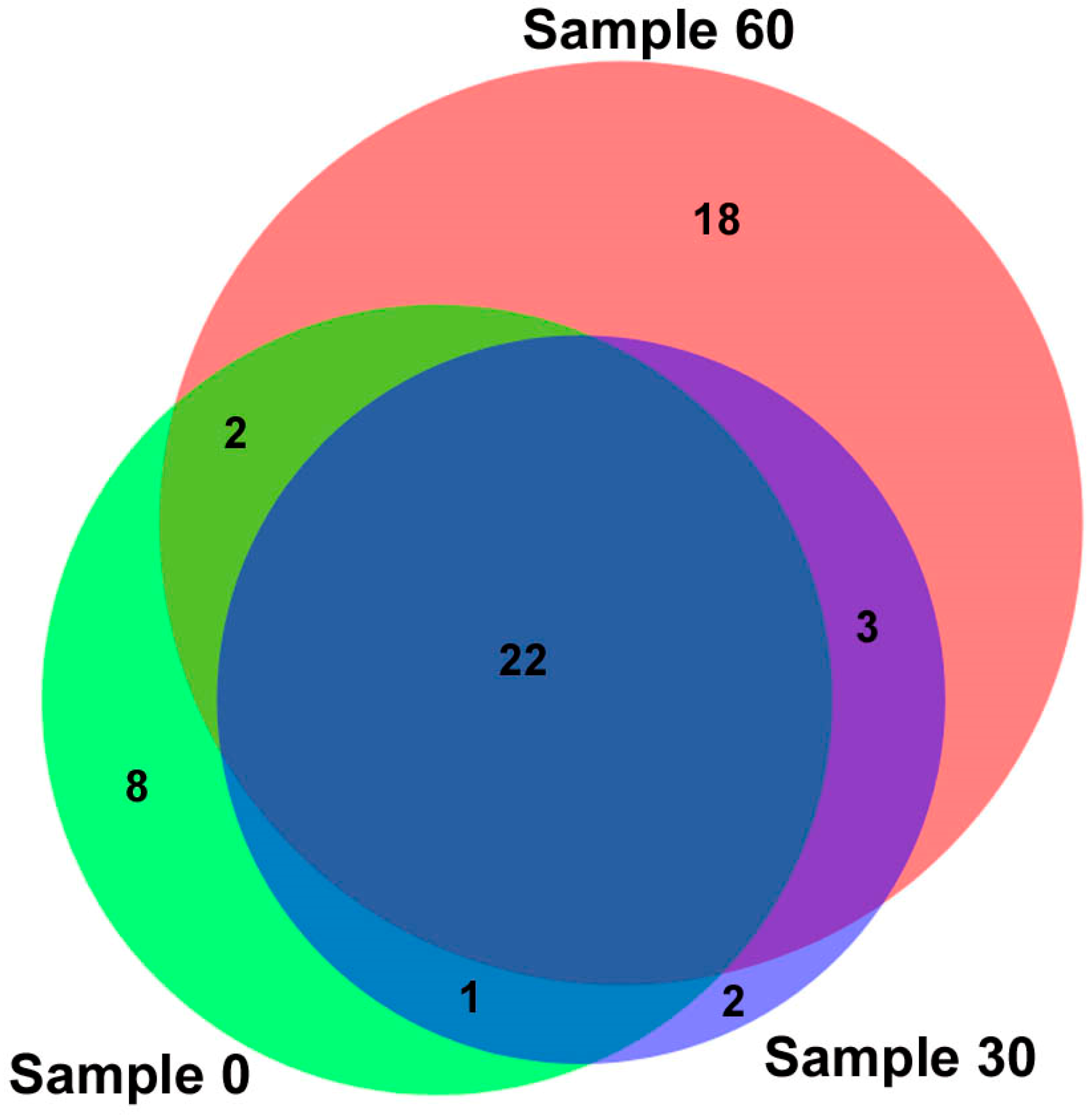
3.4. Analysis of the Microbial Response of an Indigenous Soil Community to Soil Contamination with Oil
3.5. Determination of the Total Number of Cultivable Microorganisms and the Number of Hydrocarbon-Oxidizing Microorganisms During a Model Soil Experiment
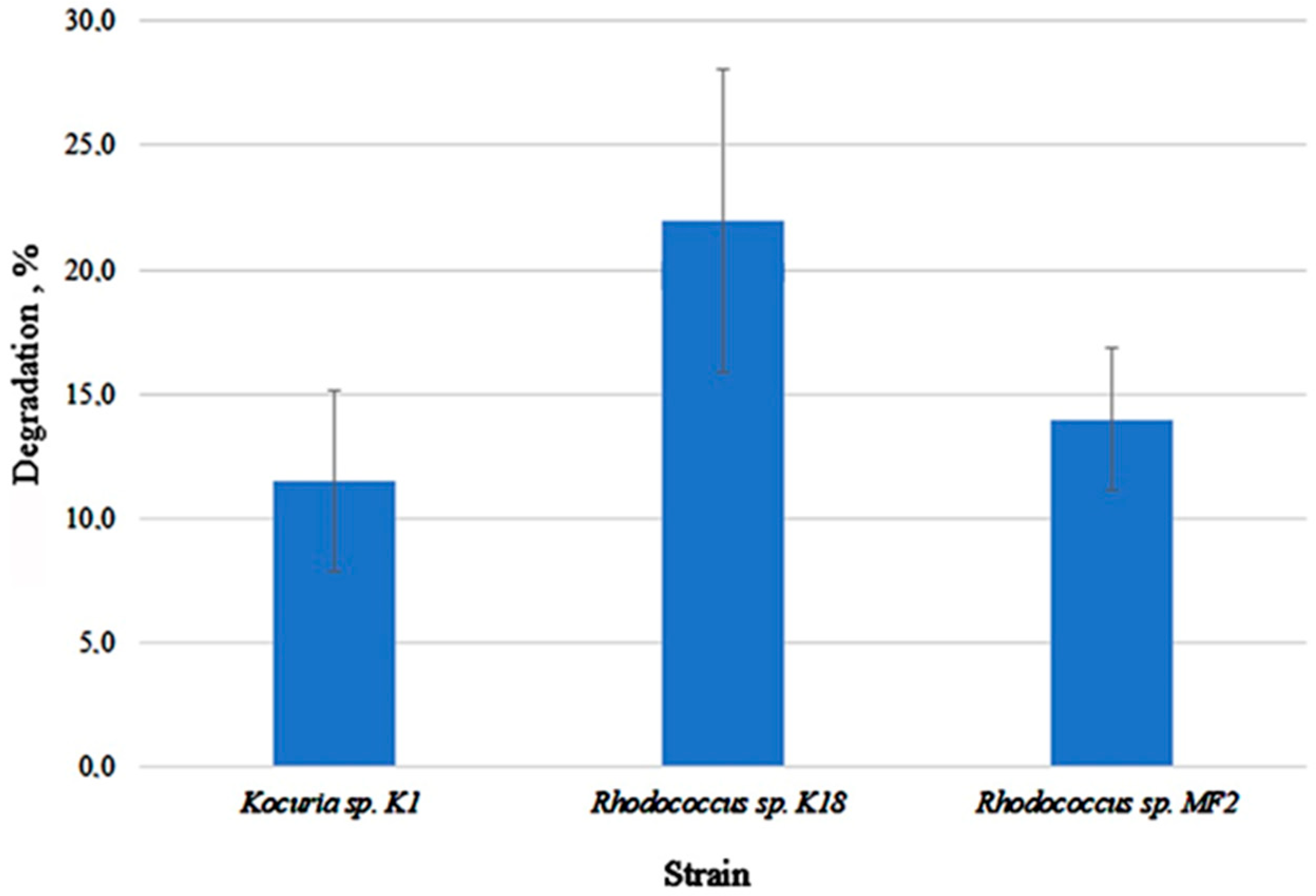
3.6. Isolation and Characterization of Cultivable Microorganisms
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
| bp | nucleotide base pairs |
| CFU | colony forming units |
| IR | infrared |
| LB | Louria-Bertany broth |
| LPSN | List of Prokaryotic Names with Standing in Nomenclature |
| Mbp | a million of nucleotide base pairs |
| NPK | nitrogen, phosphorus, potassium |
| OD600 | the value of the optical density of the medium measured at a light wavelength of 600 nm |
| PAH | polycyclic aromatic hydrocarbons |
| PBS | phosphate-buffered saline |
References
- Khozhanepesova, F.; Dadrasnia, A.; Serikbaeva, A.; Abdibattaeva, M.; Myrzabekova, A. Assessment of the influence of ambient temperature and soil salinity on the degree of oil destruction by free and immobilized microorganisms. Euras. J. Ecol. 2022, 72, 50–58. [Google Scholar] [CrossRef]
- Vasilyev, A.V.; Zabolotskikh, V.V.; Tupitsyna, O.V.; Shterenberg, A.M. Ecological monitoring of toxicity polluttion of the soils by oil products by using of biological testing methods. Oil Gas. Bus. 2012, 4, 242–249. [Google Scholar]
