Abstract
Specifically designed co-solvent mixtures are an efficient way to enhance the kinetics of enzyme-catalyzed reactions without compromising enzyme stability; among them, several deep eutectic solvents have emerged as exciting co-solvent mixtures for biocatalytic reactions. DESs nature allows one to tailor the enzyme-co-solvent interactions by using DESs constituents of diverse functional groups. In this work, the influence of co-solvents (betaine, glycerol, and sorbitol) and two DESs (betaine:glycerol and betaine:sorbitol) on the kinetics of candida boidinii Formate dehydrogenase was evaluated. The results showed a 30% increase in catalytic efficiency by adding 15 wt.-% of betaine to the buffered aqueous reaction media. Further, cbFDH folded-state stability was evaluated using differential scanning fluorimetry to finally obtain the binding affinity, unfolding curves, and thermodynamic unfolding parameters. The addition of glycerol, sorbitol, and DESs increased cbFDH protection against thermal stress, and this effect could be improved by increasing co-solvent concentrations. Moreover, DESs showed the ability to reduce the irreversibility of the unfolding process. Betaine was the only co-solvent that had a negative stability effect, which was offset by using betaine-based DESs. The latter was a result of the additivity of certain individual co-solvent effects on thermal stability. Non-monotonous stability effects were obtained by adding sorbitol to the buffer solutions, probably because hydrogen bond dynamics between cbFDH/co-solvent/water change dramatically with the amount of water present. Finally, DESs improved NAD binding affinity with cbFDH interestingly without direct correlation with the results obtained for kinetics.
1. Introduction
Liquid systems are pivotal in various industrial processes across fields such as chemistry, biotechnology, food, and pharmaceuticals, just like they are in crowded biological systems such as cells. These fluid systems notably influence the effectiveness of reactions, separations, and purification stages within biological and industrial processes [1]. It is acknowledged that the solvent itself plays a critical role in establishing the yield, speed, and even the viability of chemical and biochemical reactions [2]. While research into enhancing biocatalytic reactions has primarily revolved around identifying mutations that enhance enzymatic performance, it is equally imperative to identify a suitable liquid environment for conducting biochemical reactions [3]. Therefore, a methodical exploration of how different liquid systems impact enzyme stability becomes imperative when constructing tailored settings that optimize biochemical reactions, and this can be achieved without resorting to extensive trial-and-error methodologies.
The interest in biocatalysis as an alternative to classical chemical processes has experienced notable growth during the past decades. There is a global tendency towards optimizing production routes to make them more sustainable without compromising productivity [4,5]. Enzyme-catalyzed reactions have proven effective in bringing biochemical reactions to an industrial scale and have already been applied in multiple fields such as the food [6], pharmaceutical [7], cosmetic [8], textile [9], and paper industries [10]. Enzymes stand out due to their several advantages, in contrast to chemical catalysts such as high selectivity toward their respective bioreaction [4], which allows for the targeted conversion of a specific product [11]. In addition, they tolerate small substrate spectra, reducing the probability of undesired side reactions. For this reason, biocatalyzed reactions tend to exhibit considerably high selectivity towards specific products. Non-biological catalysts lead to produce racemic mixtures of two enantiomers, negatively affecting the yield of a desired product and making the separation process more difficult [12]. Because of the high stereospecificity that enzymes offer, it is possible to avoid this problem by applying a biocatalytic route within a specific industrial process [13].
The enzyme studied within this work is an NAD-dependent dehydrogenase, an essential enzyme class in the industry [14,15]. These enzymes facilitate key metabolic processes and belong to one of the most extensively studied protein families [16]. Among these, Candida boidinii formate dehydrogenase (cbFDH) stands out as a particularly suitable and straightforward model enzyme for investigating the fundamental mechanisms of catalysis involving hydride ion transfer [17]. cbFDH, which typically forms homodimers and lacks metal ions or prosthetic groups [18], plays a catalytic role in converting formate anions into carbon dioxide while concurrently reducing NAD to NADH, as depicted in Figure 1. Noteworthy industrial applications of cbFDH encompass CO fixation [19] and nicotinamide recycling systems [20]. The substantial usage of cofactors like NADH or NADPH in other industrially relevant enzyme systems incurs elevated expenses [21]. To mitigate these costs or even transform them into economic gains, extensive research has been dedicated to comprehending the cbFDH reaction in recent years [18,22,23]. Nevertheless, cbFDH encounters certain limitations in terms of its chemical, thermal, and long-term stability, along with the costly production of native cbFDHs and their limited enzymatic efficiency [24]. These challenges restrict their practical use in commercial production. Consequently, diverse strategies, with a strong emphasis on protein engineering, have been employed to ameliorate one or more of these drawbacks [25].
