Utilizing Machine Learning Models with Molecular Fingerprints and Chemical Structures to Predict the Sulfate Radical Rate Constants of Water Contaminants
Abstract
1. Introduction
2. Data Collection and Model Construction
2.1. Data Sets and MF
2.2. Hyperparameter Optimization
2.3. ML Models and Validation
2.4. Model Interpretation
3. Results and Discussion
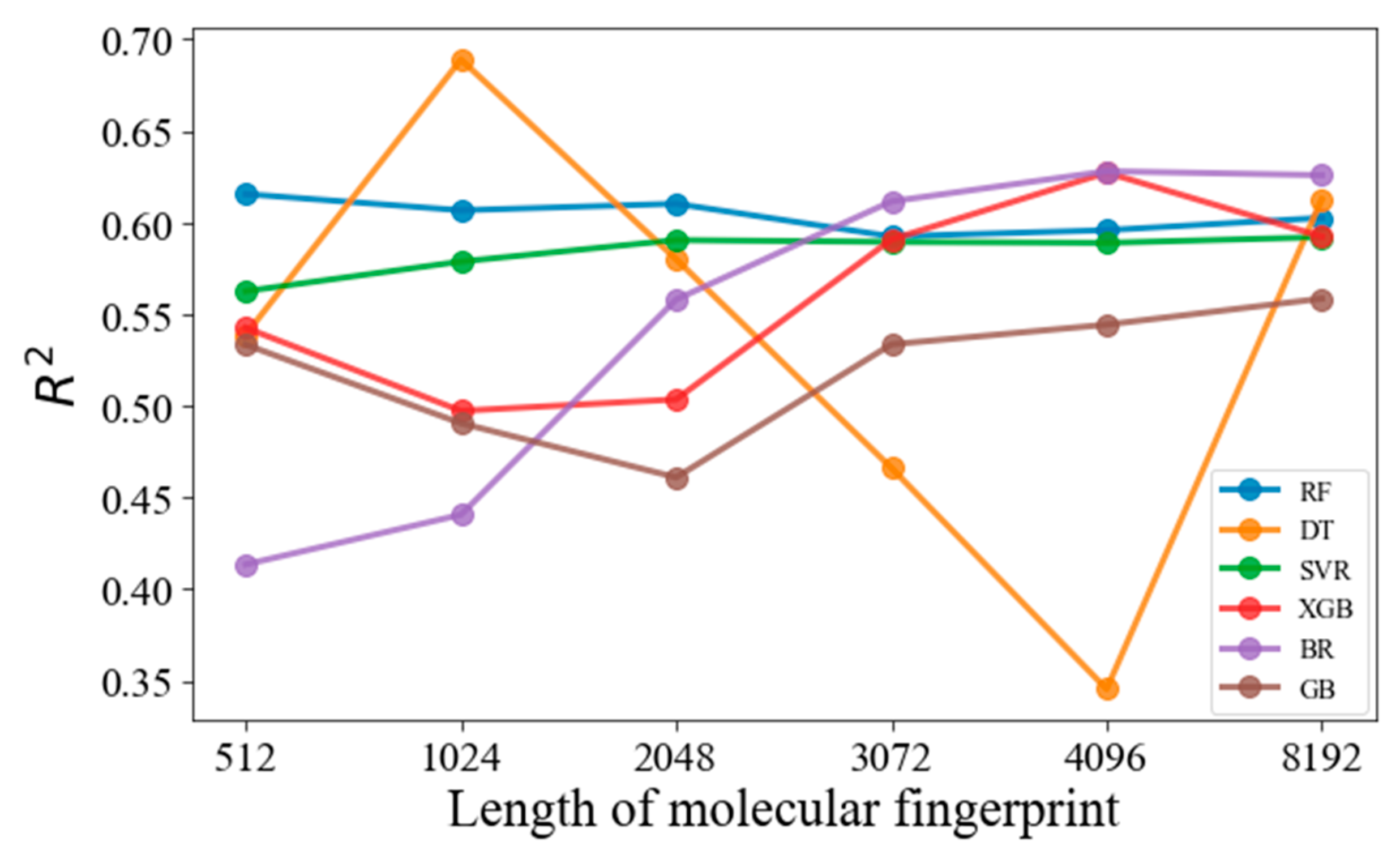
3.1. Effects of the MF Length
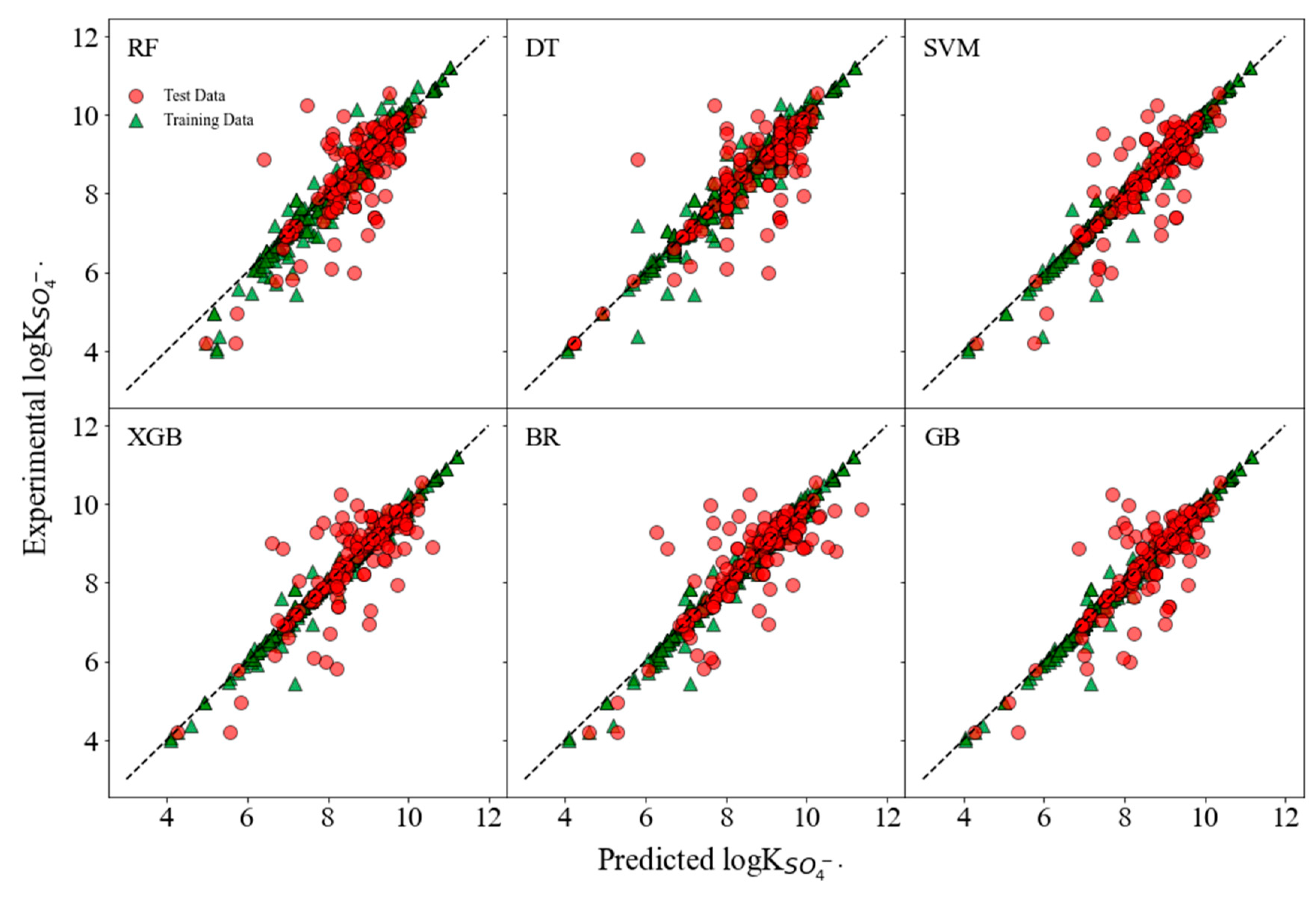
3.2. Internal and External Validation
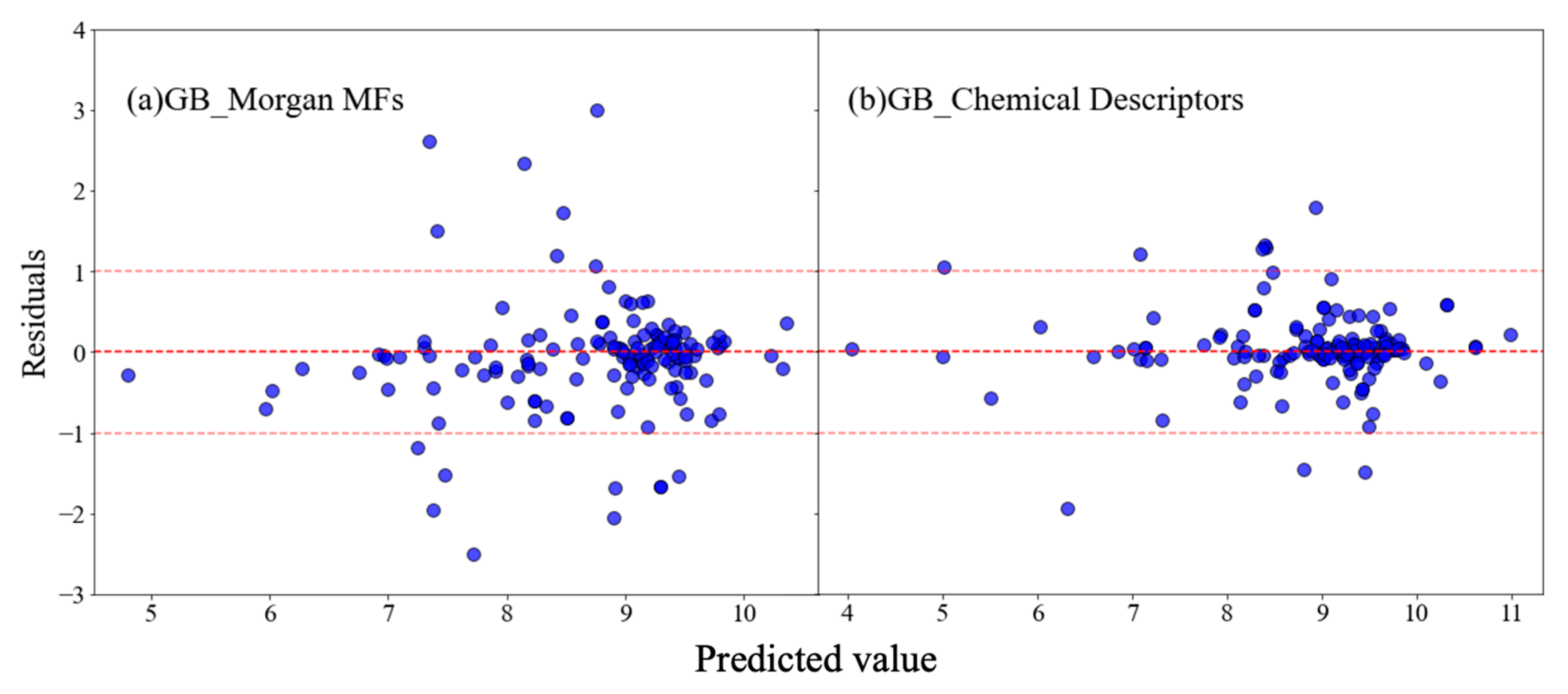
3.3. SHapley Additive exPlanations (SHAP) Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Giannakis, S.; Lin, K.-Y.A.; Ghanbari, F. A review of the recent advances on the treatment of industrial wastewaters by Sulfate Radical-based Advanced Oxidation Processes (SR-AOPs). Chem. Eng. J. 2021, 406, 127083. [Google Scholar] [CrossRef]
- Hassani, A.; Scaria, J.; Ghanbari, F.; Nidheesh, P. Sulfate radicals-based advanced oxidation processes for the degradation of pharmaceuticals and personal care products: A review on relevant activation mechanisms, performance, and perspectives. Environ. Res. 2023, 217, 114789. [Google Scholar] [CrossRef] [PubMed]
- Lian, L.; Yao, B.; Hou, S.; Fang, J.; Yan, S.; Song, W. Kinetic study of hydroxyl and sulfate radical-mediated oxidation of pharmaceuticals in wastewater effluents. Environ. Sci. Technol. 2017, 51, 2954–2962. [Google Scholar] [CrossRef] [PubMed]
- Nfodzo, P.; Choi, H. Sulfate radicals destroy pharmaceuticals and personal care products. Environ. Eng. Sci. 2011, 28, 605–609. [Google Scholar] [CrossRef]