- Filonov, A.E.; Akhmetov, L.I.; Vetrova, A.A.; Ivanova, A.A.; Sazonova, O.I.; Puntus, I.F.; Chaika, N.Y.; Boronin, A.M. Current status and trends in environmental biotechnology. Biol. Bioteh. 2024, 1, 2. [Google Scholar] [CrossRef]
- Idrisova, D.T.; Mukhamedova, N.S.; Zhumadilova, Z.S.; Abdieva, K.M.; Shorabaev, E.Z.; Sadanov, A.K. Investigation of the processes of bioremediation of soils with different degrees of oil pollution in the Kyzylorda region in field conditions. Fundam. Stud. 2014, 12, 1669–1671. [Google Scholar]
- Idrisova, D.T.; Mukhamedova, N.S.; Zhumadilova, Z.S.; Shorabaev, E.Z.; Sadanov, A.K. Influence of organo-mineral fertilizers on bioremediation of oil-contaminated soils. Fundam. Stud. 2014, 9, 2246–2249. [Google Scholar]
- ASTM D4052-22; Standard Test Method for Density, Relative Density, and Api Gravity of Liquids by Digital Density Meter. ASTM: West Conshohocken, PA, USA, 2022. Available online: https://store.astm.org/d4052-22.html (accessed on 30 September 2025).
- ASTM D4377-00e1; Standard Test Method for Water in Crude Oils by Potentiometric Karl Fischer Titration. ASTM: West Conshohocken, PA, USA, 2020. Available online: https://store.astm.org/d4377-00e01.html (accessed on 30 September 2025).
- ASTM D4807-05; Standard Test Method for Sediment in Crude Oil by Membrane Filtration. ASTM: West Conshohocken, PA, USA, 2020. Available online: https://store.astm.org/d4807-05r20.html (accessed on 30 September 2025).
- ASTM D664-24; Standard Test Method for Acid Number of Petroleum Products by Potentiometric Titration. ASTM: West Conshohocken, PA, USA, 2024. Available online: https://store.astm.org/standards/d664 (accessed on 30 September 2025).
