Design of Portable Water Quality Spectral Detector and Study on Nitrogen Estimation Model in Water
Abstract
1. Introduction
2. Materials and Methods
2.1. Design of Portable Water Quality Spectral Detector
2.2. Experimental Design
2.3. Data Analysis Methods
3. Results
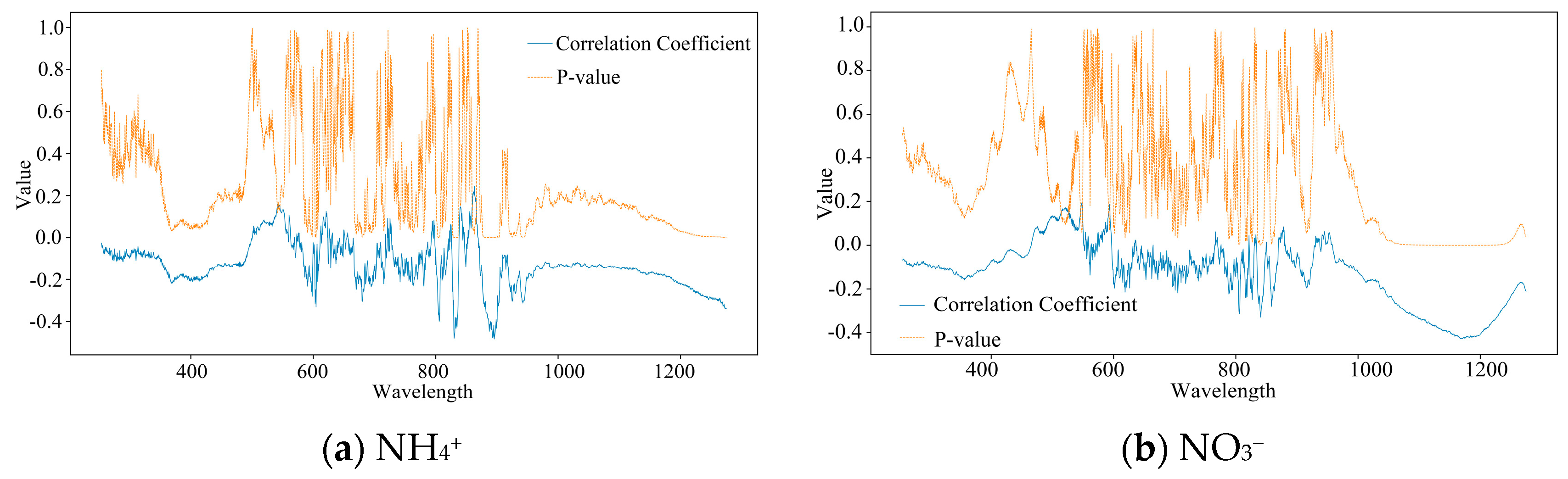
3.1. Correlation Analysis Between Nitrogen Indicators and Spectral Bands
3.2. Comparison of Full Band Raw Data Models and Spectral Screening Results
3.2.1. PCA Results
3.2.2. Analysis of Full Band Modeling Results
3.2.3. Selecting 1000 Main Bands for Modeling Result Analysis
3.2.4. Selecting 400 Main Bands for Modeling Result Analysis
3.3. Comparison of Spectral Models Under Different Preprocessing Methods
3.3.1. PCA Results After Preprocessing Using Three Methods
- (1)
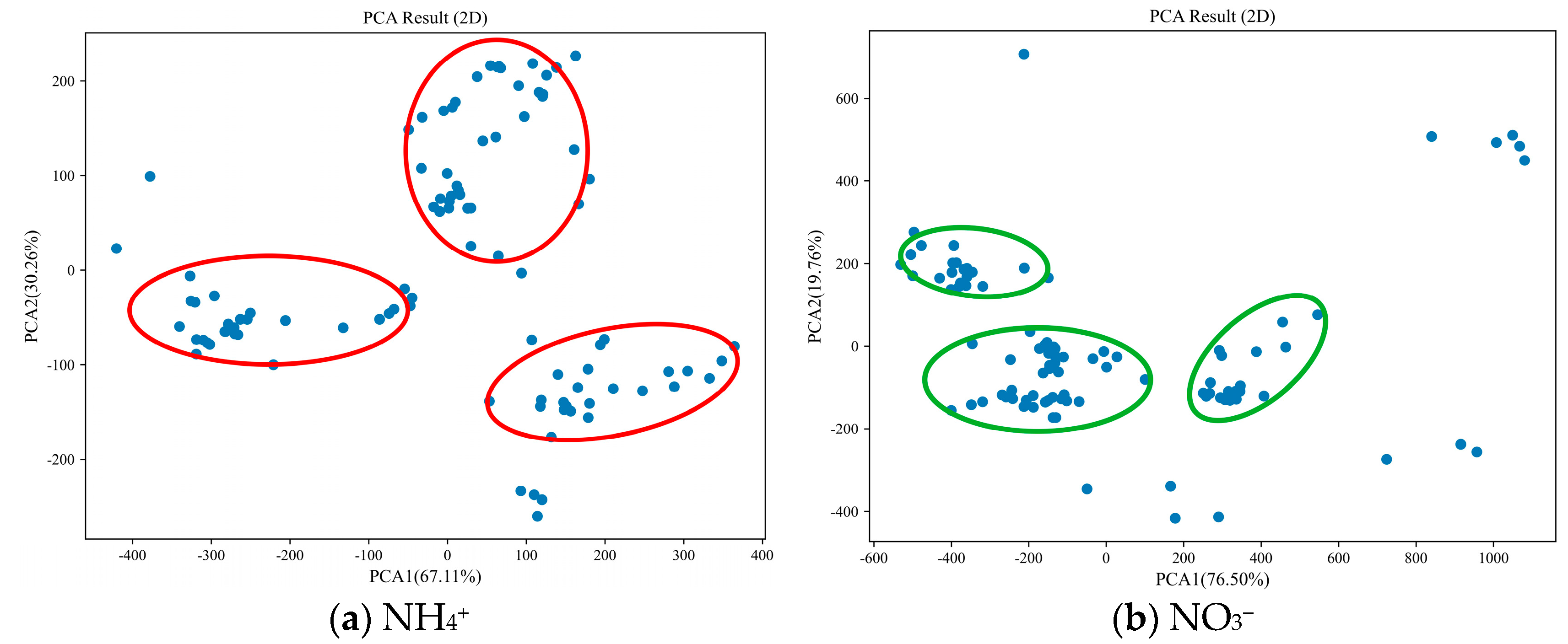
- MSC. From Figure 5, it can be seen that the PCA of NH4+ spectra explains 51.20% and 45.75% of the data changes in the first and second principal axes, totaling 96.95%. The samples are mainly distributed in three concentrated areas (red circles in Figure 5a), with significant differences between the areas. The PCA of NO3− spectra explained 80.70% and 14.67% of the data changes in the first and second principal axes, totaling 95.37%. The samples were mainly distributed in three concentrated small areas (green circles in Figure 5b), with some differences between the areas.