- Li, W.; Orozco, R.; Camargos, N.; Liu, H. Mechanisms on the impacts of alkalinity, pH, and chloride on persulfate-based groundwater remediation. Environ. Sci. Technol. 2017, 51, 3948–3959. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Ren, X.; Hu, L.; Guo, W.; Zhan, J. Facet-controlled activation of persulfate by goethite for tetracycline degradation in aqueous solution. Chem. Eng. J. 2021, 412, 128628. [Google Scholar] [CrossRef]
- Ji, Y.; Wang, L.; Jiang, M.; Lu, J.; Ferronato, C.; Chovelon, J.-M. The role of nitrite in sulfate radical-based degradation of phenolic compounds: An unexpected nitration process relevant to groundwater remediation by in-situ chemical oxidation (ISCO). Water Res. 2017, 123, 249–257. [Google Scholar] [CrossRef]
- Lai, J.; Tang, T.; Du, X.; Wang, R.; Liang, J.; Song, D.; Dang, Z.; Lu, G. Oxidation of 1, 3-diphenylguanidine (DPG) by goethite activated persulfate: Mechanisms, products identification and reaction sites prediction. Environ. Res. 2023, 232, 116308. [Google Scholar] [CrossRef]
- Pardue, H.L. Kinetic aspects of analytical chemistry. Anal. Chim. Acta 1989, 216, 69–107. [Google Scholar] [CrossRef]
- Neta, P.; Madhavan, V.; Zemel, H.; Fessenden, R.W. Rate constants and mechanism of reaction of sulfate radical anion with aromatic compounds. J. Am. Chem. Soc. 1977, 99, 163–164. [Google Scholar] [CrossRef]
- Tran, L.N.; Abellar, K.A.; Cope, J.D.; Nguyen, T.B. Second-Order Kinetic Rate Coefficients for the Aqueous-Phase Sulfate Radical (SO4•–) Oxidation of Some Atmospherically Relevant Organic Compounds. J. Phys. Chem. A 2022, 126, 6517–6525. [Google Scholar] [CrossRef] [PubMed]