- ASTM D445-24; Standard Test Method for Kinematic Viscosity of Transparent and Opaque Liquids (and Calculation of Dynamic Viscosity). ASTM: West Conshohocken, PA, USA, 2024. Available online: https://store.astm.org/standards/d445 (accessed on 30 September 2025).
- ASTMD 1160-06; Standart Test Method for Distillation of Petroleum Products at Reduced Pressure, IDT. ASTM: West Conshohocken, PA, USA, 2010. Available online: http://gost.gtsever.ru/Data2/1/4293738/4293738703.pdf (accessed on 30 September 2025).
- ISO 3016:2019; Petroleum and Related Products from Natural or Synthetic Sources—Determination of Pour Point, MOD. ISO: Geneve, Switzerland, 2019. Available online: https://www.iso.org/standard/73386.html (accessed on 30 September 2025).
- PNDF 16.1.41-04; Quantitative Chemical Analysis of Soils. Methodology for Measuring the Mass Concentration of Petroleum Products in Soil Samples Using the Gravimetric Method. The Ministry of Natural Resources and Environment of the Russian Federation: Moscow, Russia, 2004. Available online: https://gostrf.com/normadata/1/4293846/4293846504.pdf (accessed on 31 August 2025).
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. Version 0.10. 2010;1. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 31 August 2025).
- Wick, R. Porechop: Adapter Trimmer for Oxford Nanopore Reads. 2017. Available online: https://github.com/rrwick/Porechop (accessed on 30 September 2025).