- (2)
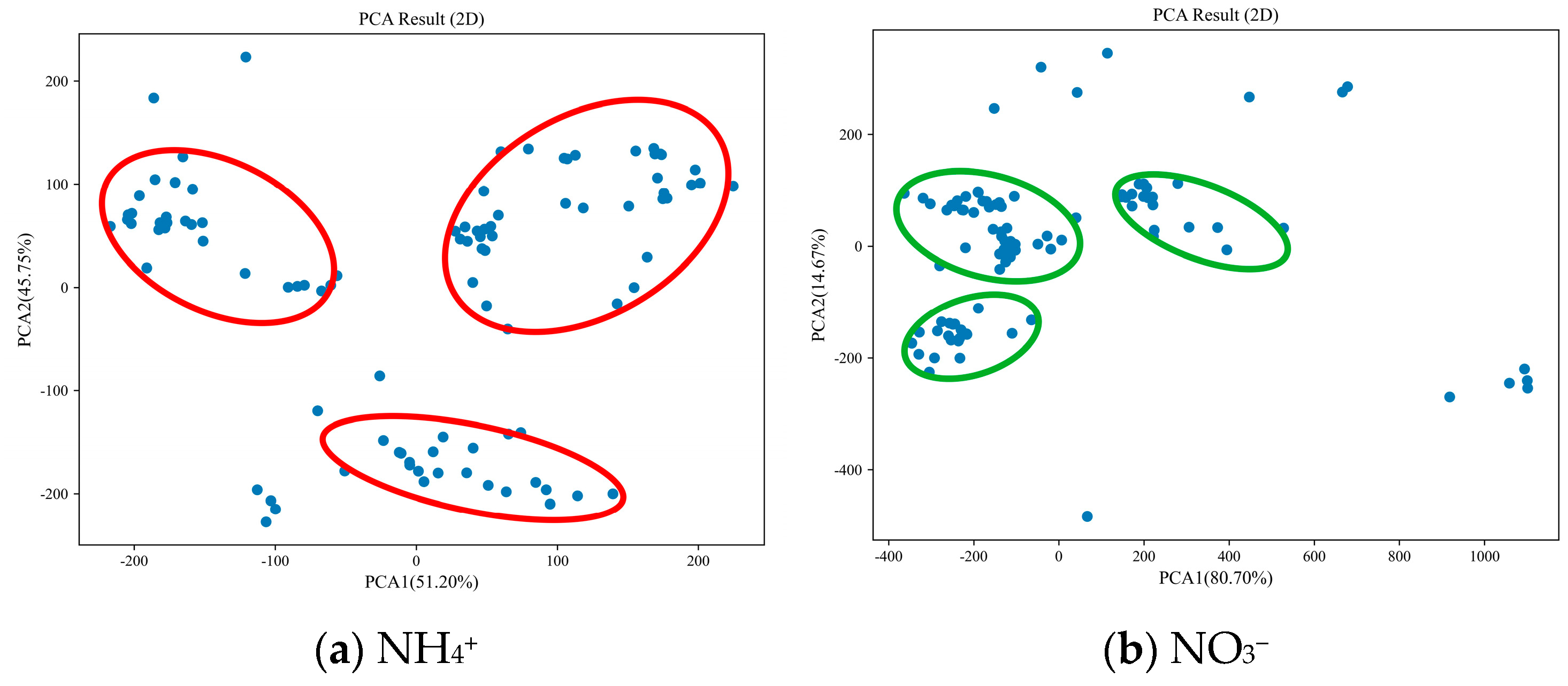
- SG. From Figure 6, it can be seen that the PCA of NH4+ spectra explains 61.76% and 30.28% of the data changes in the first and second principal axes, totaling 92.04%. The samples are mainly distributed in three concentrated areas (red circles in Figure 6a), with significant differences between the areas. The PCA of NO3− spectra explained 76.51% and 19.76% of the data changes in the first and second principal axes, totaling 96.27%. The samples were mainly distributed in three concentrated small areas (green circles in Figure 6b), with some differences between the areas.
- (3)
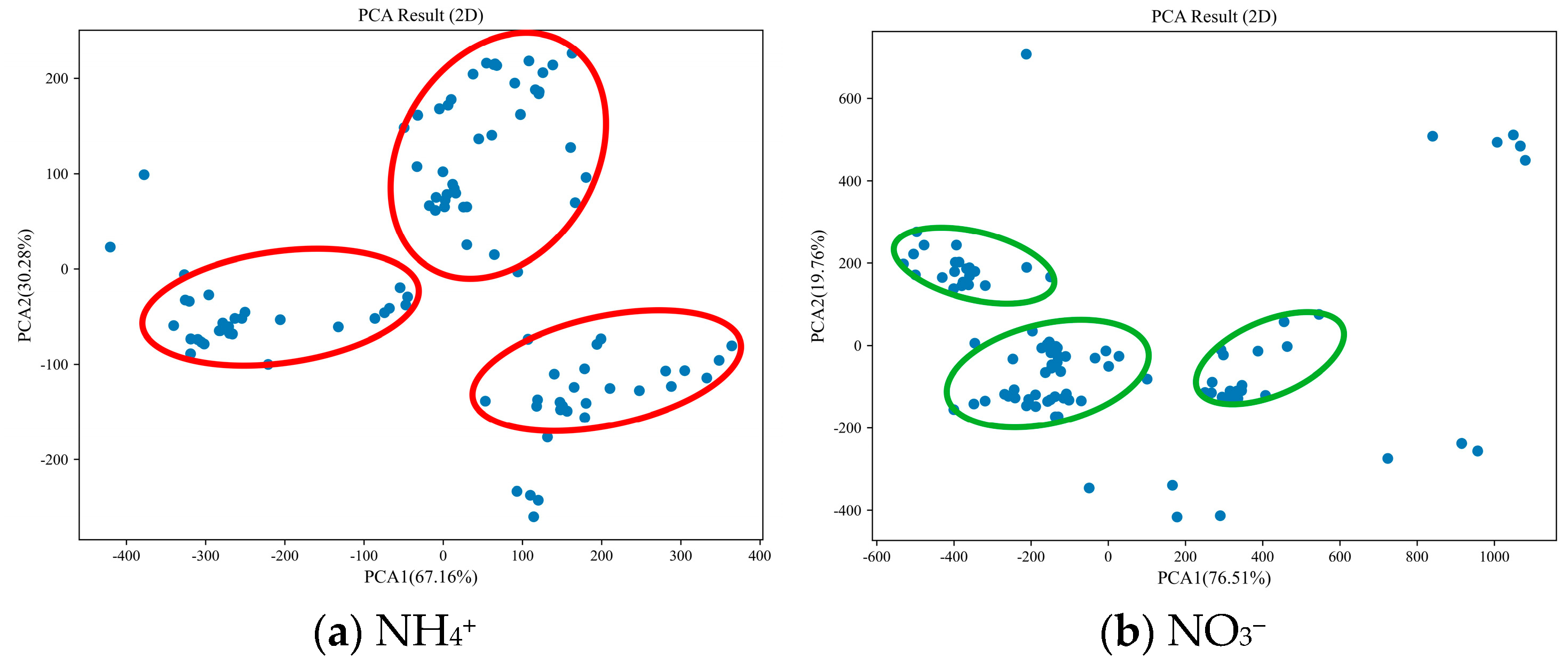
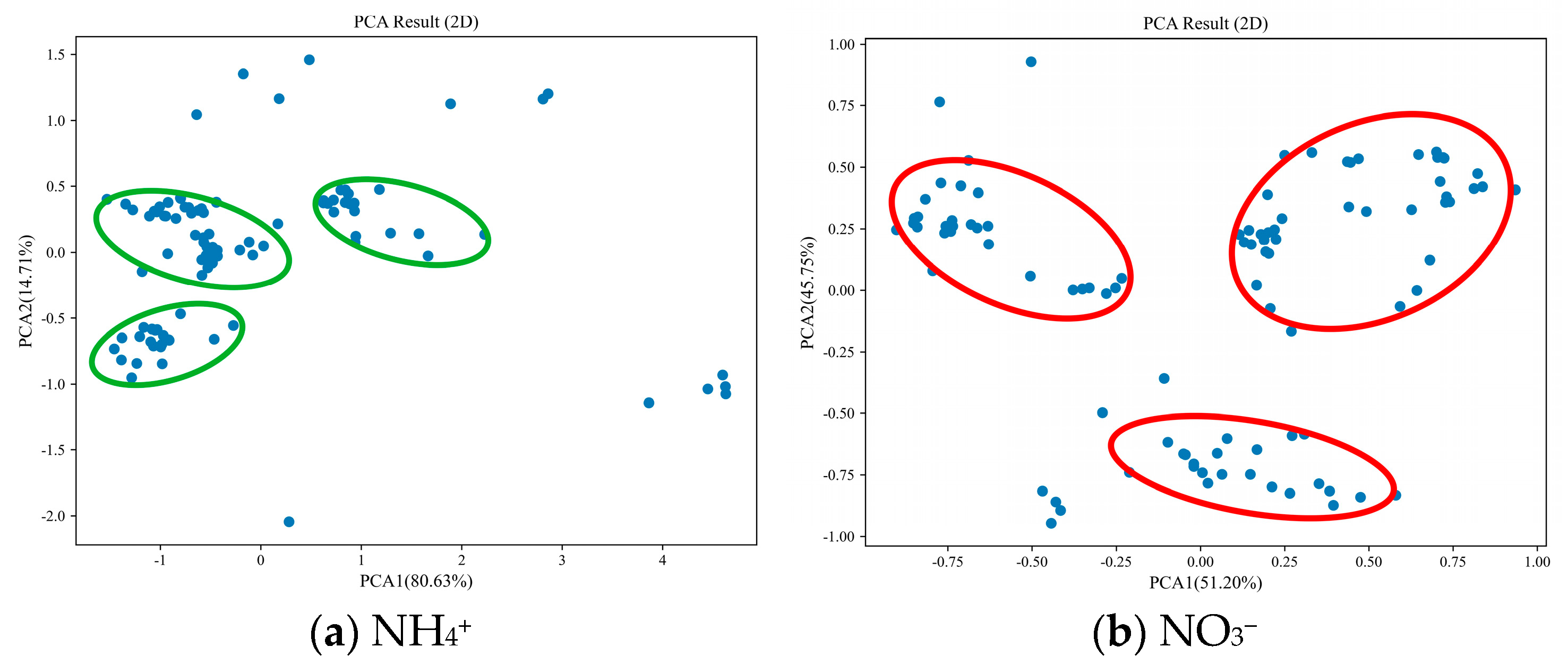
- SS. From Figure 7, it is evident that the PCA of NH4+ spectra accounts for 80.63% and 14.71% of the data variability along the first and second principal axes, respectively, summing up to 95.34%. The samples are predominantly clustered in three distinct regions (highlighted by red circles in Figure 7a), with notable differences among these areas. Similarly, the PCA of NO3− spectra explains 51.20% and 45.75% of the data variability along the first and second principal axes, respectively, totaling 96.95%. The samples are primarily concentrated in three small, distinct regions (indicated by green circles in Figure 7b), with some discernible differences among these areas.
3.3.2. Spectral Model Results After MSC Processing
3.3.3. Spectral Model Results After SG Processing
3.3.4. Spectral Model Results After SS Processing
3.4. Comparison Between the Predicted and Measured Values of the Optimal Model
4. Discussion
4.1. Water Quality Indicators Are Closely Related to Different Spectral Bands
4.2. Impact of PCA Dimensionality Reduction on Model Accuracy
4.3. Impact of Data Preprocessing on Model Accuracy
4.4. Research Shortcomings and Future Research Directions
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
| MSC | Multiple Scattering Correction |
| SG | Savitzky–Golay Filtering |
| SS | Standardization |
| SVM | Support Vector Machine |
| MLP | Multilayer Perceptron |
| SVR | Support Vector Regression |
| RF | Random Forest |
| PLSR | Partial Least Squares Regression |
| PCA | Principal Component Analysis |
| RMSE | Root Mean Square Error |
| 1DCNN | 1D Convolutional Neural Network |
| UAVs | Unmanned Aerial Vehicles |
| LSTM | Long Short-Term Memory |
| KF | Kalman Filtering |
| CODMn | Permanganate index |
| SNV | Standard Normal Variate Transformation |
| XGBoost | eXtreme Gradient Boosting |
References
- Smith, J. Nitrogen dynamics in eutrophic freshwater systems. Environ. Sci. Technol. 2020, 12, 7213–7222. [Google Scholar]
- Johnson, M.; Lee, K. Challenges in environmental monitoring infrastructure development. Glob. Environ. Change 2018, 52, 54–63. [Google Scholar]
- Imaizumi, M. Time Series Analysis of Influence of Water Cycle on Nitrate Contamination in Miyako Island Ryukyu Limestone Aquifer. Water 2025, 18, 2723. [Google Scholar] [CrossRef]
- Mitchell, V.; Gong, J.; Moon, E.; Wu, W. Bushfire Impact on Drinking Water Distribution Networks and Investigation Methods: A Review. Water Resour. Res. 2025, 3, e2024WR038225. [Google Scholar] [CrossRef]
- Rogers, K.M.; Tschritter, C.; Bradshaw, D.; Abel, S.; Pannell, J.L.; Thern, J.; Heath, T.; Tio, P.; Liu, X.; Buckthought, L.; et al. A New Zealand freshwater nitrate data quality validation study using commercial laboratories and portable testing instruments. Chemosphere 2025, 381, 144472. [Google Scholar] [CrossRef]
- Zhang, X.; Ma, L.; Xu, Y. Design of Aquaculture Water Quality Monitoring System Based on Wireless Communication. In Proceedings of the International Conference on Information Control, Electrical Engineering and Rail Transit, Shanghai, China, 17–19 November 2023; Springer Nature: Singapore, 2025. [Google Scholar] [CrossRef]