- Nirmalakhandan, N.N.; Speece, R.E. QSAR model for predicting Henry’s constant. Environ. Sci. Technol. 1988, 22, 1349–1357. [Google Scholar] [CrossRef]
- Agrawal, V.; Khadikar, P. QSAR prediction of toxicity of nitrobenzenes. Bioorg. Med. Chem. 2001, 9, 3035–3040. [Google Scholar] [CrossRef] [PubMed]
- Du, Q.-S.; Huang, R.-B.; Chou, K.-C. Recent advances in QSAR and their applications in predicting the activities of chemical molecules, peptides and proteins for drug design. Curr. Protein Pept. Sci. 2008, 9, 248–259. [Google Scholar] [CrossRef]
- Xiao, R.; Ye, T.; Wei, Z.; Luo, S.; Yang, Z.; Spinney, R. Quantitative Structure-Activity Relationship (QSAR) for the Oxidation of Trace Organic Contaminants by Sulfate Radical. Environ. Sci. Technol. 2015, 49, 13394–13402. [Google Scholar] [CrossRef]
- Sudhakaran, S.; Amy, G.L. QSAR models for oxidation of organic micropollutants in water based on ozone and hydroxyl radical rate constants and their chemical classification. Water Res. 2013, 47, 1111–1122. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Belle, J.H.; Meng, X.; Wildani, A.; Waller, L.A.; Strickland, M.J.; Liu, Y. Estimating PM2. 5 concentrations in the conterminous United States using the random forest approach. Environ. Sci. Technol. 2017, 51, 6936–6944. [Google Scholar] [CrossRef]
- Gupta, P.; Christopher, S.A. Particulate matter air quality assessment using integrated surface, satellite, and meteorological products: Multiple regression approach. J. Geophys. Res. Atmos. 2009, 114, D20205. [Google Scholar] [CrossRef]