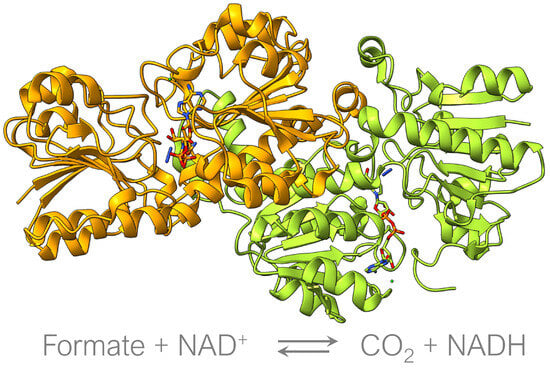
Figure 1.
Candida boidinii Formate dehydrogenase complexed with NAD (PDB: 5DN9). Oxidation of formate to CO using the NAD/NADH cofactor.
Nonetheless, the complete realization of the potential inherent in biochemical reactions, including those catalyzed by cbFDH, remains an ongoing pursuit. In nature, enzymes function remarkably well under mild conditions (such as ambient temperature, neutral pH, and normal pressure), which often results in their limited thermal and chemical stability [18]. cbFDH follows this pattern and further displays a relatively modest specific activity, suggesting room for enhancement. However, large-scale industrial processes typically necessitate more rigorous conditions to ensure commercial viability and profitability [26]. Consequently, numerous investigations in biocatalysis are dedicated to gaining a profound comprehension of enzymatic mechanisms and interactions, with the overarching aim of ameliorating these constraints [5].
Incorporating co-solvents and co-solvent mixtures into the classical aqueous reaction media for biocatalysis has emerged as a successful strategy for augmenting enzymatic attributes [27]. Remarkably, deep eutectic solvents (DESs) have garnered noteworthy attention in recent times as promising reaction environments for biocatalysis [28,29]. DESs are complex mixtures composed of two or more Lewis acids and bases, each serving distinct roles as hydrogen-bond acceptors (HBA) or hydrogen-bond donors (HBD). On one hand, the HBA typically comprises a halogenated quaternary salt with a notably high melting point, while HBD consists of compounds characterized by hydroxyl or carboxyl groups, often sourced from natural compounds or primary metabolites [30,31]. The DESs formation process involves the blending of these HBA and HBD components, with the aim of creating uniform liquid DESs with properties comparable to conventional industrial solvents slated for replacement. Consequently, an expansive array of DESs exists, resulting from different combinations of HBA and HBD. The large number of HBA and HBs available allows for the tailoring of the thermophysical properties of the DESs, with different molar proportions or the addition of water, to generate specific solvents for certain industrial applications [32]. These solvents possess distinctive attributes compared to traditional organic solvents, rendering them more suitable for industrial use and aligning them with the prevailing trend towards sustainable large-scale processes [33]. The appeal of these co-solvent mixtures lies in their unique qualities such as easy preparation, tailored composition, enhanced solubility, and environmentally friendly biodegradability [34,35].
Numerous investigations have already considered their efficiency in enhancing enzyme performance, especially using choline chloride and betaine as hydrogen bond acceptors. Their effects on numerous enzymes have been thoroughly explored, including their effects on alcohol dehydrogenase [28,36], -chymotrypsin [37,38], horseradish peroxidase [39,40], laccase [41,42], and lysozyme [43,44], yielding promising outcomes and indications of substantial efficiency and stability improvements. Importantly, these solvents’ bespoke attributes and compositional flexibility offer the potential for an extensive range of component combinations encompassing diverse functional groups. This versatility entails the prospect of optimizing catalytic performance across various enzyme types [45].