- Nygaard, A.B.; Charnock, C. The bacterial composition of ventilation alter dust in Norwegian pre-school nurseries. Indoor Built. Environ. 2017, 27, 1392–1404. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. Fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef] [PubMed]
- Wood, D.E.; Lu, J.; Langmead, B. Improved metagenomic analysis with Kraken 2. Genome Biol. 2019, 20, 257. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peples, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2012, 41, D590–D596. [Google Scholar] [CrossRef]
- McMurdie, P.J.; Holmes, S. Phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef]
- Wickham, H. Getting started with ggplot2. In Ggplot2: Elegant Graphics for Data Analysis, 2nd ed.; Gentleman, R., Hornik, K., Parmigiani, G., Eds.; Springer International Publishing AG: Cham, Switzerland, 2016; pp. 11–31. [Google Scholar] [CrossRef]
- Wickham, H.; Averick, M.; Bryan, J.; Chang, W.; McGowan, L.D.; François, R.; Grolemund, G.; Hayes, A.; Henry, L.; Hester, J.; et al. Welcome to the Tidyverse. J. Open Source Softw. 2019, 4, 1686. [Google Scholar] [CrossRef]
- Willis, A.; Bunge, J.; Whitman, T. Improved detection of changes in species richness in high diversity microbial communities. J. Roy. Stat. Soc. Ser. C Appl. Stat. 2017, 66, 963–977. [Google Scholar] [CrossRef]
- Lahti, L.; Shetty, S. Microbiome R Package; Bioconductor: Boston, MA, USA, 2017. [Google Scholar] [CrossRef]
- Dixon, P. VEGAN, a package of R functions for community ecology. J. Veg. Sci. 2003, 14, 927–930. [Google Scholar] [CrossRef]
- Breitwieser, F.P.; Salzberg, S.L. Pavian: Interactive analysis of metagenomics data for microbiome studies and pathogen identification. Bioinformatics 2020, 36, 1303–1304. [Google Scholar] [CrossRef]
- Sambrook, J.; Fritsch, E.R.; Maniatis, T. Molecular Cloning: A Laboratory Manual, 2nd ed.; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 1989; pp. 154–196. [Google Scholar]
- Carhart, G.; Hegeman, G. Improved method of selection for mutants of Pseudomonas putida. Appl. Microbiol. 1975, 30, 1046. [Google Scholar] [CrossRef]
- PND F 14.1:2:4.168-2000 FR.1.31.2017.26183 Methodology (Method) for Measuring the Mass Concentration of Petroleum Products in Samples of Drinking, Natural and Treated Wastewater by IR Spectrophotometry Using KN Series Concentrator Meters. Available online: https://gostrf.com/normadata/1/4293846/4293846475.pdf (accessed on 30 September 2025).
- Marchesi, J.R.; Weightman, A.J.; Cragg, B.A.; Parkes, R.J.; Fry, J.C. Methanogen and bacterial diversity and distribution in deep gas hydrate sediments from the Cascadia Margin as revealed by 16S rRNA molecular analysis. FEMS Microbiol. Ecol. 2001, 34, 221–228. [Google Scholar] [CrossRef] [PubMed]
- Available online: https://technelysium.com.au/wp/chromas (accessed on 30 September 2025).
- Available online: https://www.megasoftware.net (accessed on 30 September 2025).
- Available online: https://blast.ncbi.nlm.nih.gov/Blast.cgi (accessed on 7 November 2025).
- Available online: https://lpsn.dsmz.de/species (accessed on 30 September 2025).