- Zhu, J.-J.; Anderson, P.R. Performance evaluation of the ISMLR package for predicting the next day’s influent wastewater flowrate at Kirie WRP. Water Sci. Technol. 2019, 80, 695–706. [Google Scholar] [CrossRef]
- Haimi, H.; Mulas, M.; Corona, F.; Vahala, R. Data-derived soft-sensors for biological wastewater treatment plants: An overview. Environ. Model. Softw. 2013, 47, 88–107. [Google Scholar] [CrossRef]
- Lu, J.; Zhang, H.; Yu, J.; Shan, D.; Qi, J.; Chen, J.; Song, H.; Yang, M. Predicting Rate Constants of Hydroxyl Radical Reactions with Alkanes Using Machine Learning. J. Chem. Inf. Model. 2021, 61, 4259–4265. [Google Scholar] [CrossRef]
- Cheng, W.; Ng, C.A. Using Machine Learning to Classify Bioactivity for 3486 Per- and Polyfluoroalkyl Substances (PFASs) from the OECD List. Environ. Sci. Technol. 2019, 53, 13970–13980. [Google Scholar] [CrossRef]
- Kavzoglu, T.; Teke, A. Predictive Performances of ensemble machine learning algorithms in landslide susceptibility mapping using random forest, extreme gradient boosting (XGBoost) and natural gradient boosting (NGBoost). Arab. J. Sci. Eng. 2022, 47, 7367–7385. [Google Scholar] [CrossRef]
- Yin, G.; Jameel Ibrahim Alazzawi, F.; Mironov, S.; Reegu, F.; El-Shafay, A.S.; Lutfor Rahman, M.; Su, C.-H.; Lu, Y.-Z.; Chinh Nguyen, H. Machine learning method for simulation of adsorption separation: Comparisons of model’s performance in predicting equilibrium concentrations. Arab. J. Chem. 2022, 15, 103612. [Google Scholar] [CrossRef]