- Huang, X.; Xiong, J.; Lin, H.; Pan, Z.; Wang, K.; Zhang, M.; Zhu, Z.; Ou, Y. Research on water quality detection integrating spectral analysis and automated control. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2025, 339, 126260. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Li, X.; Cheng, W.; Zhao, G.; Tang, L.; Yang, Y.; Wu, Y.; Zhang, P.; Wang, Q. Data fusion strategy based on ultraviolet–visible spectra and near-infrared spectra for simultaneous and accurate determination of key parameters in surface water. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2023, 302, 123007. [Google Scholar] [CrossRef]
- Liu, M. Application of fluorescence spectrum detection technology in COD detection of water quality. Hydro Sci. Cold Zone Eng. 2024, 10, 75–78. [Google Scholar] [CrossRef]
- Gao, B.-C.; Li, R.-R.; Montes, M.J.; Mccarthy, S.C. Combining Cirrus and Aerosol Corrections for Improved Reflectance Retrievals over Turbid Waters from Visible Infrared Imaging Radiometer Suite Data. Oceans 2025, 2, 28. [Google Scholar] [CrossRef]
- Thompson, R. Hyperspectral imaging for contaminant detection. Anal. Chem. 2019, 3, 2149–2157. [Google Scholar]
- Harvey, E.T.; Kratzer, S.; Philipson, P. Satellite-based water quality monitoring for improved spatial and temporal retrieval of chlorophyll-a in coastal waters. Remote Sens. Environ. 2015, 158, 417–430. [Google Scholar] [CrossRef]
- Wang, M.; Pang, C.; Shi, B.; Schang, C.; Nolan, M.; Poon, R.; Catsamas, S.; Zhu, W.; Mccarthy, D. A Low-Cost, Open-Source, 3D-Printed, Compact, In Situ, Automatic Water Sampler for Environmental Surveillance. ACS EST Water 2025, 7, 4067–4078. [Google Scholar] [CrossRef]
- Moon, J.; Jung, S.; Suh, S.; Pyo, J. Development of deep learning quantization framework for remote sensing edge device to estimate inland water quality in South Korea. Water Res. 2025, 283, 123760. [Google Scholar] [CrossRef]
- Obořilová, R.; Buzík, J.; Skládal, P.; Farka, Z.; Lacina, K. Portable turbidimetric device for in-time monitoring of bacterial contamination in drinking water. J. Water Process Eng. 2025, 74, 107832. [Google Scholar] [CrossRef]
- Jekel Könnel, E.; Nonno, S.D.; Ulber, R. Low-cost and easy-to-use: A portable photometer for simple and comprehensive analysis of critical water quality parameters. Microchem. J. 2025, 214, 113946. [Google Scholar] [CrossRef]
- Liu, G.; Gong, Y.; Zhang, H.; Liang, H. Infrared Spectral Noise Reduction Algorithm Based on Wavelet Transform Optimized EEMD Combined with SG. Infrared Technol. 2024, 12, 1453–1458. [Google Scholar]
- Li, K.; Guo, Y.; Zhong, H.; Jin, Y.; Li, B.; Fang, H.; Yao, L.; Zhao, C. Rapid Identification of Dendrobium Species Using Near-Infrared Hyperspectral Imaging Technology. Sensors 2025, 18, 5625. [Google Scholar] [CrossRef]
- Souza, L.L.D.; Candeias, D.N.C.; Moreira, E.D.T.; Diniz, P.H.G.D.; Springer, V.H.; Fernandes, D.D.D.S. UV–Vis spectralprint-based discrimination and quantification of sugar syrup adulteration in honey using the Successive Projections Algorithm (SPA) for variable selection. Chemom. Intell. Lab. Syst. 2025, 257, 105314. [Google Scholar] [CrossRef]
- Zhou, H.; Xu, H.; Long, L.; Ji, D.; Han, Y.; Ji, X.; Cui, Y. A Comparative Study of Hyperspectral Estimation Models for Total Phosphorus and Total Nitrogen in the Midstream of the Xiangxi River. China Rural. Water Hydropower 2024, 11, 125–132. [Google Scholar] [CrossRef]
- Gao, Z.; Wang, G.; Chen, J.; Fang, L.; Ren, S.; Yinglan, A.; Ji, S.; Liu, R.; Wang, Q. Kalman filtering assimilated machine learning methods significantly improve the prediction performance of water quality parameters. Ecol. Inform. 2025, 90, 103337. [Google Scholar] [CrossRef]
- Zhang, L. Deep learning for spectral analysis. IEEE Trans. Geosci. Remote Sens. 2021, 6, 5041–5052. [Google Scholar]