- Adenan, N.H.; Ting, A.S.Y. Actinobacteria from soils and their applications in environmental bioremediation. In Microbial Biotechnology: Role in Ecological Sustainability and Research; Chowdhary, P., Mani, S., Chaturvedi, P., Eds.; John Wiley & Sons: London, UK, 2022; pp. 313–333. [Google Scholar] [CrossRef]
- Krivoruchko, A.; Kuyukina, M.; Peshkur, T.; Cunningham, C.J.; Ivshina, I. Rhodococcus strains from the specialized collection of alkanotrophs for biodegradation of aromatic compounds. Molecules 2023, 28, 2393. [Google Scholar] [CrossRef]
- Ivshina, I.; Bazhutin, G.; Tyumina, E. Rhodococcus strains as a good biotool for neutralizing pharmaceutical pollutants and obtaining therapeutically valuable products: Through the past into the future. Front. Microbiol. 2022, 13, 967127. [Google Scholar] [CrossRef] [PubMed]
- Makarani, N.; Kaushal, R.S. Advances in actinobacteria-based bioremediation: Mechanistic insights, genetic regulation, and emerging technologies. Biodegradation 2025, 36, 24. [Google Scholar] [CrossRef]
- Chen, Q.; Li, J.; Liu, M.; Sun, H.; Bao, M. Study on the biodegradation of crude oil by free and immobilized bacterial consortium in marine environment. PLoS ONE 2017, 12, e0174445. [Google Scholar] [CrossRef] [PubMed]
- Rocca, J.D.; Simonin, M.; Blaszczak, J.R.; Ernakovich, J.G.; Gibbons, S.M.; Midani, F.S.; Washburne, A.D. The microbiome stress project: Toward a global meta-analysis of environmental stressors and their effects on microbial communities. Front. Microbiol. 2019, 9, 3272. [Google Scholar] [CrossRef]
- Park, C.; Park, W. Survival and energy producing strategies of alkane degraders under extreme conditions and their biotechnological potential. Front. Microbiol. 2018, 9, 1081. [Google Scholar] [CrossRef]
- Liu, J.; Zhen, B.; Qiu, H.; Zhou, X.; Zhang, H. Impact of waterlogging and heat stress on rice rhizosphere microbiome assembly and potential function in carbon and nitrogen transformation. Arch. Agron. Soil. Sci. 2022, 69, 1920–1932. [Google Scholar] [CrossRef]
- Elufisan, T.; Rodríguez-Luna, I.; Oyedara, O.; Sánchez-Varela, A.; Hernández-Mendoza, A.; González, E.; Paz-González, A.; Muhammad, K.; Rivera, G.; Villalobos-López, M.; et al. The polycyclic aromatic hydrocarbon (PAH) degradation activities and genome analysis of a novel strain Stenotrophomonas sp. Pemsol Isol. Mexico Peer J. 2020, 8, e8102. [Google Scholar] [CrossRef]
- Akhmetov, L.I.; Puntus, I.F.; Narmanova, R.A.; Appazov, N.O.; Funtikova, T.V.; Regepova, A.A.; Filonov, A.E. Recent advances in creating biopreparations to fight oil spills in soil ecosystems in sharply continental climate of Republic of Kazakhstan. Processes 2022, 10, 549. [Google Scholar] [CrossRef]
- Funtikova, T.V.; Akhmetov, L.I.; Puntus, I.F.; Mikhailov, P.A.; Appazov, N.O.; Narmanova, R.A.; Filonov, A.E.; Solyanikova, I.P. Bioremediation of oil-contaminated soil of the Republic of Kazakhstan using a new biopreparation. Microorganisms 2023, 11, 522. [Google Scholar] [CrossRef]
- Narmanova, R.; Tapalova, A.; Zhapparbergenov, R.; Appazov, N. Biological products for soil and water purification from oil and petroleum products. Evergr. Jt. J. Nov. Carb Res. Sci. Green. Asia Strat. 2023, 10, 688–695. [Google Scholar] [CrossRef]
- Nzila, A.; Ramirez, C.O.; Musa, M.M.; Sankara, S.; Basheer, C.; Li, Q.X. Pyrene biodegradation and proteomic analysis in Achromobacter xylosoxidans, PY4 strain. Int Biodeter Biodegr 2018, 130, 40–47. [Google Scholar] [CrossRef]