- Ding Han, H.j. Prediction of Second-Order Rate Constants of Sulfate Radical with Aromatic Contaminants Using Quantitative Structure-Activity Relationship Mode. Water 2021, 14, 766. [Google Scholar] [CrossRef]
- Sanches-Neto, F.O.; Dias-Silva, J.R.; Keng Queiroz Junior, L.H.; Carvalho-Silva, V.H. “pySiRC”: Machine Learning Combined with Molecular Fingerprints to Predict the Reaction Rate Constant of the Radical-Based Oxidation Processes of Aqueous Organic Contaminants. Environ. Sci. Technol. 2021, 55, 12437–12448. [Google Scholar] [CrossRef]
- Fabio Mercado, D.; Bracco, L.L.B.; Argues, A.; Gonzalez, M.C.; Caregnato, P. Reaction kinetics and mechanisms of organosilicon fungicide flusilazole with sulfate and hydroxyl radicals. Chemosphere 2018, 190, 327–336. [Google Scholar] [CrossRef] [PubMed]
- Gabet, A.; Metivier, H.; Brauer, C.d.; Mailhot, G.; Brigante, M. Hydrogen peroxide and persulfate activation using UVA-UVB radiation: Degradation of estrogenic compounds and application in sewage treatment plant waters. J. Hazard. Mater. 2021, 405, 124693. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Shao, Y.; Gao, N.; Lu, X.; An, N. Degradation of diethyl phthalate (DEP) by UV/persulfate: An experiment and simulation study of contributions by hydroxyl and sulfate radicals. Chemosphere 2018, 193, 602–610. [Google Scholar] [CrossRef] [PubMed]
- Rickman, K.A.; Mezyk, S.P. Kinetics and mechanisms of sulfate radical oxidation of β-lactam antibiotics in water. Chemosphere 2010, 81, 359–365. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.; Basant, N. Modeling the reactivity of ozone and sulphate radicals towards organic chemicals in water using machine learning approaches. RSC Adv. 2016, 6, 108448–108457. [Google Scholar] [CrossRef]