- Wei, Y.; Zhong, R.; Yang, Y. Groundwater Fluoride Prediction for Sustainable Water Management: A Comparative Evaluation of Machine Learning Approaches Enhanced by Satellite Embeddings. Sustainability 2025, 18, 8505. [Google Scholar] [CrossRef]
- Das, A. Surface water quality evaluation, apportionment of pollution sources and aptness testing for drinking using water quality indices and multivariate modelling in Baitarani River basin, Odisha. HydroResearch 2025, 8, 244–264. [Google Scholar] [CrossRef]
- Sattley, W.M.; Burchell, B.M.; Conrad, S.D.; Madigan, M.T. Design, Construction, and Application of an Inexpensive, High-Resolution Water Sampler. Water 2017, 8, 578. [Google Scholar] [CrossRef]
- Sánchez-Saquín, C.H.; Soto-Cajiga, J.A.; Barrera-Fernández, J.M.; Gómez-Hernández, A.; Rodríguez-Olivares, N.A. Identification, Control, and Characterization of Peristaltic Pumps in Hemodialysis Machines. Appl. Syst. Innov. 2025, 2, 44. [Google Scholar] [CrossRef]
- Brown, A. UV—Vis spectroscopy of nitrogen compounds. J. Phys. Chem. A 2021, 18, 3987–3995. [Google Scholar]
- Tatartchenko, V. Infrared radiation during phase transitions of water. J. Phys. Chem. B 2012, 12, 3726–3734. [Google Scholar]
- Schmidt, M. Quantum mechanical modeling of NH4+ spectra. J. Chem. Phys. 2023, 158, 4. [Google Scholar]
- Stagni, A. Experimental and kinetic modeling study of ammonia oxidation. React. Chem. Eng. 2020, 3, 456–467. [Google Scholar] [CrossRef]
- Liu, X.; Zhou, L.; Xia, X.; Han, R.; Lyu, Q.; Xie, R.; Yi, S. Predicting Citrus Leaf Nitrogen Content Based on Hybrid Bat Algorithm Optimized PLSR. J. Southwest Univ. (Nat. Sci. Ed.) 2025, 2, 160–170. [Google Scholar] [CrossRef]
- Puttipipatkajorn, A.; Puttipipatkajorn, A. Development of low-cost portable spectrometer equipped with 18-band spectral sensors using deep learning model for evaluating moisture content of rubber sheets. Smart Agric. Technol. 2024, 9, 100562. [Google Scholar] [CrossRef]
- Wong, C.; Gesmundo, A. Transfer Learning to Learn with Multitask Neural Model Search. arXiv 2017, arXiv:1710.10776. [Google Scholar] [CrossRef]
- Lee, L.C.; Liong, C.-Y.; Jemain, A.A. A contemporary review on Data Preprocessing (DP) practice strategy in ATR-FTIR spectrum. Chemom. Intell. Lab. Syst. 2017, 163, 64–75. [Google Scholar] [CrossRef]
- Pishini, K.; Abdolazimi, O.; Shishebori, D.; Rezaee, M.J.; Sepehrifar, M. Evaluating efficiency in water and sewerage services: An integrated DEA approach with DOE and PCA. Sci. Total Environ. 2025, 959, 178288. [Google Scholar] [CrossRef]
- Karpagalakshmi, R.C.; Selvam, P.; Sakthivel, S.; Kalaivanan, K.; Sharma, R.; Sungheetha, A.; Mahapatra, S. Analysis of Water Quality in Cauvery River by Using PCA with Various Classifications Technique. Procedia Comput. Sci. 2025, 258, 1391–1403. [Google Scholar] [CrossRef]
- Ivakhno, S.; Armstrong, J.D. Non-linear dimensionality reduction of signaling networks. BMC Syst. Biol. 2007, 1, 27. [Google Scholar] [CrossRef] [PubMed]
- Rinnan, A.; Van Den, B.F.; Engelsen, S.B. Review of the most common pre-processing techniques for Near-Infrared spectra. Trends Anal. Chem. 2009, 28, 1201–1222. [Google Scholar] [CrossRef]
- Savitzky, A.; Golay, M.J.E. Smoothing and Differentiation of Data by Simplified Least Squares Procedures. Anal. Chem. 1964, 8, 1627–1639. [Google Scholar] [CrossRef]
- Barnes, R.J.; Dhanoa, M.S.; Lister, S.J. Standard Normal Variate Transformation and De-trending of Near-Infrared Diffuse Reflectance Spectra. Appl. Spectrosc. 1989, 5, 772–777. [Google Scholar] [CrossRef]
- Engel, J.; Gerretzen, J.; Szymańska, E.; Jansen, J.J.; Downey, G.; Blanchet, L.; Buydens, L.M.C. Breaking with trends in pre-processing? TrAC Trends Anal. Chem. 2013, 50, 96–106. [Google Scholar] [CrossRef]
- Wold, S.; Sjöström, M.; Eriksson, L. PLS-regression: A basic tool of chemometrics. Chemom. Intell. Lab. Syst. 2001, 2, 109–130. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, C.; Wang, H. Simulated Estimation of BOD Content in Water Bodies Based on PCA Transmission Spectrum Reconstruction With Noise Reduction. Spectrosc. Spectr. Anal. 2025, 2, 386–393. [Google Scholar]