The main focus of this work is to study the co-solvent effects on the kinetics, thermal stability, and unfolding reversibility of cbFDH, as well as the impact on the binding affinity of cbFDH with NAD and unfolding thermodynamics parameters. The co-solvents under consideration were betaine, sorbitol, and glycerol, as well as two DESs (betaine:glycerol and betaine:sorbitol) that have been extensively characterized in the literature [46,47], including their use in the enzymatic reaction [48,49], at three different co-solvent concentrations in water (5, 10, and 15 wt.%). All of the investigations were carried out at 25 °C and 1 bar in a pH 8.5 Tris buffer. These results will unlock opportunities for expanding the operational parameters, enhancing the operational stability and catalytic efficiency of the cbFDH-catalyzed reaction.
2. Materials and Methods
2.1. Chemicals and Reagents
The necessary chemicals for the characterization of cbFDH stability and kinetics experiments were purchased as follows: sodium formate (>99.0) and glycerol(>99.5) were purchased from VWR Chemicals (Westchester, USA). NAD(>96.5), betaine(>99.0), D-sorbitol(>99.0), Trisma HCl(>99.0), and Base(>99.9) were obtained from Sigma-Aldrich (St. Louis, MO, USA). The chemicals used in this work are listed in Table S1. All of the materials were used as received without any further purification.
2.2. Expression, Purification, and Storage of cbFDH
The enzyme used in this work is a codon-optimized version of the formate dehydrogenase (GeneArt Gene Synthesis, ThermoFisher Scientific, Waltham, MA, USA) from Candida bodinii (cbFDH) (UniProt: O13437 (FDH-CANBO)) obtained from recombinant protein expression in Escherichia coli. Electrocompetent E. coli strain BL21 (DE3) Rosetta (CmR) cells were transformed with pET21b-cbFDH (AmpR) plasmid containing cbFDH that is linked through a serine-alanine linker to a N-terminal strepII-tag. An overnight preculture with the transformant was grown in lysogeny broth (LB, Sigma Aldrich, St. Louis, MO, USA) medium containing ampicillin (100 mg·L) and chloramphenicol (15 mg·L) at 37 °C and 130 rpm. A LB-MOPS (0.1 M 3 (N morpholino) propanesulfonic acid), a pH 7.4 main culture supplemented with ampicillin (100 mg·L), chloramphenicol (15 mg·L), and glucose (5 g·L), was inoculated with the preculture to an OD of 0.05. Cultures were grown at 37 °C, 130 rpm until an OD of 0.35–0.5 was reached. Protein expression was induced by the addition of 0.1 mM IPTG (Isopropyl -d-1-thiogalactopyranoside), and the temperature was decreased to 23 °C. After 16 h, cells were harvested by centrifugation (6000 rpm, 20 min, 4 °C (Avanti JXN 26, Beckman Coulter, Brea, CA, USA)). The resulting cell pellet was dissolved in 0.1 M Tris HCl pH 8.5 buffer, and cell disruption was carried out using ultrasonication (Sonifier 250, Branson, Brookfield, CT, USA) with four cycles of 45 s treatment with an output of 40 % and 45 s of resting time between each cycle. The cell lysate was centrifuged again (42,000 rpm, 1 h, 4 °C, L8 80M Ultracentrifuge, Beckman Coulter), and the supernatant, containing the recombinant protein, was filtrated using sterile syringe filters (0.2 mm pore size, Sarstedt, Numbrecht, Germany). The filtrate was purified with affinity chromatography using Strep-Tactin Superflow high-capacity gravity flow columns (IBA Lifesciences, Gottingen, Germany), according to the manufacturer’s recommendations. Washing and elution steps were performed with 0.1 M Tris HCl pH 8.5 buffer. The protein was concentrated using Amicon Ultracel filters (Merck Millipore, Burlington, MA, USA) with a 30 kDa cut off, and the final protein concentration was determined by Bradford assay. The purified proteins were stored at −80 °C. The SDS-PAGE of purified cbFDH is shown in Figure S1.
2.3. DES Preparation
The DESs were prepared gravimetrically. DES1 is composed of BET and GLY in the molar ratio of 1:2, while DES2 is made of BET and SOR in the molar ratio of 1:1. After the respective osmolytes were weighed, they were put together in 50 mL Falcon® tubes and constantly agitated at 60 °C for around 6 hours until a homogeneous solution formed. When a homogeneous liquid was ensured, the stock solution was diluted in a buffer solution.