- Real, F.J.; Acero, J.L.; Benitez, J.F.; Roldan, G.; Casas, F. Oxidation of the emerging contaminants amitriptyline hydrochloride, methyl salicylate and 2-phenoxyethanol by persulfate activated by UV irradiation. J. Chem. Technol. Biotechnol. 2016, 91, 1004–1011. [Google Scholar] [CrossRef]
- Cvetnic, M.; Stankov, M.N.; Kovacic, M.; Ukic, S.; Bolanca, T.; Kusic, H.; Rasulev, B.; Dionysiou, D.D.; Loncaric-Bozic, A. Key structural features promoting radical driven degradation of emerging contaminants in water. Environ. Int. 2019, 124, 38–48. [Google Scholar] [CrossRef]
- Cvetnic, M.; Tomic, A.; Sigurnjak, M.; Stankov, M.N.; Ukic, S.; Kusic, H.; Bolanca, T.; Bozic, A.L. Structural features of contaminants of emerging concern behind empirical parameters of mechanistic models describing their photooxidative degradation. J. Water Process Eng. 2020, 33, 101053. [Google Scholar] [CrossRef]
- Shi, Y.; Yan, F.; Jia, Q.; Wang, Q. Normindex for predicting the rate constants of organic contaminants oxygenated with sulfate radical. Environ. Sci. Pollut. Res. 2020, 27, 974–982. [Google Scholar] [CrossRef]
- Wojnárovits, L.; Takács, E. Rate constants of sulfate radical anion reactions with organic molecules: A review. Chemosphere 2019, 220, 1014–1032. [Google Scholar] [CrossRef] [PubMed]
- RDKit: Open-Source Cheminformatics. Available online: http://www.rdkit.org (accessed on 10 February 2024).