- Xiao, M.; Zhu, Y.; Gao, W.; Zeng, Y.; Li, H.; Chen, S.; Liu, P.; Huang, H. Comparative Study of Water Quality Prediction Methods Based on Different Artificial Neural Network. Environ. Sci. 2024, 10, 5761–5767. [Google Scholar] [CrossRef]
- Sarpal, D.; Sinha, R.; Jha, M.; Tn, P. AgriWealth: IoT based farming system. Microprocess. Microsyst. 2022, 89, 104447. [Google Scholar] [CrossRef]
- Li, Y.; Guo, J.; Liu, G.; Zhang, J.; Guo, Z.; Wang, W. Design of high speed miniature spectrometer based on FPGA. Proc. SPIE 2024, 13070, 130700J. [Google Scholar] [CrossRef]
- Irwan, D.; Ibrahim, S.L.; Latif, S.D.; Winston, C.A.; Ahmed, A.N.; Sherif, M.; El-Shafie, A.H.; El-Shafie, A. River water quality monitoring using machine learning with multiple possible in-situ scenarios. Environ. Sustain. Indic. 2025, 26, 100620. [Google Scholar] [CrossRef]
- Clark, R.N.; Roush, T.L. Reflectance spectroscopy: Quantitative analysis techniques for remote sensing applications. J. Geophys. Res. Solid Earth 1984, B7, 6329–6340. [Google Scholar] [CrossRef]
- Li, F.L.; Chang, Q.R. Estimation of winter wheat leaf nitrogen content based on continuum removed spectra. J. Agric. Mach. 2017, 7, 174–179. [Google Scholar]
- Rinnan, S. Pre-processing in vibrational spectroscopy—When, why and how. Anal. Methods 2014, 18, 7124. [Google Scholar] [CrossRef] [PubMed]
- Valladares-Castellanos, M.; De Jesús Crespo, R.; Douthat, T. Using machine learning for long-term calibration and validation of water quality ecosystem service models in data-scarce regions. Sci. Total Environ. 2025, 1000, 180388. [Google Scholar] [CrossRef] [PubMed]
- Xiao, F.; Zhang, R.; Jian, Z.; Liu, W.; Sun, T.; Pang, W.; Han, L.; Qin, H. Using ensemble machine learning to predict and understand spatiotemporal water quality variations across diverse watersheds in coastal urbanized areas. Ecol. Indic. 2025, 178, 113976. [Google Scholar] [CrossRef]
- Chen, W.; Ren, T.; Zhao, C.; Wen, Y.; Gu, Y.; Zhou, M.; Wang, P. Transformer-Based Fast Mole Fraction of CO2 Retrievals from Satellite-Measured Spectra. J. Remote Sens. 2025, 5, 0470. [Google Scholar] [CrossRef]
- Li, W.; Ren, M.; Zhang, H.; Duan, Y.; Chen, D.; Li, S.; Xu, M.; Wang, L.; Yang, X. Estimation of water quality in coastal aquaculture waters using the combination of machine learning and unmanned aerial vehicle multispectral imagery. Aquaculture 2026, 611, 743002. [Google Scholar] [CrossRef]




| Measure Day | NH4+ (mol/L) | NO3− (mol/L) |
|---|---|---|
| DAY1 | 0, 0.02, 0.04, 0.06, 0.08, 0.1, 0.16, 0.2, 0.24, 0.28, 0.3, 0.36, 0.4, 0.44, 0.48, 0.5, 0.56, 0.6, 0.64, 0.68, 0.7, 0.76, 0.78, 0.8, 0.86, 0.9, 0.94, 1 | 0, 0.02, 0.04, 0.06, 0.08, 0.1, 0.16, 0.2, 0.24, 0.28, 0.3, 0.36, 0.4, 0.44, 0.48, 0.5, 0.56, 0.6, 0.64, 0.68, 0.7, 0.76, 0.78, 0.8, 0.86, 0.9, 0.94, 0.96, 1 |
| DAY2 | 1, 0.94, 0.9, 0.86, 0.8, 0.78, 0.76, 0.7, 0.68, 0.64, 0.6, 0.56, 0.5, 0.48, 0.44, 0.4, 0.36, 0.3, 0.28, 0.24, 0.2, 0.16, 0.1, 0.08, 0.06, 0.04, 0.02, 0 | 1, 0.96, 0.94, 0.9, 0.86, 0.8, 0.78, 0.76, 0.7, 0.68, 0.64, 0.6, 0.56, 0.5, 0.48, 0.44, 0.4, 0.36, 0.3, 0.28, 0.24, 0.2, 0.16, 0.1, 0.08, 0.06, 0.04, 0.02, 0 |
| DAY3 | 0, 0.12, 0.14, 0.18, 0.22, 0.26, 0.32, 0.34, 0.38, 0.42, 0.46, 0.52, 0.54, 0.58, 0.62, 0.66, 0.72, 0.74, 0.82, 0.84, 0.88 | 0, 0.12, 0.14, 0.18, 0.22, 0.26, 0.32, 0.34, 0.38, 0.42, 0.46, 0.52, 0.54, 0.58, 0.6, 0.62, 0.66, 0.72, 0.74, 0.82 |
| DAY4 | 0.88, 0.84, 0.82, 0.74, 0.72, 0.66, 0.62, 0.58, 0.54, 0.52, 0.46, 0.42, 0.38, 0.34, 0.32, 0.26, 0.22, 0.18, 0.14, 0.12, 0 | 0.82, 0.74, 0.72, 0.66, 0.62, 0.58, 0.54, 0.52, 0.46, 0.42, 0.38, 0.34, 0.32, 0.26, 0.22, 0.18, 0.14, 0.12, 0 |
| Index | Model | R2 | MSE | RMSE | MAE | Time/s |
|---|---|---|---|---|---|---|
| NH4+ | 1DCNN | 0.2815 | 0.0497 | 0.223 | 0.1781 | 357.1088 |