- Deng, M.C.; Li, J.; Liang, F.R.; Yi, M.; Xu, X.M.; Yuan, J.P.; Peng, J.; Wu, C.-F.; Wang, J.H. Isolation and characterization of a novel hydrocarbon-degrading bacterium Achromobacter sp. HZ01 from the crude oil-contaminated seawater at the Daya Bay, southern China. Mar. Pollut. Bull. 2014, 83, 79–86. [Google Scholar] [CrossRef] [PubMed]
- Hassanshahian, M.; Zeynalipour, M.S.; Musa, F.H. Isolation and characterization of crude oil degrading bacteria from the Persian Gulf (Khorramshahr provenance). Mar. Pollut. Bull. 2014, 82, 39–44. [Google Scholar] [CrossRef] [PubMed]
- Ishaya, S.; Usman, S.; Nweke, O.D.; Adams, N.H.; Umar, R.; Ilyasu, N.S.; Jagaba, A.H.; Atangwho, I.; Yakasai, H.M. Degradation of used engine oil by Alcaligenes sp. strain isolated from oil contaminated site: Isolation, identification, and optimization of the growth parameters. Case Stud. Chem. Environ. Eng. 2023, 8, 100516. [Google Scholar] [CrossRef]
- Mahjoubi, M.; Aliyu, H.; Cappello, S.; Naifer, M.; Souissi, Y.; Cowan, D.A.; Cherif, A. The genome of Alcaligenes aquatilis strain BU33N: Insights into hydrocarbon degradation capacity. PLoS ONE 2019, 14, e0221574. [Google Scholar] [CrossRef]
- Wang, X.; Wang, X.; Liu, M.; Zhou, L.; Gu, Z.; Zhao, J. Bioremediation of marine oil pollution by Brevundimonas diminuta: Effect of salinity and nutrients. Desalin Water Treat. 2016, 57, 19768–19775. [Google Scholar] [CrossRef]
- Fayidh, M.A.; Kalleary, S.; Kasirajan, S.; Muthusamy, S. Isolation of a unique phenol degrading bacterial strain Escherichia coli moh 1 from effluent of an edible oil industry in Chennai, India. J. Biotechnol. 2015, 10, 36. [Google Scholar]
- McDonald, I.R.; Miguez, C.B.; Rogge, G.; Bourque, D.; Wendlandt, K.D.; Groleau, D.; Murrell, J.C. Diversity of soluble methane monooxygenase-containing methanotrophs isolated from polluted environments. FEMS Microbiol. Lett. 2006, 255, 225–232. [Google Scholar] [CrossRef]
- Sun, W.; Ali, I.; Liu, J.; Dai, M.; Cao, W.; Jiang, M.; Saren, G.; Yu, X.; Peng, C.; Naz, I. Isolation, identification, and characterization of diesel-oil-degrading bacterial strains indigenous to Changqing oil field, China. J. Basic. Microbiol. 2019, 59, 723–734. [Google Scholar] [CrossRef]
- Bhattacharya, M.; Biswas, D.; Sana, S.; Datta, S. Biodegradation of waste lubricants by a newly isolated Ochrobactrum sp. C1. 3 Biotech 2015, 5, 807–817. [Google Scholar] [CrossRef]
- Chang, S.; Zhang, G.; Chen, X.; Long, H.; Wang, Y.; Chen, T.; Liu, G. The complete genome sequence of the cold adapted crude-oil degrader: Pedobacter steynii DX4. Stand. Genom Sci. 2017, 12, 45. [Google Scholar] [CrossRef] [PubMed]
- Rabodonirina, S.; Rasolomampianina, R.; Krier, F.; Drider, D.; Merhaby, D.; Net, S.; Ouddane, B. Degradation of fluorene and phenanthrene in PAHs-contaminated soil using Pseudomonas and Bacillus strains isolated from oil spill sites. J. Environ. Manag. 2019, 232, 1–7. [Google Scholar] [CrossRef]
- Santos, E.C.; Jacques, R.J.; Bento, F.M.; Peralba, M.D.C.R.; Selbach, P.A.; Sá, E.L.; Camargo, F.A. Anthracene biodegradation and surface activity by an iron-stimulated Pseudomonas sp. Biores Technol. 2008, 99, 2644–2649. [Google Scholar] [CrossRef]
- Zhao, H.P.; Wu, Q.S.; Wang, L.; Zhao, X.T.; Gao, H.W. Degradation of phenanthrene by bacterial strain isolated from soil in oil refinery fields in Shanghai China. J. Haz Mater. 2009, 164, 863–869. [Google Scholar] [CrossRef]
