Adaptive Lag Binning and Physics-Weighted Variograms: A LOOCV-Optimised Universal Kriging Framework with Trend Decomposition for High-Fidelity 3D Cryogenic Temperature Field Reconstruction
Abstract
1. Introduction
2. Related Works
2.1. Cryogenic Temperature Field Reconstruction Challenges
2.2. Kriging for Spatial Interpolation
2.3. Domain-Specific Kriging Adaptations
2.4. Lag-Binning Improvements
2.5. Research Gap and Contribution
3. Experimental Methodology
3.1. Adaptive Lag-Binning Strategies for Robust Empirical Variogram Calculation
3.1.1. The Baseline Algorithm: Equal-Distance Binning
3.1.2. Distribution-Adaptive Binning Strategies: Quantile-Based and Logarithmic-Based
- (1)
- Quantile-Based Binning and Cluster-Enhanced Hybridisation
- Initial Quantile Partitioning—Compute initial bin boundaries using the quantile-based method described above. This ensures a minimum count of point pairs per bin and provides a statistically stable starting point.
- Boundary Refinement via Clustering—Refine bin boundaries using k-means clustering in an augmented feature space defined as follows:
- (2)
- Logarithmic-Based Binning and Threshold-Controlled Hybridisation
3.1.3. Operations Research-Inspired Approaches
- (1)
- Greedy Binning with Residual Merging
- (2)
- Dynamic Programming-Based Binning Methods