- Bouwmeester, R.; Martens, L.; Degroeve, S. Comprehensive and Empirical Evaluation of Machine Learning Algorithms for Small Molecule LC Retention Time Prediction. Anal. Chem. 2019, 91, 3694–3703. [Google Scholar] [CrossRef]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V. Scikit-learn: Machine learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Hodson, T.O. Root-mean-square error (RMSE) or mean absolute error (MAE): When to use them or not. Geosci. Model Dev. 2022, 15, 5481–5487. [Google Scholar] [CrossRef]
- OECD. Guidance Document on the Validation of (Quantitative) Structure-Activity Relationship [(Q) SAR] Models; Organisation for Economic Co-Operation and Development: Paris, France, 2014. [Google Scholar]
- Lundberg, S.; Lee, S. A Unified Approach to Interpreting Model Predictions. ArXiv 2017, arXiv:1705.07874. [Google Scholar]
- Lundberg, S.M.; Erion, G.; Chen, H.; DeGrave, A.; Prutkin, J.M.; Nair, B.; Katz, R.; Himmelfarb, J.; Bansal, N.; Lee, S.-I. From local explanations to global understanding with explainable AI for trees. Nat. Mach. Intell. 2020, 2, 56–67. [Google Scholar] [CrossRef] [PubMed]
- Bourel, M.; Cugliari, J.; Goude, Y.; Poggi, J.-M. Boosting diversity in regression ensembles. Stat. Anal. Data Min. ASA Data Sci. J. 2020, 1–17. [Google Scholar] [CrossRef]
- Odegua, R. An empirical study of ensemble techniques (bagging, boosting and stacking). In Proceedings of the Deep Learning IndabaX. 2019. Available online: https://www.researchgate.net/publication/338681864_An_Empirical_Study_of_Ensemble_Techniques_Bagging_Boosting_and_Stacking (accessed on 10 February 2024).
- Cai, J.; Xu, K.; Zhu, Y.; Hu, F.; Li, L. Prediction and analysis of net ecosystem carbon exchange based on gradient boosting regression and random forest. Appl. Energy 2020, 262, 114566. [Google Scholar] [CrossRef]
- Mosavi, A.; Sajedi Hosseini, F.; Choubin, B.; Goodarzi, M.; Dineva, A.A.; Rafiei Sardooi, E. Ensemble Boosting and Bagging Based Machine Learning Models for Groundwater Potential Prediction. Water Resour. Manag. 2021, 35, 23–37. [Google Scholar] [CrossRef]
- Boldini, D.; Grisoni, F.; Kuhn, D.; Friedrich, L.; Sieber, S.A. Practical guidelines for the use of gradient boosting for molecular property prediction. J. Cheminform. 2023, 15, 73. [Google Scholar] [CrossRef]


| Models | Hyperparameters | ||
|---|---|---|---|
| Range | Molecular Fingerprints | Chemical Descriptors | |
| RF | n_estimators:rage (1, 200, 20) max_depth: range (2, 40, 4) | n_estimators = 21 max_depth = 26 | n_estimators = 40 max_depth = 20 |
| DT | max_depth: range (1, 100, 10) min_samples_split: range (1, 10, 2) min_samples_leaf: range (1, 10, 2) | max_depth = 15 min_samples_split = 6 min_samples_leaf = 1 | max_depth = 60 min_samples_split = 2 min_samples_leaf = 4 |
| SVM | kernel: [‘linear’,‘rbf’] C:range (1, 10, 2) gamma: [0.005, 0.01, 0.05] | Kernel = rbf, C = 5 gamma = 0.05 | Kernel = rbf, C = 4 gamma = 0.005 |
| XGB | n_estimators: rage (10, 300, 50) min_child_weight: range (1, 5) max_depth: range (20, 100, 10) | n_estimators = 60 min_child_weight = 4 max_depth = 70 | n_estimators = 200 min_child_weight = 4 max_depth = 20 |
| GB | n_estimators:range (20, 100, 20) learning_rate’: [0.01, 0.05, 0.1] max_depth’:range (2, 10, 2) | n_estimators = 80 learning_rate = 0.1 max_depth = 8 | n_estimators = 80 learning_rate = 0.1 max_depth = 6 |