- Zhu, X.; Ni, X.; Waigi, M.G.; Liu, J.; Sun, K.; Gao, Y. Biodegradation of mixed PAHs by PAH-degrading endophytic bacteria. Int. J. Environ. Res. Publ. Health 2016, 13, 805. [Google Scholar] [CrossRef] [PubMed]
- Osadebe, A.U.; Chukwu, C.B. Degradation properties of Rhizobium petrolearium on different concentrations of crude oil and its derivative fuels. Environ. Exp. Biol. 2023, 21, 83–92. [Google Scholar] [CrossRef]
- Larkin, M.J.; Allen, C.C.; Kulakov, L.A.; Lipscomb, D.A. Purification and characterization of a novel naphthalene dioxygenase from Rhodococcus sp. strain NCIMB12038. J. Bacteriol. 1999, 181, 6200–6204. [Google Scholar] [CrossRef]
- Kulakov, L.A.; Allen, C.C.; Lipscomb, D.A.; Larkin, M.J. Cloning and characterization of a novel cis-naphthalene dihydrodiol dehydrogenase gene (narB) from Rhodococcus sp. NCIMB12038. FEMS Microbiol. Lett. 2000, 182, 327–331. [Google Scholar] [CrossRef]
- Alegbeleye, O.O.; Opeolu, B.O.; Jackson, V.A. Polycyclic aromatic hydrocarbons: A critical review of environmental occurrence and bioremediation. Environ. Manag. 2017, 60, 758–783. [Google Scholar] [CrossRef]
- Mallick, S. Biodegradation of Acenaphthene by Sphingobacterium sp. strain RTSB involving trans-3-carboxy-2-hydroxybenzylidenepyruvic acid as a metabolite. Chemosphere 2019, 219, 748–755. [Google Scholar] [CrossRef]
- Mikolasch, A.; Omirbekova, A.; Schumann, P.; Reinhard, A.; Sheikhany, H.; Berzhanova, R.; Mukasheva, T.; Schauer, F. Enrichment of aliphatic, alicyclic and aromatic acids by oil-degrading bacteria isolated from the rhizosphere of plants growing in oil-contaminated soil from Kazakhstan. Appl. Microbiol. Biotechnol. 2015, 99, 4071–4084. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Zhao, R.; Xiao, A.; Li, L.; Jiang, Z.; Chen, F.; Ni, H. Characterization of an alkaline β-agarase from Stenotrophomonas sp. NTa Enzym. hydrolysates. Int. J. Biol. Macromolec. 2016, 86, 525–534. [Google Scholar] [CrossRef] [PubMed]

| Stage | Cycle Quantity | Temperature, °C | Duration, min |
|---|---|---|---|
| initial denaturation | 1 | 95 | 2 |
| product synthesis | 27 | 95 | 1 |
| 60 | 1 | ||
| 72 | 3 | ||
| final elongation | 1 | 72 | 2 |
| cooling | 1 | 4 | 10 |
| Primer | Sequence | Annealing Temperature, °C | Amplicon Size, bp | Ref. |
|---|---|---|---|---|
| 63F | CAG GCC TAA CAC ATG CAA GTC | 55 | 1300 | [30] |
| 1387R | GGG CGG WGT GTA CAA GGC |
| Trait | Value | |
|---|---|---|
| Soil type | Medium loam (sierozem) | |
| Water extract pH | 7.5–8.3 | |
| Anion content, % | Cl− | 0.06–0.14 |
| SO42− | 0.18–0.52 | |
| CO32− | 0.03–0.06 | |
| NPK content, mg/kg soil | P2O5 | 2.6–33.6 |
| Total nitrogen | 7–21 | |
| K2O | 50–60 | |
| Cation content, % | Na+ | 0.11–0.12 |
| Ca2+ | 0.05–0.07 | |
| Mg2+ | 0.003–0.021 | |
| Humus content, % | 0.102 | |
| Trait | Value | |
|---|---|---|
| Density (t = 20 °C), g/cm3 | 0.821 | |
| Water content, % w/w | 0.46 | |
| Mechanical impurities, % w/w | 0.02 | |
| Water extract pH | 6 | |
| Freezing point, °C | −4.8 | |
| Viscosity characteristics (t = 20 °C) | Kinematic viscosity, mm2/s | 18.065 |
| Dynamic viscosity, Pa·s | 14.831·10−3 | |
| Proportion (% w/w) of Various Fractions in Oil | ||||
|---|---|---|---|---|
| Alkanes | Cycloalkanes | Alkenes | Arenes | PAHs |