- DP with Range Minimisation (Basic Formulation)
- DP with Adjusted Range Cost (Hybrid Objective)
- DP with Variance Minimisation (Statistical Precision)
DP Binning with Range Minimisation
DP Binning with Adjusted Range Cost
DP Binning with Variance Minimisation
3.2. Trend Modeling
3.3. Computational Implementation
- PyKrige 1.7.2 [25]: Core framework for universal kriging, including variogram estimation (e.g., adaptive lag binning) and spatial prediction;
- NumPy 2.0.1: Foundation for numerical operations (e.g., coordinate storage and distance calculations);
- Pandas 2.2.2: Manages structured data (e.g., reading time-series temperature data and storing optimisation results);
- SciPy 1.14.0: Supports trend fitting (nonlinear regression) and spatial distance calculations (e.g., pdist for point-pair distances);
- Joblib 1.4.2: Parallelizes batch processing (e.g., LOOCV for parameter optimisation) to reduce runtime by ~60%.
- binning_methods_latest.py: Adaptive lag binning (equal, greedy, logarithmic, dynamic programming);
- detrend_latest.py: Physics-guided trend decomposition (linear, quadratic, and exponential models);
- empirical_variogram_latest.py: Empirical variogram calculation with weighting schemes.
4. Setup
5. Results and Discussion
5.1. Binning Method Comparison: Structural and Statistical
5.2. Sensitivity Analysis