| CAS-PLSR | 0.7274 | 0.0189 | 0.1376 | 0.1146 | 31.4315 | |
| PLSR | 0.8584 | 0.0098 | 0.0991 | 0.0818 | 0.0307 | |
| RF | 0.5033 | 0.0345 | 0.1847 | 0.1458 | 2.6102 | |
| RF-Lasso | 0.5408 | 0.0319 | 0.1785 | 0.1368 | 2.6316 | |
| SVM-MLP | −6.8377 | 0.544 | 0.7376 | 0.5914 | 0.1046 | |
| SVR | −0.0027 | 0.0696 | 0.2638 | 0.225 | 0.006 | |
| NO3− | 1DCNN | 0.1735 | 0.0799 | 0.2827 | 0.2187 | 345.1332 |
| CAS-PLSR | 0.5624 | 0.0421 | 0.2052 | 0.1647 | 30.4024 | |
| PLSR | 0.7376 | 0.0252 | 0.1589 | 0.1313 | 0.0203 | |
| RF | 0.4557 | 0.0524 | 0.2288 | 0.1872 | 2.4340 | |
| RF-Lasso | 0.4400 | 0.0539 | 0.2321 | 0.1928 | 2.5284 | |
| SVM-MLP | −0.8598 | 0.179 | 0.423 | 0.336 | 0.1129 | |
| SVR | −0.0230 | 0.0984 | 0.3137 | 0.2736 | 0.006 |
| Index | Model | R2 | MSE | RMSE | MAE | Time/s |
|---|---|---|---|---|---|---|
| NH4+ | 1DCNN | 0.7429 | 0.0182 | 0.1351 | 0.1144 | 348.3281 |
| CAS-PLSR | 0.7489 | 0.0174 | 0.132 | 0.1075 | 18.3215 | |
| PLSR | 0.8428 | 0.0109 | 0.1045 | 0.0899 | 0.021 | |
| RF | 0.4890 | 0.0355 | 0.1883 | 0.1474 | 1.4893 | |
| RF-Lasso | 0.5416 | 0.0318 | 0.1784 | 0.1374 | 1.4782 | |
| SVM-MLP | 0.7324 | 0.0186 | 0.1363 | 0.1176 | 0.3458 | |
| SVR | −0.0027 | 0.0696 | 0.2638 | 0.225 | 0.0050 | |
| NO3− | 1DCNN | 0.6935 | 0.0295 | 0.1718 | 0.1513 | 315.3012 |
| CAS-PLSR | 0.6777 | 0.0310 | 0.1761 | 0.1481 | 18.5225 | |
| PLSR | 0.7243 | 0.0265 | 0.1629 | 0.1337 | 0.0324 | |
| RF | 0.4762 | 0.0504 | 0.2245 | 0.1839 | 1.3831 | |
| RF-Lasso | 0.4353 | 0.0543 | 0.2331 | 0.1948 | 1.3941 | |
| SVM-MLP | 0.5688 | 0.0415 | 0.2037 | 0.1673 | 0.3500 | |
| SVR | −0.0230 | 0.0984 | 0.3137 | 0.2736 | 0.0040 |
| Index | Model | R2 | MSE | RMSE | MAE | Time/s |
|---|---|---|---|---|---|---|
| NH4+ | 1DCNN | 0.7483 | 0.0175 | 0.1322 | 0.1121 | 288.6113 |
| CAS-PLSR | 0.8106 | 0.0131 | 0.1146 | 0.0979 | 11.602 | |
| PLSR | 0.7918 | 0.0145 | 0.1202 | 0.1011 | 0.0136 | |
| RF | 0.5815 | 0.029 | 0.1704 | 0.1369 | 0.6371 | |
| RF-Lasso | 0.5828 | 0.029 | 0.1702 | 0.1353 | 0.06743 | |
| SVM-MLP | 0.8726 | 0.0088 | 0.094 | 0.0754 | 0.2111 | |
| SVR | −0.0027 | 0.0696 | 0.2683 | 0.225 | 0.002 | |
| NO3− | 1DCNN | 0.7462 | 0.0244 | 0.1563 | 0.1346 | 310.2073 |
| CAS-PLSR | 0.6152 | 0.037 | 0.1924 | 0.1599 | 11.6084 | |
| PLSR | 0.7098 | 0.0279 | 0.1671 | 0.1376 | 0.0137 | |
| RF | 0.5836 | 0.0401 | 0.2002 | 0.1648 | 0.6357 | |
| RF-Lasso | 0.04879 | 0.0493 | 0.222 | 0.1859 | 0.632 | |
| SVM-MLP | 0.524 | 0.0458 | 0.214 | 0.1522 | 0.1488 | |
| SVR | −0.023 | 0.0984 | 0.3137 | 0.2736 | 0.002 |
| Index | Model | R2 | MSE | RMSE | MAE | Time/s |
|---|---|---|---|---|---|---|
| NH4+ | 1DCNN | 0.4484 | 0.0383 | 0.1957 | 0.1553 | 319.682 |
| CAS-PLSR | 0.7283 | 0.0189 | 0.1373 | 0.115 | 34.8485 | |
| PLSR | 0.7735 | 0.0157 | 0.1254 | 0.1089 | 0.0374 | |
| RF | 0.6678 | 0.0231 | 0.1518 | 0.1149 | 2.9428 | |
| RF-Lasso | 0.6186 | 0.0265 | 0.1627 | 0.1283 | 2.9298 | |
| SVM-MLP | 0.7315 | 0.0186 | 0.1365 | 0.1024 | 0.9335 | |
| SVR | −0.0027 | 0.0696 | 0.2638 | 0.225 | 0.0061 | |
| NO3− | 1DCNN | 0.6425 | 0.0344 | 0.1855 | 0.148 | 290.132 |
| CAS-PLSR | 0.6124 | 0.0373 | 0.1931 | 0.1442 | 36.8181 | |
| PLSR | 0.5753 | 0.0409 | 0.2021 | 0.1682 | 0.027 | |
| RF | 0.3639 | 0.0612 | 0.2474 | 0.1924 | 2.9295 | |
| RF-Lasso | 0.387 | 0.059 | 0.2429 | 0.1985 | 2.9928 | |
| SVM-MLP | 0.5263 | 0.0456 | 0.2135 | 0.1578 | 0.9122 | |
| SVR | −0.023 | 0.0984 | 0.3137 | 0.2736 | 0.005 |
| Index | Model | R2 | MSE | RMSE | MAE | Time/s |
|---|---|---|---|---|---|---|
| NH4+ | 1DCNN | 0.7564 | 0.0169 | 0.13 | 0.1049 | 309.2728 |
| CAS-PLSR | 0.7273 | 0.0189 | 0.1376 | 0.1127 | 18.9339 | |
| PLSR | 0.7447 | 0.0177 | 0.1331 | 0.1146 | 0.021 | |
| RF | 0.6902 | 0.0215 | 0.1467 | 0.1109 | 1.5722 | |
| RF-Lasso | 0.6414 | 0.0249 | 0.1578 | 0.1288 | 1.6452 | |
| SVM-MLP | 0.8263 | 0.0121 | 0.1098 | 0.0932 | 0.4056 | |
| SVR | −0.0027 | 0.0696 | 0.2638 | 0.225 | 0.004 | |