| BR | alpha_1: [1 × 10−6, 1 × 10−7, 1 × 10−8] lambda_1: [1 × 10−4, 5 × 10−3, 1 × 10−3] n_iter: [1, 30, 2] | alpha_1 = 1 × 10−8 lambda_1 = 0.005 n_iter = 20 | alpha_1 = 1 × 10−7 lambda_1 = 0.005 n_iter = 1 |
| Morgan (3072 Bits) | ||||||
|---|---|---|---|---|---|---|
| Models | Training Set | Test Set | ||||
| R2 | MAE | RMSE | R2 | MAE | RMSE | |
| RF | 0.949 | 0.380 | 0.609 | 0.577 | 0.474 | 0.751 |
| DT | 0.922 | 0.416 | 0.633 | 0.532 | 0.500 | 0.790 |
| SVM | 0.974 | 0.340 | 0.547 | 0.712 | 0.379 | 0.620 |
| XGB | 0.988 | 0.338 | 0.600 | 0.641 | 0.404 | 0.691 |
| BR | 0.982 | 0.392 | 0.645 | 0.597 | 0.449 | 0.732 |
| GB | 0.972 | 0.348 | 0.583 | 0.620 | 0.429 | 0.712 |
| Chemical Descriptors | ||||||
|---|---|---|---|---|---|---|
| Models | Training Set | Test Set | ||||
| R2 | MAE | RMSE | R2 | MAE | RMSE | |
| RF | 0.957 | 0.357 | 0.597 | 0.806 | 0.345 | 0.519 |
| DT | 0.955 | 0.429 | 0.759 | 0.724 | 0.366 | 0.618 |
| SVM | 0.984 | 0.570 | 0.849 | 0.540 | 0.496 | 0.799 |
| XGB | 0.994 | 0.346 | 0.651 | 0.788 | 0.309 | 0.542 |
| BR | 0.776 | 0.526 | 0.775 | 0.663 | 0.491 | 0.684 |
| GB | 0.992 | 0.329 | 0.594 | 0.844 | 0.275 | 0.465 |
| Feature Name | Description [37] |
|---|---|
| SMR_VSA7 | MOE-type descriptors using MR contributions and surface area contributions |
| Chi3v | Third-order valence molecular connectivity index for topology analysis |
| Balaban J | A topological descriptor that quantifies the branching and complexity of cycles within a molecule’s structure |
| BCUT2D_MRHI | Burden eigenvalues (BCUT) calculated from a 2D molecular graph, related to the highest eigenvalue of the molecular refractivity-weighted connectivity matrix |
| Slogp_VSA6 | Log P partition coefficient, used to understand hydrophobic or lipophilic properties of the molecule |
| BCUT2D_MRLOW | The lowest eigenvalue of a molecule’s Burden connectivity matrix |
| Slogp_VSA1 | Volume descriptor for logP partition, smallest value bin |
| VSA_EState5 | Distribution of electronic properties over the molecular surface |
| Fpdensitymorgan3 | Density of features in a Morgan fingerprint with a radius of 3 |
| RingCount | Total number of ring structures present in a molecule |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tang, T.; Song, D.; Chen, J.; Chen, Z.; Du, Y.; Dang, Z.; Lu, G. Utilizing Machine Learning Models with Molecular Fingerprints and Chemical Structures to Predict the Sulfate Radical Rate Constants of Water Contaminants. Processes 2024, 12, 384. https://doi.org/10.3390/pr12020384
Tang T, Song D, Chen J, Chen Z, Du Y, Dang Z, Lu G. Utilizing Machine Learning Models with Molecular Fingerprints and Chemical Structures to Predict the Sulfate Radical Rate Constants of Water Contaminants. Processes. 2024; 12(2):384. https://doi.org/10.3390/pr12020384
Chicago/Turabian StyleTang, Ting, Dehao Song, Jinfan Chen, Zhenguo Chen, Yufan Du, Zhi Dang, and Guining Lu. 2024. "Utilizing Machine Learning Models with Molecular Fingerprints and Chemical Structures to Predict the Sulfate Radical Rate Constants of Water Contaminants" Processes 12, no. 2: 384. https://doi.org/10.3390/pr12020384
APA StyleTang, T., Song, D., Chen, J., Chen, Z., Du, Y., Dang, Z., & Lu, G. (2024). Utilizing Machine Learning Models with Molecular Fingerprints and Chemical Structures to Predict the Sulfate Radical Rate Constants of Water Contaminants. Processes, 12(2), 384. https://doi.org/10.3390/pr12020384