5.2.1. IDW Baseline Performance Analysis
5.2.2. Phase-Specific Method Preferences: Frequency Analysis
- (1)
- Binning Strategy Preferences: Structural vs. Statistical Trade-Offs
- Cool-Down Phase:
- Steady-State Phase:
- Warm-up Phase:
- (2)
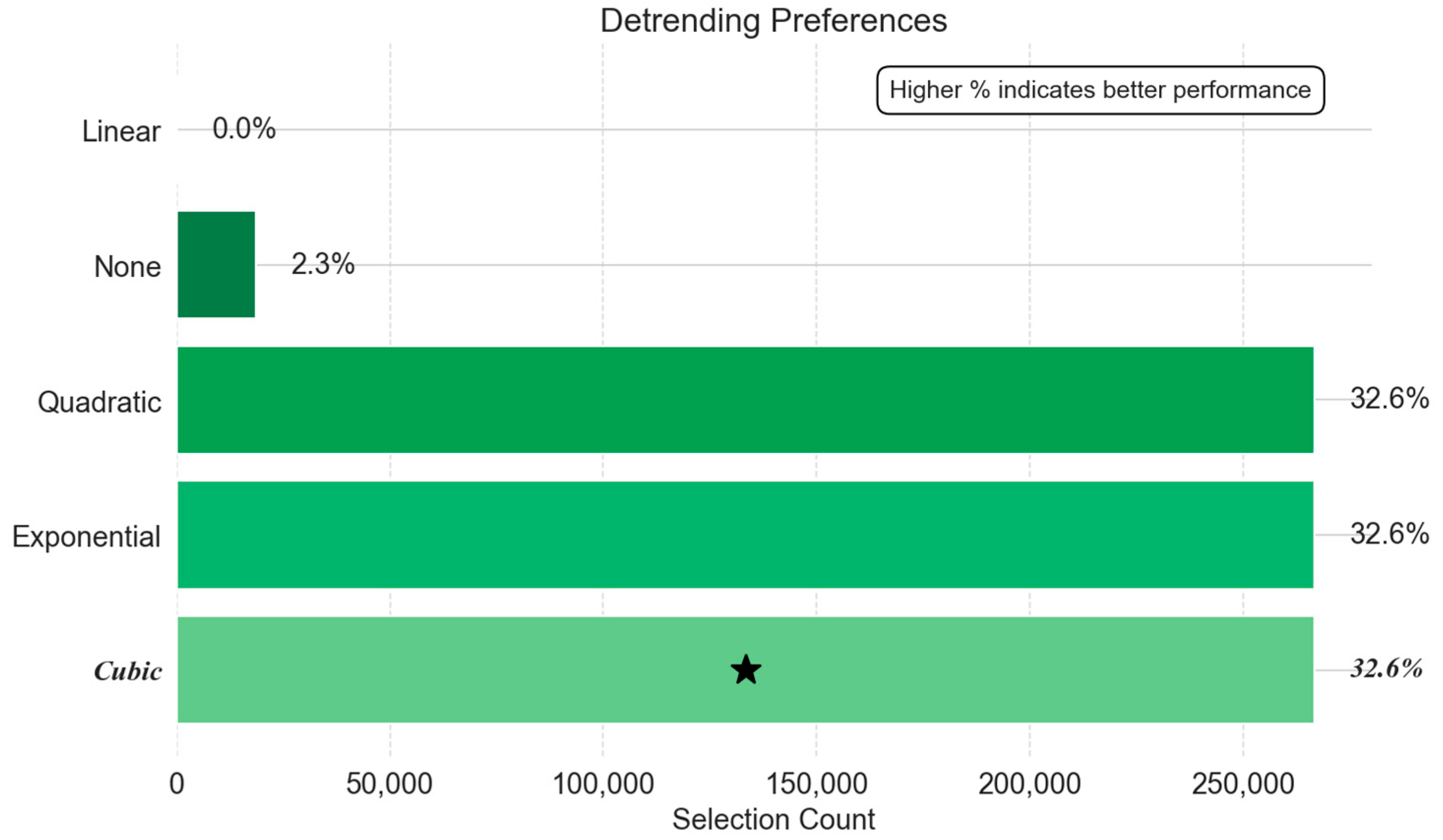
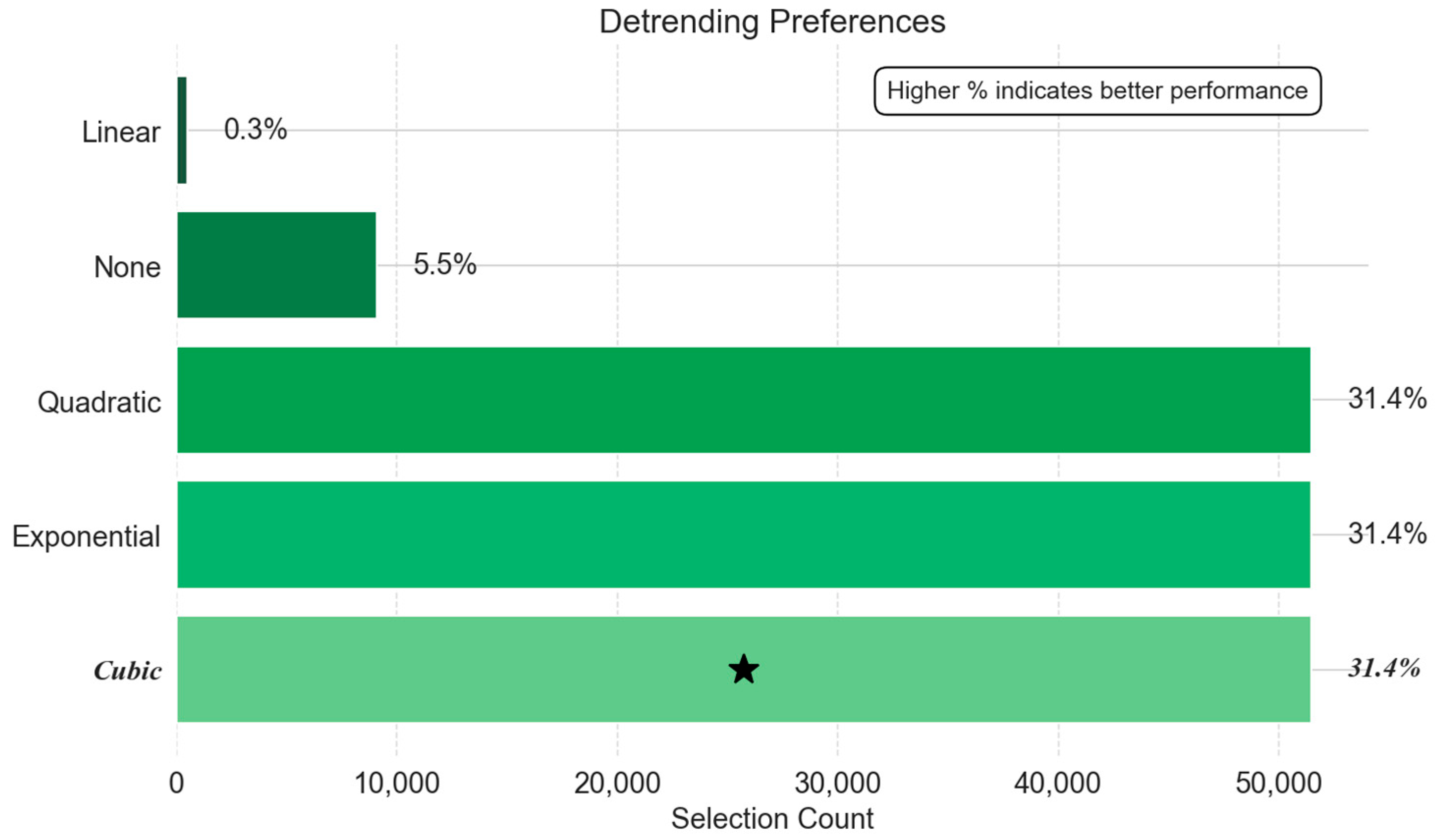
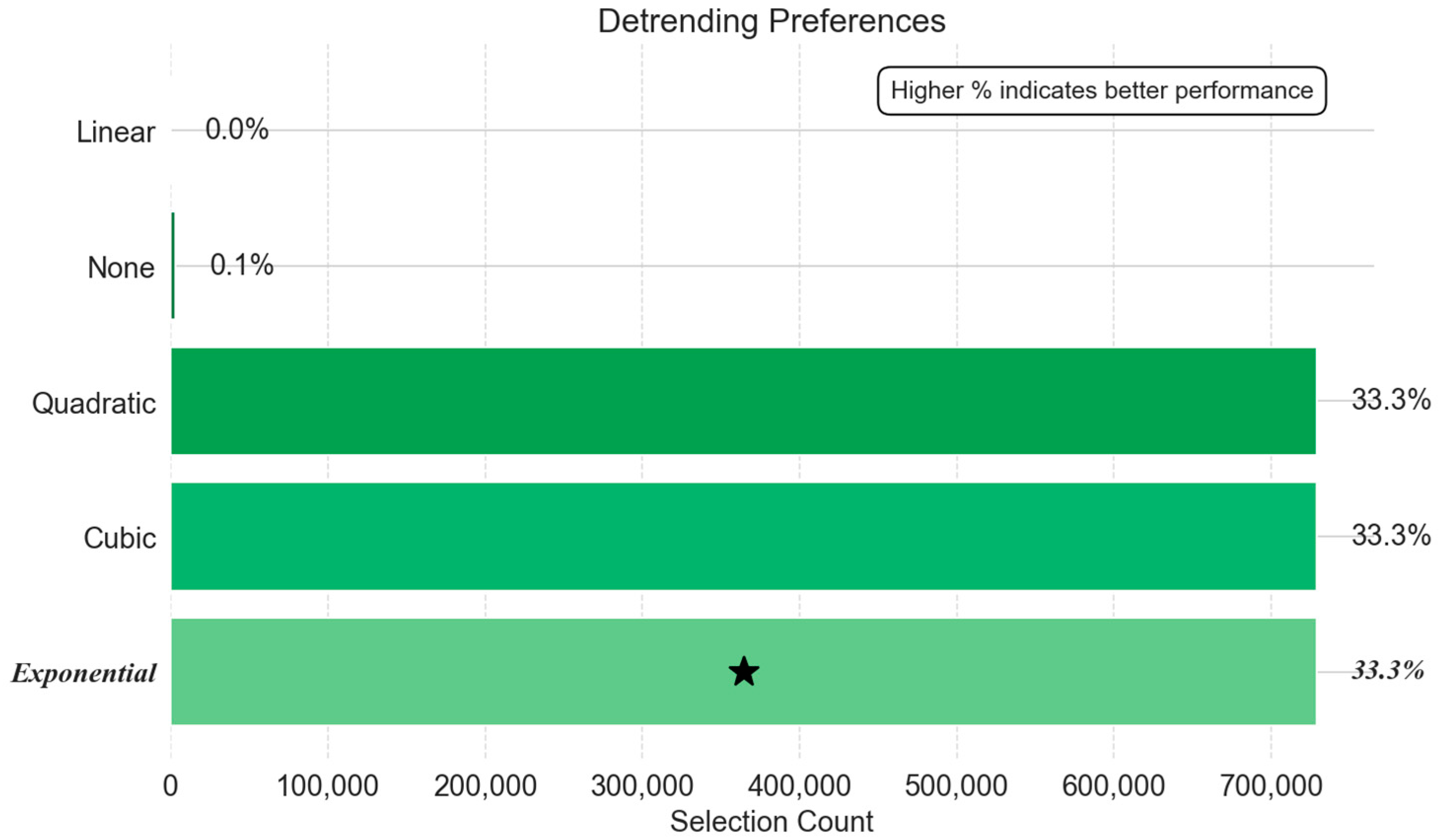
- Detrending Method Preferences
- (3)
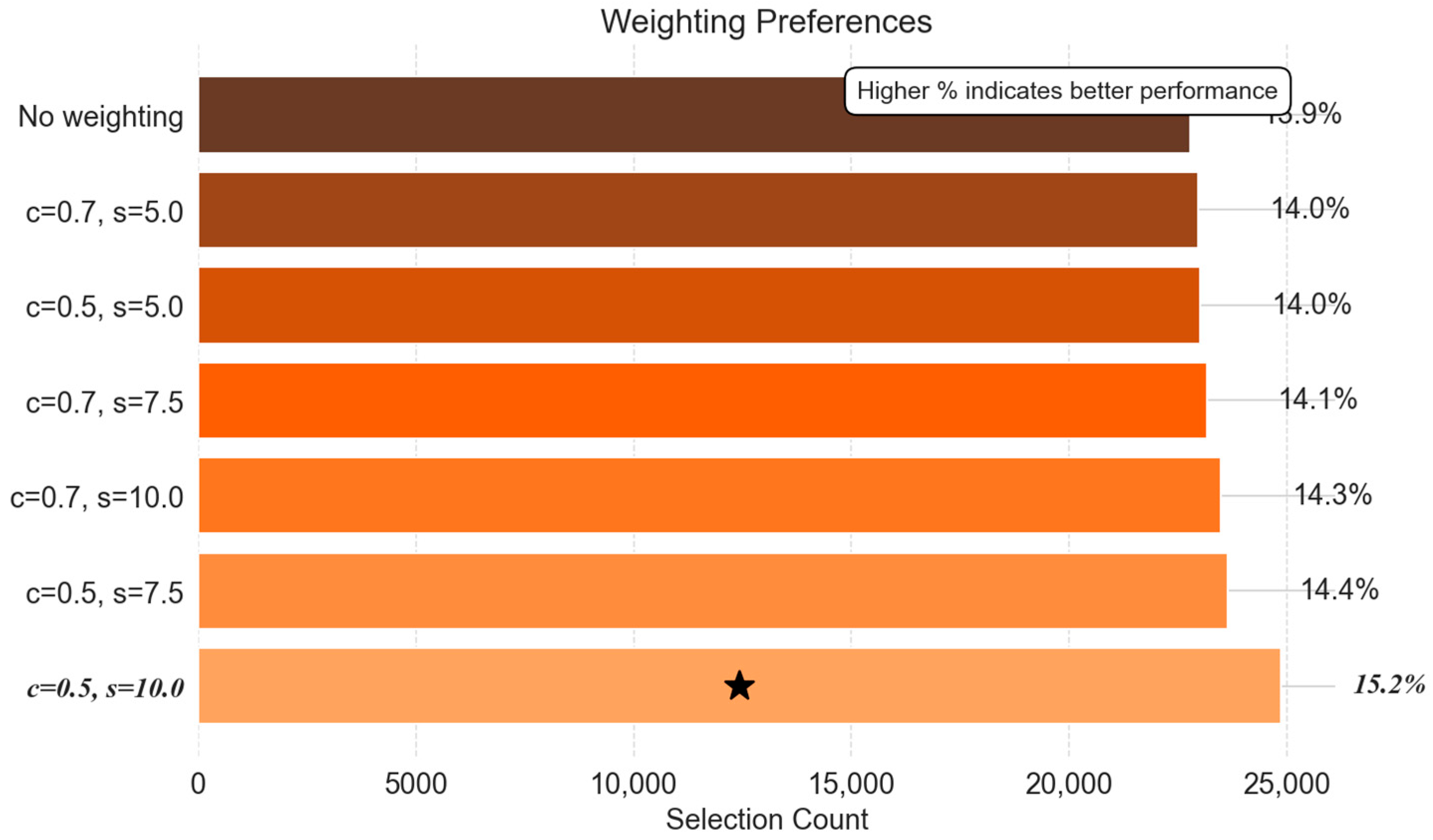
- Weighting Scheme Preferences: Spatial Sensitivity Control
- (4)
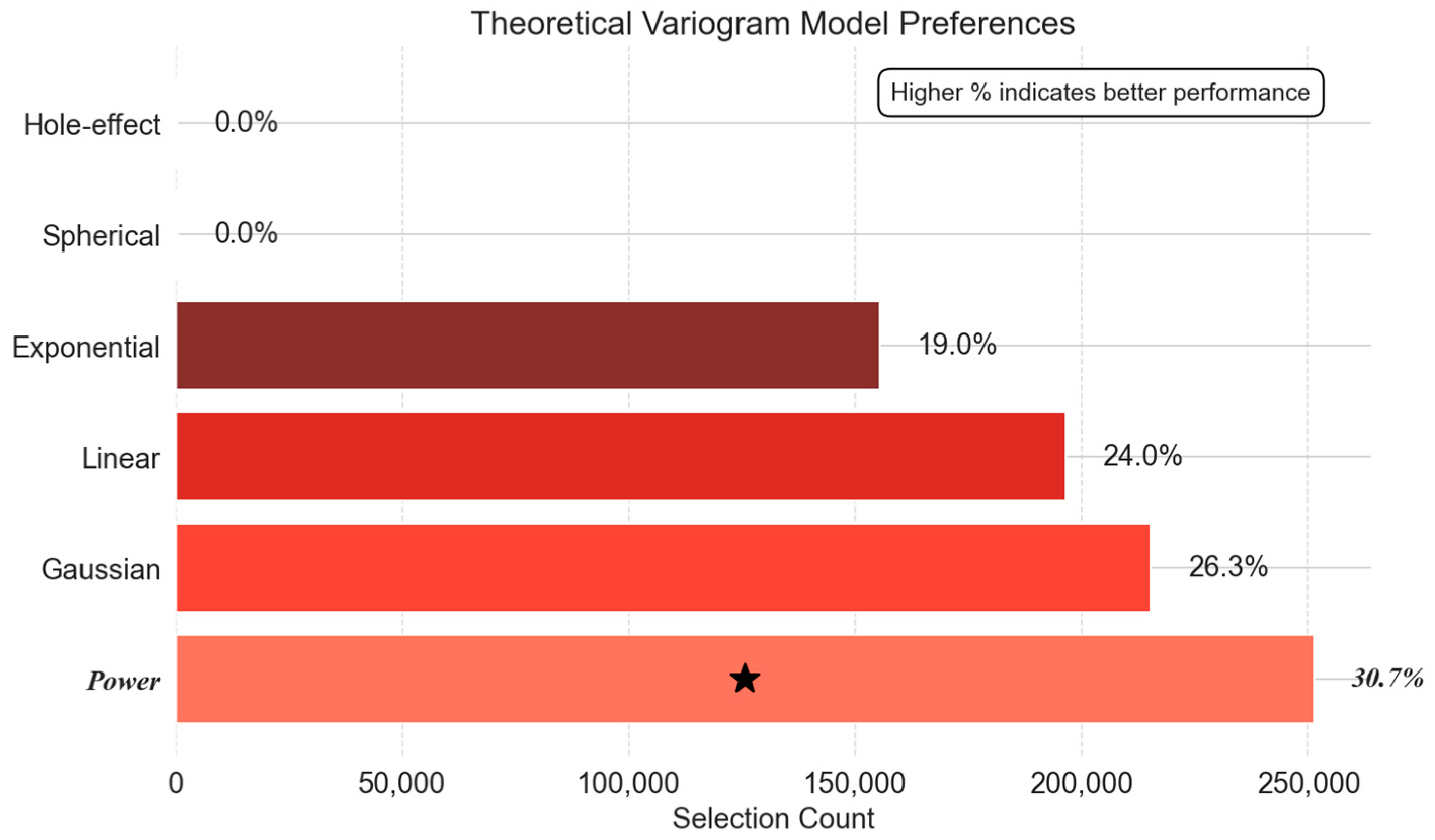
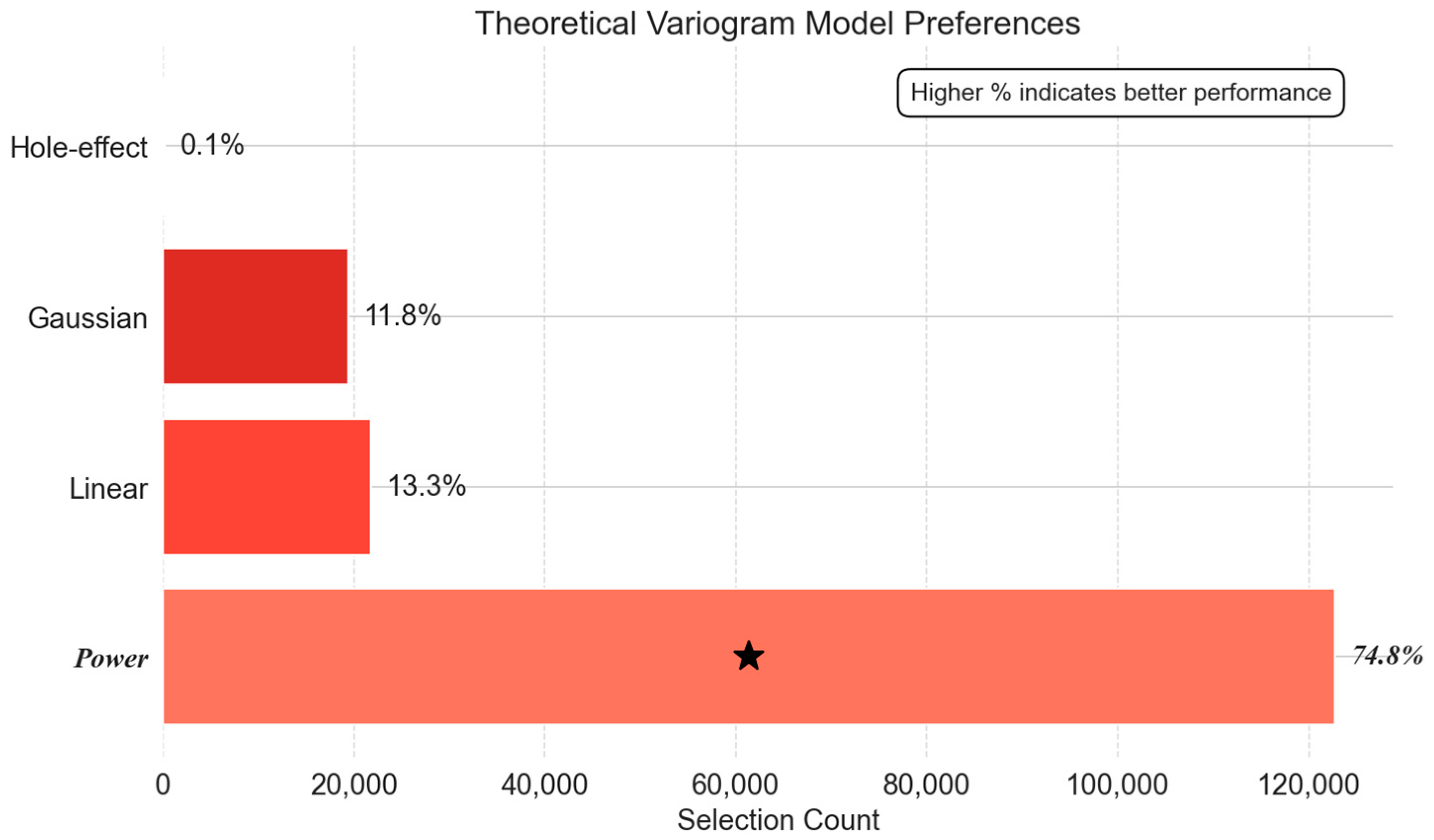
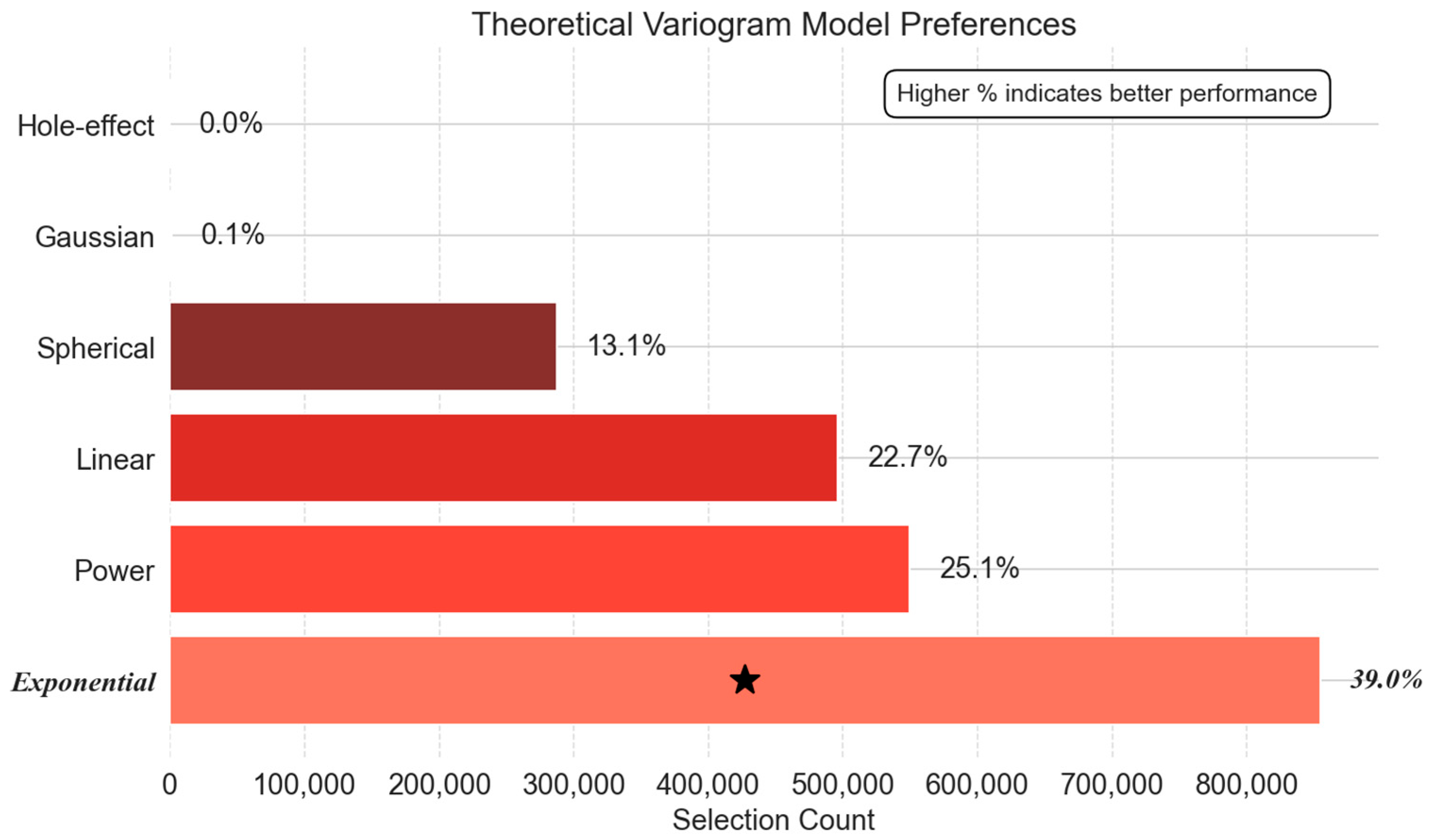
- Theoretical Variogram Model Preferences: Bridging Statistical and Physical Models
- Cooling-Down Phase:
- Steady-State Phase:
- Warm-Up Phase:
5.3. Interpolation Error Comparison and Statistical Significance
5.4. Summary of Key Findings
6. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hirtzlin, I.; Dubreuil, C.; Préaubert, N.; Duchier, J.; Jansen, B.; Simon, J.; Lobato De Faria, P.; Perez-Lezaun, A.; Visser, B.; Williams, G.D.; et al. An empirical survey on biobanking of human genetic material and data in six EU countries. Eur. J. Hum. Genet. EJHG 2003, 11, 475–488. [Google Scholar] [CrossRef] [PubMed]
- Saleh, J.; Mercier, B.; Xi, W. Bioengineering methods for organoid systems. Biol. Cell 2021, 113, 475–491. [Google Scholar] [CrossRef] [PubMed]