| NO3− | 1DCNN | 0.4781 | 0.0502 | 0.2241 | 0.1913 | 308.5333 |
| CAS-PLSR | 0.4469 | 0.0532 | 0.2307 | 0.1909 | 18.2978 | |
| PLSR | 0.5347 | 0.0448 | 0.2116 | 0.1748 | 0.0207 | |
| RF | 0.3689 | 0.0607 | 0.2464 | 0.1904 | 1.5886 | |
| RF-Lasso | 0.3953 | 0.0582 | 0.2412 | 0.1975 | 1.5744 | |
| SVM-MLP | 0.6068 | 0.0378 | 0.1945 | 0.1639 | 0.3851 | |
| SVR | −0.023 | 0.0984 | 0.3137 | 0.2736 | 0.003 |
| Index | Model | R2 | MSE | RMSE | MAE | Time/s |
|---|---|---|---|---|---|---|
| NH4+ | 1DCNN | 0.6851 | 0.0219 | 0.1478 | 0.094 | 303.1746 |
| CAS-PLSR | 0.6925 | 0.0213 | 0.1461 | 0.1171 | 11.4432 | |
| PLSR | 0.6566 | 0.0238 | 0.1544 | 0.01186 | 0.0141 | |
| RF | 0.6587 | 0.0237 | 0.1539 | 0.1189 | 0.7015 | |
| RF-Lasso | 0.6187 | 0.0265 | 0.1627 | 0.1334 | 0.6741 | |
| SVM-MLP | 0.7754 | 0.0156 | 0.1249 | 0.1018 | 0.165 | |
| SVR | −0.0026 | 0.0696 | 0.2638 | 0.225 | 0.002 | |
| NO3− | 1DCNN | 0.2561 | 0.0716 | 0.2675 | 0.2158 | 291.2573 |
| CAS-PLSR | 0.2989 | 0.0675 | 0.2597 | 0.2205 | 11.3699 | |
| PLSR | 0.2066 | 0.0763 | 0.02763 | 0.2128 | 0.013 | |
| RF | 0.3672 | 0.0609 | 0.2467 | 0.1917 | 0.7021 | |
| RF-Lasso | 0.3684 | 0.0608 | 0.2465 | 0.2012 | 0.6886 | |
| SVM-MLP | 0.6465 | 0.034 | 0.1844 | 0.1495 | 0.1513 | |
| SVR | −0.0229 | 0.0984 | 0.3137 | 0.2736 | 0.002 |
| Index | Model | R2 | MSE | RMSE | MAE | Time/s |
|---|---|---|---|---|---|---|
| NH4+ | 1DCNN | 0.78 | 0.0151 | 0.1227 | 0.1019 | 369.9942 |
| CAS-PLSR | 0.7282 | 0.0189 | 0.1373 | 0.1146 | 30.5211 | |
| PLSR | 0.8099 | 0.0132 | 0.1149 | 0.0937 | 0.024 | |
| RF | 0.4123 | 0.0408 | 0.202 | 0.1549 | 3.0994 | |
| RF-Lasso | 0.5009 | 0.0346 | 0.1861 | 0.1419 | 3.1997 | |
| SVM-ML | −6.7767 | 0.5398 | 0.7374 | 0.6131 | 0.1014 | |
| SVR | −0.0027 | 0.0696 | 0.2638 | 0.225 | 0.007 | |
| NO3− | 1DCNN | 0.63 | 0.0351 | 0.1875 | 0.1507 | 340.6888 |
| CAS-PLSR | 0.5594 | 0.0424 | 0.2059 | 0.1673 | 30.6061 | |
| PLSR | 0.7136 | 0.0276 | 0.166 | 0.1394 | 0.0202 | |
| RF | 0.504 | 0.0477 | 0.2185 | 0.1835 | 2.8402 | |
| RF-Lasso | 0.4655 | 0.0514 | 0.2268 | 0.1906 | 2.8933 | |
| SVM-ML | −9.9729 | 1.0558 | 1.0275 | 0.9029 | 0.101 | |
| SVR | −0.023 | 0.0984 | 0.3137 | 0.2736 | 0.0061 |
| Index | Model | R2 | MSE | RMSE | MAE | Time/s |
|---|---|---|---|---|---|---|
| NH4+ | 1DCNN | 0.56 | 0.0308 | 0.1756 | 0.1293 | 301.032 |
| CAS-PLSR | 0.7904 | 0.0145 | 0.1206 | 0.0911 | 18.7946 | |
| PLSR | 0.6973 | 0.021 | 0.145 | 0.1216 | 0.0231 | |
| RF | 0.4041 | 0.0414 | 0.2034 | 0.16 | 1.5359 | |
| RF-Lasso | 0.5128 | 0.0338 | 0.1839 | 0.1434 | 1.656 | |
| SVM-MLP | 0.5349 | 0.0323 | 0.1797 | 0.1611 | 0.1077 | |
| SVR | −0.0027 | 0.0696 | 0.2638 | 0.225 | 0.003 | |
| NO3− | 1DCNN | 0.6 | 0.0387 | 0.1969 | 0.1425 | 291.031 |
| CAS-PLSR | 0.5454 | 0.0437 | 0.2091 | 0.1647 | 18.9481 | |
| PLSR | 0.7173 | 0.0272 | 0.1649 | 0.1382 | 0.0204 | |
| RF | 0.494 | 0.0487 | 0.2206 | 0.1878 | 1.471 | |
| RF-Lasso | 0.4285 | 0.055 | 0.2345 | 0.1986 | 1.4995 | |
| SVM-MLP | 0.6391 | 0.0347 | 0.1863 | 0.1407 | 0.3935 | |
| SVR | −0.023 | 0.0984 | 0.3137 | 0.2736 | 0.003 |
| Index | Model | R2 | MSE | RMSE | MAE | Time/s |
|---|---|---|---|---|---|---|
| NH4+ | 1DCNN | 0.3768 | 0.0433 | 0.208 | 0.1571 | 294.1064 |
| CAS-PLSR | 0.797 | 0.0141 | 0.1187 | 0.0897 | 11.4601 | |
| PLSR | 0.644 | 0.0247 | 0.1572 | 0.1324 | 0.013 | |
| RF | 0.554 | 0.031 | 0.1759 | 0.1348 | 0.6708 | |
| RF-Lasso | 0.5527 | 0.031 | 0.1762 | 0.1387 | 0.6644 | |
| SVM-MLP | −0.2118 | 0.0841 | 0.29 | 0.2268 | 0.0301 | |
| SVR | −0.0026 | 0.0696 | 0.2638 | 0.225 | 0.002 | |
| NO3− | 1DCNN | 0.4931 | 0.0488 | 0.2209 | 0.1868 | 304.7842 |
| CAS-PLSR | 0.6332 | 0.0353 | 0.1879 | 0.141 | 11.3003 | |
| PLSR | 0.5668 | 0.0417 | 0.2042 | 0.1625 | 0.0131 | |
| RF | 0.4457 | 0.05323 | 0.2309 | 0.1877 | 0.634 | |
| RF-Lasso | 0.4087 | 0.0569 | 0.2385 | 0.1982 | 0.6793 | |
| SVM-MLP | 0.5193 | 0.0463 | 0.2151 | 0.1671 | 0.194 | |