| 70.580 | 17.402 | 2.817 | 2.040 | 7.161 |
| No | Sampling Date | Oil-Free Control | Oil Content, % |
|---|---|---|---|
| 1 | 2 August 2024 (initial point) | 0.00 | 3.26 |
| 2 | 15 August 2024 (two weeks) | 0.00 | 3.12 |
| 3 | 30 August 2024 (four weeks) | 0.00 | 3.04 |
| 4 | 16 September 2024 (six weeks) | 0.00 | 2.99 |
| 5 | 30 September 2024 (eight weeks) | 0.00 | 2.98 |
| 6 | 15 October 2024 (ten weeks) | 0.00 | 2.69 |
| 7 | 1 November 2024 (twelve weeks) | 0.00 | 2.02 |
| No | Before Filtering | After Filtering | Total Classified Sequences | Total Unclassified Sequences | ||
|---|---|---|---|---|---|---|
| Total Sequences | Total Bases, Mbp | Total Sequences | Total Bases, Mbp | |||
| sample 0 | 12,413 | 17.1 | 10,576 | 15.6 | 10,235 | 341 |
| sample 30 | 8,236 | 11.2 | 6,861 | 10.0 | 6,604 | 257 |
| sample 60 | 2,080 | 2.8 | 1,759 | 2.6 | 1,697 | 62 |
| No | Chao1 | Shannon | Simpson |
|---|---|---|---|
| sample 0 | 80.1 | 1.97 | 0.68 |
| sample 30 | 77.1 | 1.91 | 0.66 |
| sample 60 | 55.9 | 2.15 | 0.71 |
| Strain | Substrate for Isolation | Tested Compounds | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Nah | Hde | Ddc | Sal | Bzt | Tol | Phe | Dsf | ||
| K1 | crude oil | + | + | − | + | + | + | + | + |
| K18(2) | crude oil | + | + | + | − | − | + | + | + |
| MF2 | crude oil | − | + | + | − | − | − | + | + |
| Strain | Growth Substrate | Conditions | ||
|---|---|---|---|---|
| Temperature Range, °C * | pH Range | NaCl, % | ||
| K1 | diesel fuel | 6–45 | 4–10 | Up to 10 |
| K18(2) | diesel fuel | 20–45 | 5–10 | Up to 3 |
| MF2 | diesel fuel | 20–37 | 6–10 | Up to 3 |
| Strain | Species | Query Cover, % | Percent Identity, % |
|---|---|---|---|
| K1 | Kocuria sp./Kocuria rosea | 99 | 96.13 |
| K18(2) | Rhodococcus corynebacterioides | 95 | 95.73 |
| MF2 | Rhodococcus corynebacterioides | 98 | 93.60 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Narmanova, R.; Delegan, Y.; Kocharovskaya, Y.; Bogun, A.; Puntus, I.; Akhmetov, L.; Vetrova, A.; Baraboshkina, A.; Chayka, N.; Kuzhamberdieva, S.; et al. Impact of Oil on the Bacterial Community of the Sierozems of the ‘Daulet Asia’ Landfill in Southern Kazakhstan. Processes 2025, 13, 3730. https://doi.org/10.3390/pr13113730
Narmanova R, Delegan Y, Kocharovskaya Y, Bogun A, Puntus I, Akhmetov L, Vetrova A, Baraboshkina A, Chayka N, Kuzhamberdieva S, et al. Impact of Oil on the Bacterial Community of the Sierozems of the ‘Daulet Asia’ Landfill in Southern Kazakhstan. Processes. 2025; 13(11):3730. https://doi.org/10.3390/pr13113730
Chicago/Turabian StyleNarmanova, Roza, Yanina Delegan, Yulia Kocharovskaya, Alexander Bogun, Irina Puntus, Lenar Akhmetov, Anna Vetrova, Angelina Baraboshkina, Nelly Chayka, Svetlana Kuzhamberdieva, and et al. 2025. "Impact of Oil on the Bacterial Community of the Sierozems of the ‘Daulet Asia’ Landfill in Southern Kazakhstan" Processes 13, no. 11: 3730. https://doi.org/10.3390/pr13113730
APA StyleNarmanova, R., Delegan, Y., Kocharovskaya, Y., Bogun, A., Puntus, I., Akhmetov, L., Vetrova, A., Baraboshkina, A., Chayka, N., Kuzhamberdieva, S., Suleimenov, N., Kanzhar, S., Niyazova, D., Yespanova, I., Alimkhan, B., Tolegenkyzy, M., Darmagambet, K., Arynova, K., Appazov, N., & Filonov, A. (2025). Impact of Oil on the Bacterial Community of the Sierozems of the ‘Daulet Asia’ Landfill in Southern Kazakhstan. Processes, 13(11), 3730. https://doi.org/10.3390/pr13113730