- Watson, P.H.; Nussbeck, S.Y.; Carter, C.; O’Donoghue, S.; Cheah, S.; Matzke, L.A.; Barnes, R.O.; Bartlett, J.; Carpenter, J.; Grizzle, W.E.; et al. A framework for biobank sustainability. Biopreserv. Biobank. 2014, 12, 60–68. [Google Scholar] [CrossRef]
- Babel, M.; Mamilos, A.; Seitz, S.; Niedermair, T.; Weber, F.; Anzeneder, T.; Ortmann, O.; Dietmaier, W.; Brochhausen, C. Compared DNA and RNA quality of breast cancer biobanking samples after long-term storage protocols in −80 °C and liquid nitrogen. Sci. Rep. 2020, 10, 14404. [Google Scholar] [CrossRef]
- Shabihkhani, M.; Lucey, G.M.; Wei, B.; Mareninov, S.; Lou, J.J.; Vinters, H.V.; Singer, E.J.; Cloughesy, T.F.; Yong, W.H. The procurement, storage, and quality assurance of frozen blood and tissue biospecimens in pathology, biorepository, and biobank settings. Clin. Biochem. 2014, 47, 258–266. [Google Scholar] [CrossRef] [PubMed]
- Weikert, J.; Mehrländer, A.; Baber, R. Keep cool! Observed temperature variations at different process stages of the biobanking workflow—Examples from the Leipzig medical biobank. J. Lab. Med. 2023, 47, 69–80. [Google Scholar] [CrossRef]
- Gunes, H.; Sirisup, S.; Karniadakis, G.E. Gappy data: To Krig or not to Krig? J. Comput. Phys. 2006, 212, 358–382. [Google Scholar] [CrossRef]