| SVR | −0.023 | 0.0984 | 0.3137 | 0.2736 | 0.001 |
| Index | Model | R2 | MSE | RMSE | MAE | Time/s |
|---|---|---|---|---|---|---|
| NH4+ | 1DCNN | −0.0344 | 0.0718 | 0.268 | 0.2164 | 315.1378 |
| CAS-PLSR | 0.7315 | 0.0186 | 0.1365 | 0.1139 | 36.4751 | |
| PLSR | 0.7684 | 0.0161 | 0.1268 | 0.1095 | 0.038 | |
| RF | 0.693 | 0.0213 | 0.146 | 0.1115 | 3.0567 | |
| RF-Lasso | 0.4513 | 0.0381 | 0.1952 | 0.1672 | 2.9991 | |
| SVM-MLP | 0.7455 | 0.0177 | 0.1329 | 0.1008 | 0.9894 | |
| SVR | 0.7396 | 0.0181 | 0.1344 | 0.1137 | 0.005 | |
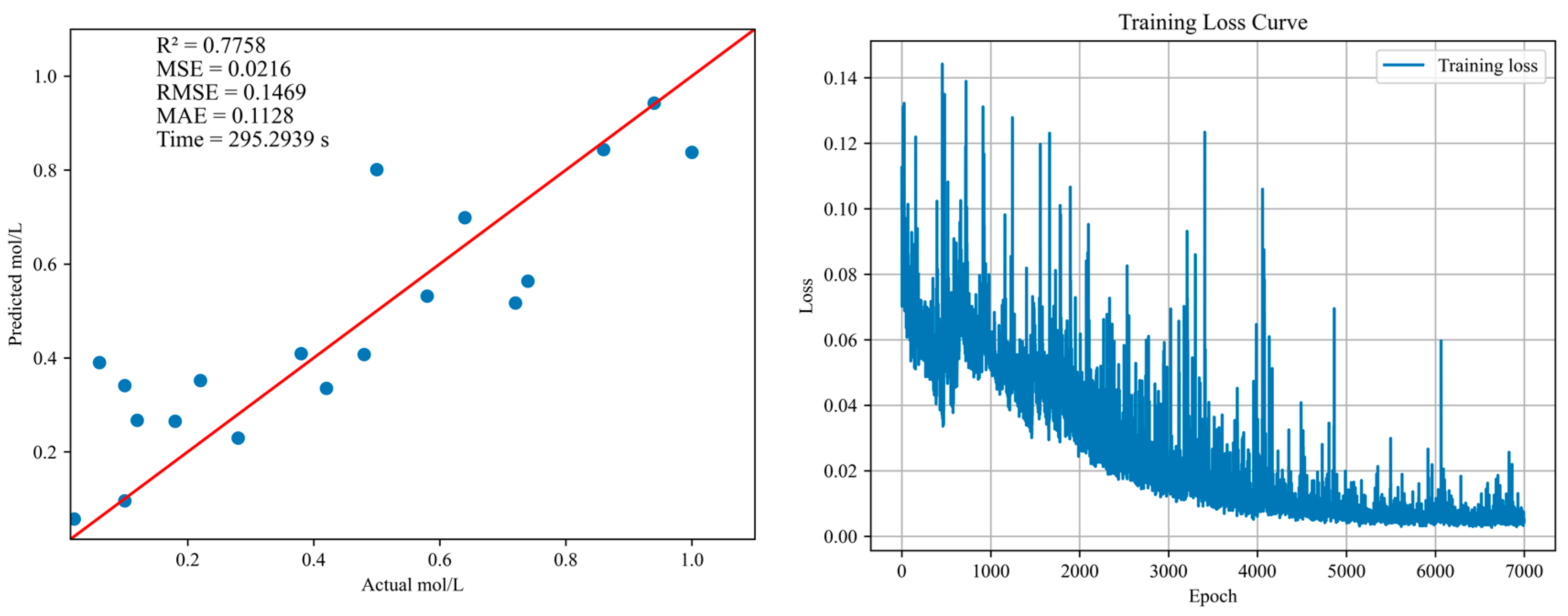
| NO3− | 1DCNN | 0.7758 | 0.0216 | 0.1469 | 0.1128 | 295.2939 |
| CAS-PLSR | 0.6027 | 0.0382 | 0.1955 | 0.1473 | 35.2799 | |
| PLSR | 0.5929 | 0.0392 | 0.1979 | 0.1659 | 0.0284 | |
| RF | 0.4095 | 0.0568 | 0.2384 | 0.1873 | 2.9513 | |
| RF-Lasso | 0.3039 | 0.067 | 0.2588 | 0.217 | 3.0252 | |
| SVM-MLP | 0.5616 | 0.0422 | 0.2054 | 0.1538 | 0.8683 | |
| SVR | 0.6754 | 0.0312 | 0.1767 | 0.1511 | 0.005 |
| Index | Model | R2 | MSE | RMSE | MAE | Time/s |
|---|---|---|---|---|---|---|
| NH4+ | 1DCNN | 0.4895 | 0.0354 | 0.1882 | 0.1544 | 307.1359 |
| CAS-PLSR | 0.7249 | 0.0191 | 0.1382 | 0.1132 | 18.8136 | |
| PLSR | 0.6645 | 0.0233 | 0.1526 | 0.1305 | 0.0213 | |
| RF | 0.6788 | 0.0223 | 0.1493 | 0.1155 | 1.5523 | |
| RF-Lasso | 0.4379 | 0.039 | 0.1975 | 0.1701 | 1.6108 | |
| SVM-MLP | −0.9531 | 0.1356 | 0.3682 | 0.2974 | 0.0565 | |
| SVR | 0.7303 | 0.0187 | 0.1368 | 0.1119 | 0.004 | |
| NO3− | 1DCNN | 0.2874 | 0.0686 | 0.2619 | 0.212 | 303.3181 |
| CAS-PLSR | 0.3982 | 0.0579 | 0.2406 | 0.2033 | 19.6413 | |
| PLSR | 0.4084 | 0.0569 | 0.2386 | 0.1893 | 0.0202 | |
| RF | 0.5002 | 0.0481 | 0.2193 | 0.1769 | 1.6467 | |
| RF-Lasso | 0.3471 | 0.0328 | 0.2506 | 0.2134 | 1.607 | |
| SVM-MLP | 0.5476 | 0.0435 | 0.2086 | 0.176 | 0.4082 | |
| SVR | 0.6394 | 0.0347 | 0.1863 | 0.1552 | 0.005 |
| Index | Model | R2 | MSE | RMSE | MAE | Time/s |
|---|---|---|---|---|---|---|
| NH4+ | 1DCNN | 0.6536 | 0.024 | 0.1551 | 0.1257 | 285.4631 |
| CAS-PLSR | 0.7509 | 0.0173 | 0.1315 | 0.1047 | 11.3829 | |
| PLSR | 0.6313 | 0.0256 | 0.16 | 0.1304 | 0.0242 | |
| RF | 0.6677 | 0.0231 | 0.1519 | 0.1155 | 0.7221 | |
| RF-Lasso | 0.4529 | 0.038 | 0.1949 | 0.1677 | 0.6825 | |
| SVM-MLP | 0.8876 | 0.00478 | 0.0883 | 0.0683 | 0.1869 | |
| SVR | 0.7736 | 0.0157 | 0.1254 | 0.1005 | 0.003 | |
| NO3− | 1DCNN | 0.5619 | 0.0422 | 0.2053 | 0.1745 | 303.5223 |
| CAS-PLSR | 0.2752 | 0.0697 | 0.2641 | 0.2195 | 11.2831 | |
| PLSR | 0.084 | 0.0881 | 0.2969 | 0.2385 | 0.0146 | |
| RF | 0.4171 | 0.0561 | 0.2368 | 0.1832 | 0.659 | |
| RF-Lasso | 0.3034 | 0.067 | 0.2589 | 0.2176 | 0.6553 | |
| SVM-MLP | 0.4668 | 0.0513 | 0.2265 | 0.1721 | 0.239 | |
| SVR | 0.5041 | 0.0477 | 0.2184 | 0.1887 | 0.003 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lu, H.; Zhou, H.; Cao, R.; Shi, D.; Xu, C.; Bai, F.; Han, Y.; Liu, S.; Wang, M.; Zhen, B. Design of Portable Water Quality Spectral Detector and Study on Nitrogen Estimation Model in Water. Processes 2025, 13, 3161. https://doi.org/10.3390/pr13103161
Lu H, Zhou H, Cao R, Shi D, Xu C, Bai F, Han Y, Liu S, Wang M, Zhen B. Design of Portable Water Quality Spectral Detector and Study on Nitrogen Estimation Model in Water. Processes. 2025; 13(10):3161. https://doi.org/10.3390/pr13103161
Chicago/Turabian StyleLu, Hongfei, Hao Zhou, Renyong Cao, Delin Shi, Chao Xu, Fangfang Bai, Yang Han, Song Liu, Minye Wang, and Bo Zhen. 2025. "Design of Portable Water Quality Spectral Detector and Study on Nitrogen Estimation Model in Water" Processes 13, no. 10: 3161. https://doi.org/10.3390/pr13103161
APA StyleLu, H., Zhou, H., Cao, R., Shi, D., Xu, C., Bai, F., Han, Y., Liu, S., Wang, M., & Zhen, B. (2025). Design of Portable Water Quality Spectral Detector and Study on Nitrogen Estimation Model in Water. Processes, 13(10), 3161. https://doi.org/10.3390/pr13103161