- Thaokar, C.; Rabin, Y. Temperature field reconstruction for minimally invasive cryosurgery with application to wireless implantable temperature sensors and/or medical imaging. Cryobiology 2012, 65, 270–277. [Google Scholar] [CrossRef]
- Powell, S.; Molinolo, A.; Masmila, E.; Kaushal, S. Real-Time Temperature Mapping in Ultra-Low Freezers as a Standard Quality Assessment. Biopreserv. Biobank. 2019, 17, 139–142. [Google Scholar] [CrossRef]
- Sun, Y.; Zhang, C.; Ji, H. A temperature field reconstruction method for spacecraft leading edge structure with optimized sensor array. J. Intell. Mater. Syst. Struct. 2021, 32, 2024–2038. [Google Scholar] [CrossRef]
- Ferrando, I.; De Rosa, P.; Federici, B.; Sguerso, D. Spatial interpolation techniques for a near real-time mapping of pressure and temperature data. PeerJ Prepr. 2016, 4, e2223v2. [Google Scholar] [CrossRef]
- Yao, Z.; Zhang, T.; Wu, L.; Wang, X.; Huang, J. Physics-Informed Deep Learning for Reconstruction of Spatial Missing Climate Information in the Antarctic. Atmosphere 2023, 14, 658. [Google Scholar] [CrossRef]
- Xiong, S. The Reconstruction Approach: From Interpolation to Regression. Technometrics 2021, 63, 225–235. [Google Scholar] [CrossRef]
- Zimmerman, D.; Pavlik, C.; Ruggles, A.; Armstrong, M.P. An Experimental Comparison of Ordinary and Universal Kriging and Inverse Distance Weighting. Math. Geol. 1999, 31, 375–390. [Google Scholar] [CrossRef]
- Wu, T.; Li, Y. Spatial interpolation of temperature in the United States using residual kriging. Appl. Geogr. 2013, 44, 112–120. [Google Scholar] [CrossRef]
- Li, S.; Griffith, D.A.; Shu, H. Temperature prediction based on a space-time regression-kriging model. J. Appl. Stat. 2020, 47, 1168–1190. [Google Scholar] [CrossRef]
- Yang, S.; Zhu, Z. Variance estimation and kriging prediction for a class of non-stationary spatial models. Stat. Sin. 2015, 25, 135–149. [Google Scholar] [CrossRef]
- Parrott, R.W.; Stytz, M.R.; Amburn, P.; Robinson, D. Statistically optimal interslice value interpolation in 3D medical imaging: Theory and implementation. In Proceedings of the Fifth Annual IEEE Symposium on Computer-Based Medical Systems, Durham, NC, USA, 14–17 June 1992; pp. 276–283. [Google Scholar] [CrossRef]
- Li, H.; Zhang, Q.; Pan, M.; Chen, D.; Yu, Z.; Xu, Y.; Ding, Z.; Liu, X.; Wan, K.; Dai, W. Enhancing Precision in Magnetic Map Interpolation for Regions with Sparse Data. Appl. Sci. 2024, 14, 756. [Google Scholar] [CrossRef]
- Yu, Z.; Wang, K.; Wu, J. Research on spatial interpolation of pile defects based on improved Kriging method. J. Ground Improv. 2023, 5, 376–382. [Google Scholar] [CrossRef]
- Cui, M.; Hou, E.; Lu, T.; Hou, P.; Feng, D. Study on Spatial Interpolation Methods for High Precision 3D Geological Modeling of Coal Mining Faces. Appl. Sci. 2025, 15, 2959. [Google Scholar] [CrossRef]
- Ng, F.; Hughes, A. Reconstructing ice-flow fields from streamlined subglacial bedforms: A kriging approach. Earth Surf. Process. Landf. 2018, 44, 861–876. [Google Scholar] [CrossRef]
- Jia, Y.; Deng, S.W.; Yao, X.M.; Cai, Y.F. Kriging interpolation algorithm based on constraint particle swarm optimization. J. Chengdu Univ. Technol. (Sci. Technol. Ed.) 2015, 42, 104–109. [Google Scholar] [CrossRef]
- Li, K.; Li, X.; Pang, C.; Zeng, X. Fast Reconstruction and Optimization of 3D Temperature Field Based-on Kriging Interpolation. In Proceedings of the 2021 3rd International Symposium on Smart and Healthy Cities (ISHC), Toronto, ON, Canada, 28–29 December 2021; pp. 198–203. [Google Scholar] [CrossRef]
- Murphy, B.; Yurchak, R.; Müller, S. GeoStat-Framework/PyKrige: v1.7.2, Version 1.7.2; Zenodo: Geneva, Switzerland, 2024.
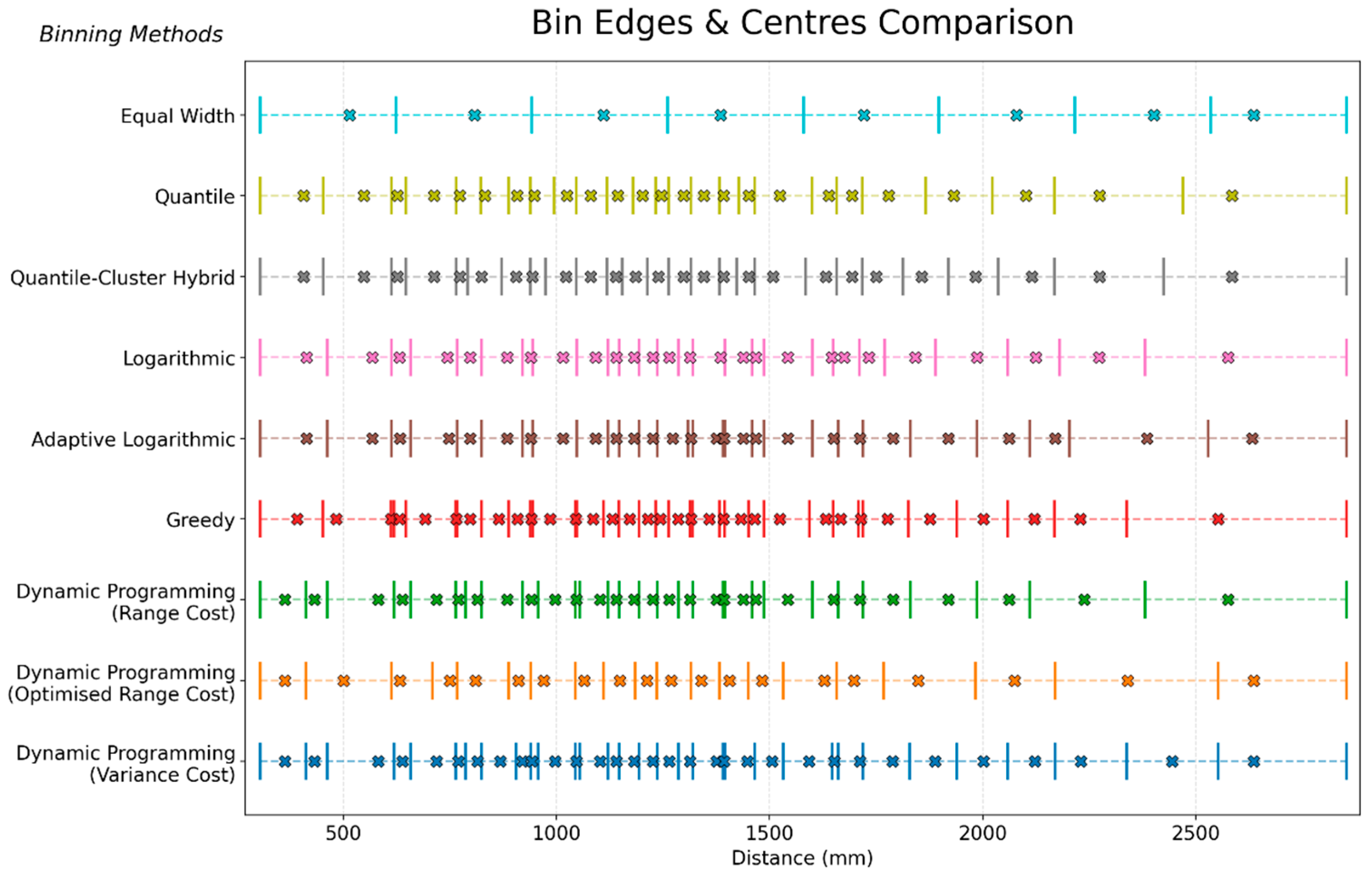
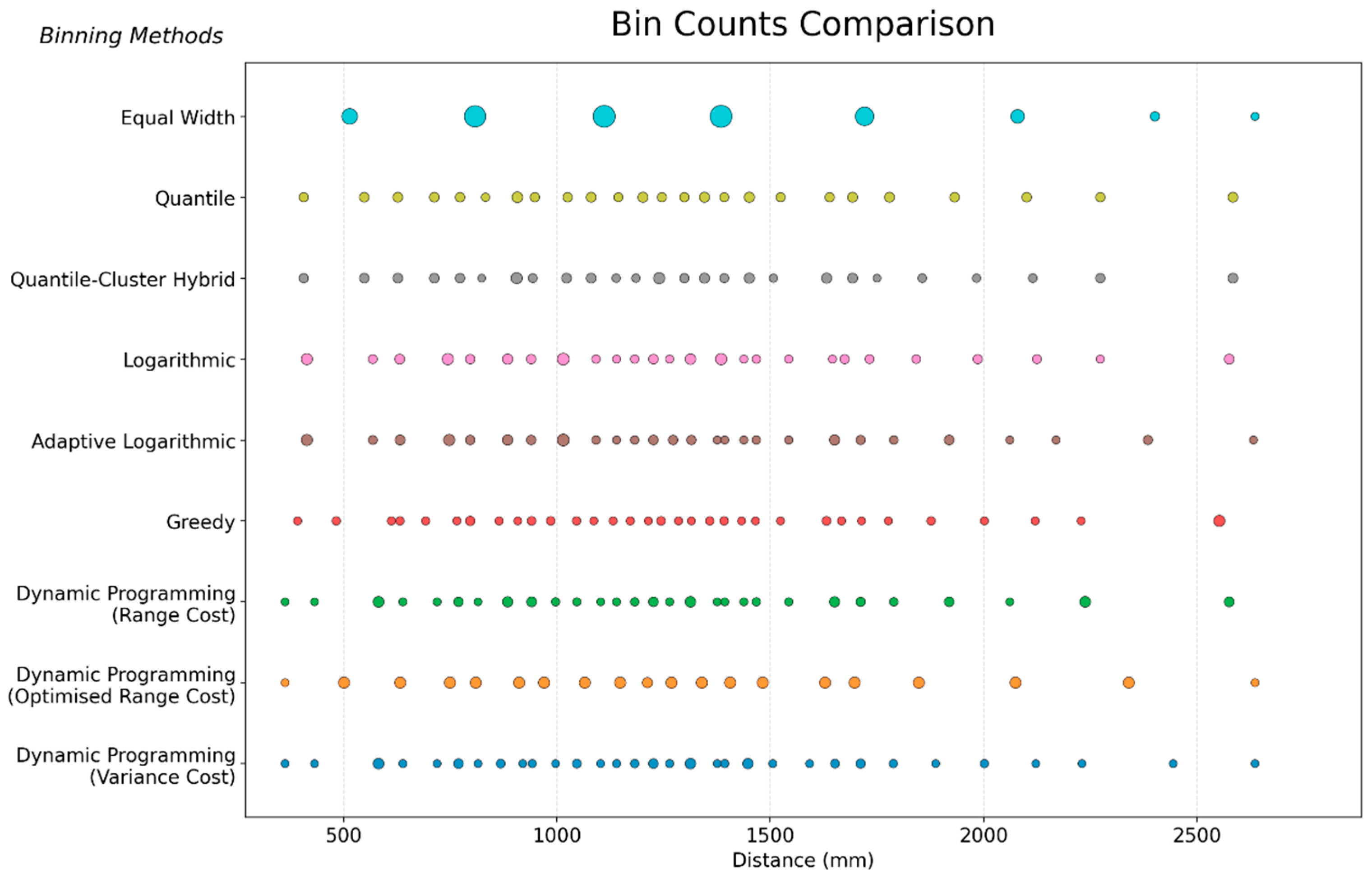
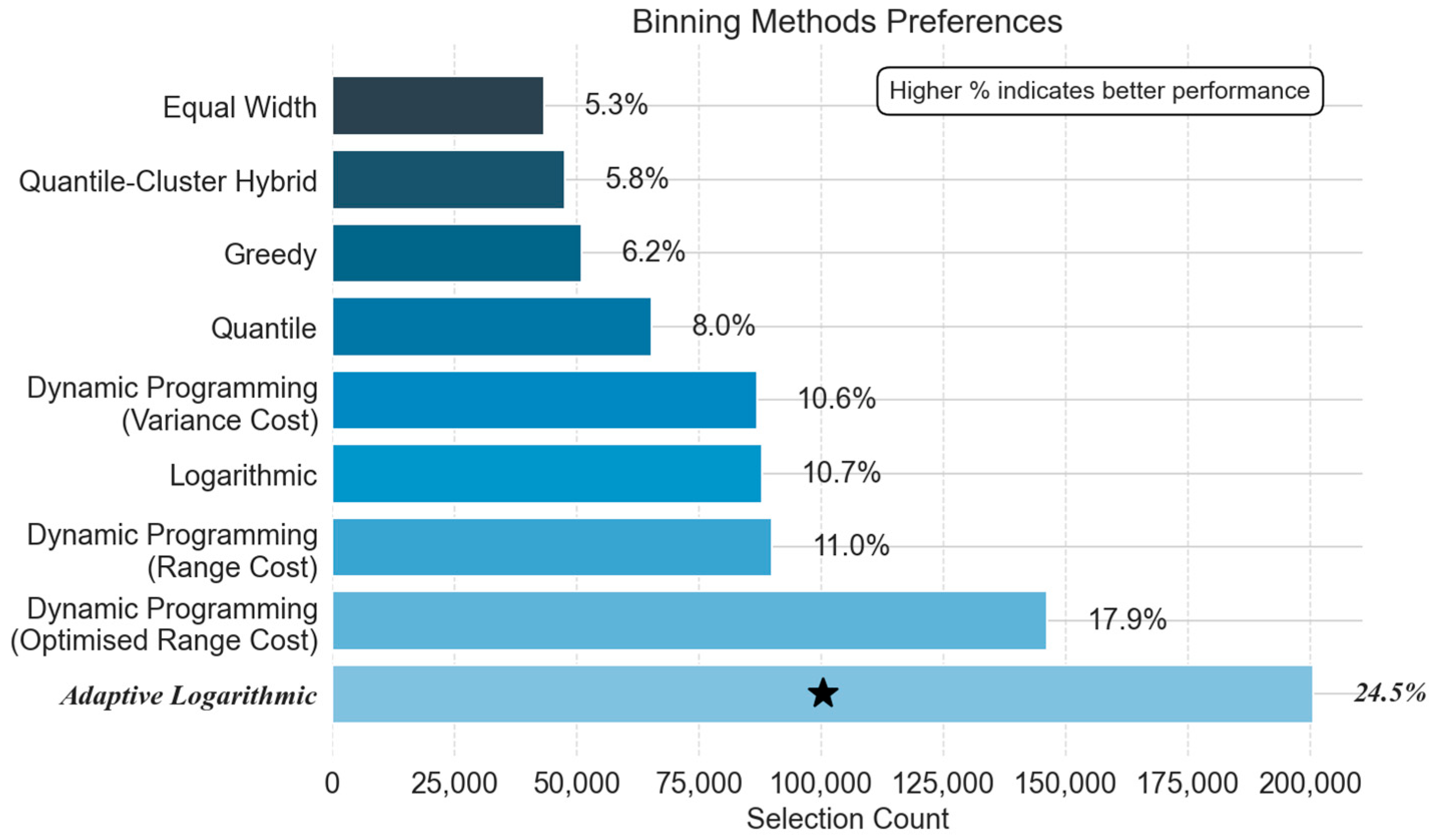

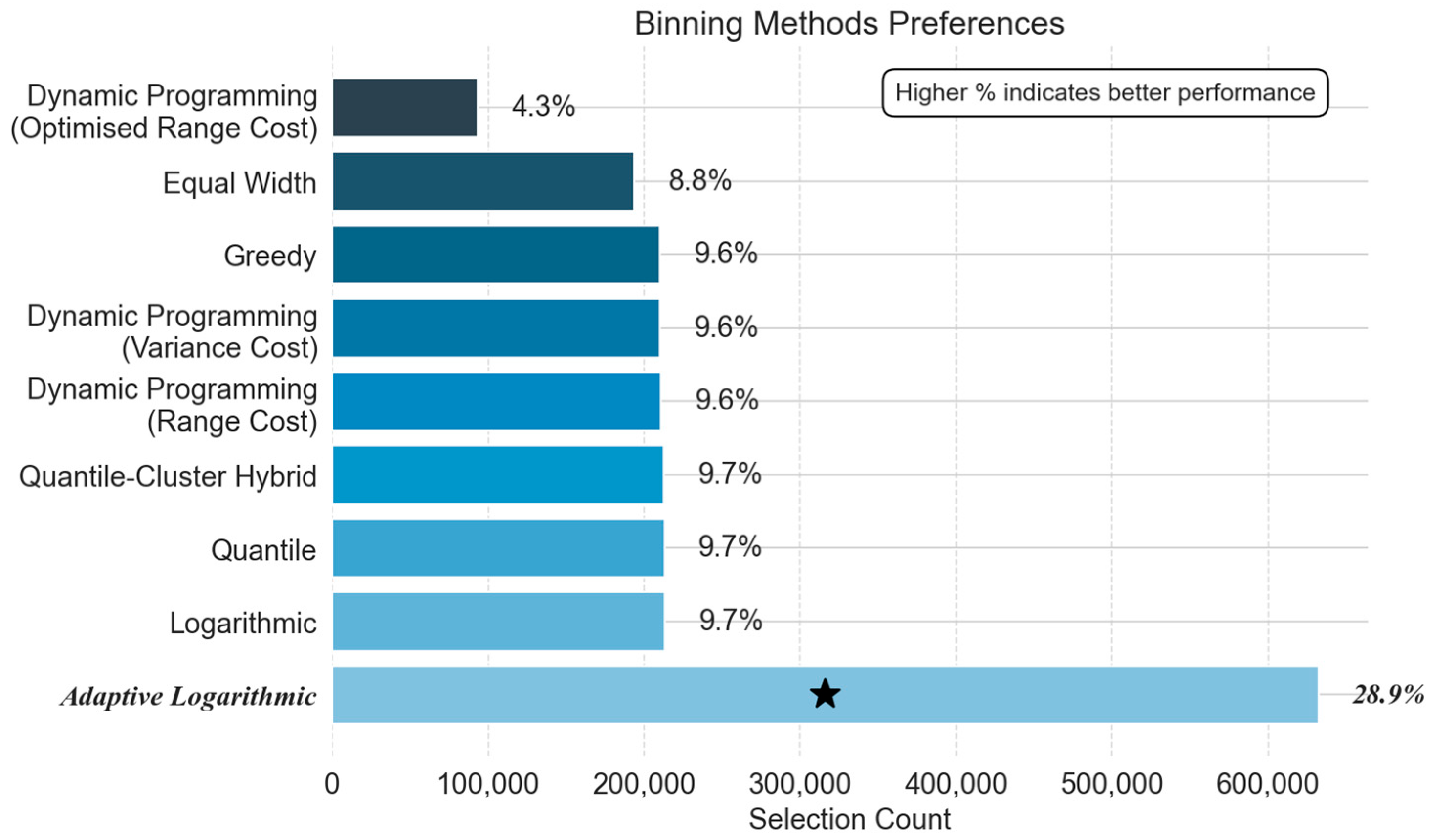


| Phase | Power Exponent (p) | Neighbours (k) | Average RMSE (°C) |
|---|---|---|---|
| Steady State | −0.6 | 3 | 1.179 |
| Cool-Down | −0.1 | 3 | 1.370 |
| Warm-Up | 5.5 | 4 | 0.936 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tang, J.; Chen, Y.; Liu, B.; Cao, J.; Wang, J. Adaptive Lag Binning and Physics-Weighted Variograms: A LOOCV-Optimised Universal Kriging Framework with Trend Decomposition for High-Fidelity 3D Cryogenic Temperature Field Reconstruction. Processes 2025, 13, 3160. https://doi.org/10.3390/pr13103160
Tang J, Chen Y, Liu B, Cao J, Wang J. Adaptive Lag Binning and Physics-Weighted Variograms: A LOOCV-Optimised Universal Kriging Framework with Trend Decomposition for High-Fidelity 3D Cryogenic Temperature Field Reconstruction. Processes. 2025; 13(10):3160. https://doi.org/10.3390/pr13103160
Chicago/Turabian StyleTang, Jiecheng, Yisha Chen, Baolin Liu, Jie Cao, and Jianxin Wang. 2025. "Adaptive Lag Binning and Physics-Weighted Variograms: A LOOCV-Optimised Universal Kriging Framework with Trend Decomposition for High-Fidelity 3D Cryogenic Temperature Field Reconstruction" Processes 13, no. 10: 3160. https://doi.org/10.3390/pr13103160
APA StyleTang, J., Chen, Y., Liu, B., Cao, J., & Wang, J. (2025). Adaptive Lag Binning and Physics-Weighted Variograms: A LOOCV-Optimised Universal Kriging Framework with Trend Decomposition for High-Fidelity 3D Cryogenic Temperature Field Reconstruction. Processes, 13(10), 3160. https://doi.org/10.3390/pr13103160