2.4. Preparation of Stock Solutions
All of the experiments within this work were carried out in aqueous buffer solutions to guarantee constant pH conditions for cbFDH. For this, an 0.1 M Trizma-Buffer was used. Solutions for three osmolytes (BET, GLY, and SOR) and two mixtures of these osmolytes (DES1 and DES2) were prepared at three different concentrations (mass fractions): 20 wt.-%, 40 wt.-%, and 60 wt.-%. The stock solutions were prepared with a higher concentration than the ones considered in this work (5 wt.-%, 10 wt.-%, and 15 wt.-%) since a four-factor dilution occurred when combining the reactants for the measurement on kinetics. For the substrate solution, a formate stock solution of 200 mmol·kg was prepared, dissolving the corresponding amount of formate in Trizma buffer. This solution was then diluted to molalities varying between 2 and 160 mmol·kg. Several stock solutions of 10 mmol·kg NAD were prepared and stored under refrigeration to avoid degradation. The enzyme stock was diluted in Trizma-Buffer to a molality of 25.5 mol·kg for the measurements on kinetics, and it was stored in several aliquotes at −20 °C to avoid decomposition. For thermal stability measurements, the enzyme was diluted to a molality of 100 mol·kg to increase the equipment readability.
2.5. Reaction Kinetics Measurements
For the analysis of the kinetic behavior of the enzyme cbFDH under the influence of co-solvents and co-solvent mixtures, an Eppendorf BioSpectrometer® (Eppendorf, Germany) was utilized. The equipment was used to measure the absorbance of the reaction’s by-product NADH at a 340 nm wavelength plotted against the specified measurement time. The resulting plots describing a linear increase in the NADH absorbance over time (due to the reduction in NAD) were used to derive the Michaelis–Menten curves. This method is based on the direct correlation between the measured absorbance and the concentration of the light-absorbing compound, namely, NADH for this case, which follows the Beer–Lambert law (Equation (1)). The change in the NADH concentration is expressed in Equation (2)
where is the absorbance difference measured between the absorbance A of the reaction media and the absorbance A of the blank media. stands for the molar absorption coefficient, l for the optical path length, and for the molality of the absorbing compound. The slope of the linear plot of the absorbance describes the reaction rate at a specific substrate concentration.
For the absorbance measurements, 10 mm cuvettes were used. First, the reactants (except the enzyme) were placed together in an Eppendorf® 1.5 mL tube: 250 L co-solute solution, 250 L formate solution, and 500 L NAD solution. The content of the tube was then mixed with a vortex mixer to obtain a homogenized reaction medium. After that, 20 L of enzyme solution was added to the cuvette, and the measurement was started. The reaction’s conditions were 25 °C and 1 atm, and the measurement time was 3 min. Reactions were measured in triplicate.
2.6. Thermal Stability Assay
The unfolding analysis of cbFDH was performed using a nanoDSF device Prometheus NT.48 (NanoTemper, Munich, Germany). The method is based on the difference in fluorescence between Trp and Tyr residues. During denaturation, the enzymes suffer conformational changes that affect the orientation of these amino acids. Hence, the fluorescence intensity is affected. The fluorescence intensity was measured at two different wavelengths—330 nm and 350 nm—and reported as the F350/F330 ratio. For this, 200 L of the studied co-solvents, each with 5 wt.-%, 10 wt.-%, and 15 wt.-%, were placed in 1.5 mL Eppendorf tubes. Next, 20 L of the enzyme solution was added. The samples were placed using thin 10 L capillaries in the device. For data collection, the software PR.ThermControl V2.1 was used. The measurement was performed between 20 and 90 °C with a temperature ramp of 0.2 °C/min. To access relevant thermodynamic properties, the open-source software package MoltenProt was used [50] (https://spc.embl-hamburg.de/app/moltenprot (accessed on 3 April 2023)).
2.7. Modulated Scanning Fluorometry
For this, the software PR.TimeControl, available in the Prometheus NT.48, was used. This method applies incremental temperature cycling and temperature stepping to derive non-reversibility curves that offer access to T. The sample preparation was carried out analogously to the one exposed in Section 2.6, and MSFAnalyzer [51] was used to perform the curve fitting (Accessed: https://github.com/CoriolisPharmaResearch/MSFAnalyser (accessed on 1 May 2023)). The measurement was performed between 20 and 80 °C with a temperature ramp of 1 °C/min, i.e., for each cycle, the maximal temperature reached was always one degree higher than the previous one; the temperature then remained constant for 1 min until it decreased back to the starting temperature of 20 °C.
2.8. Binding Affinity
The procedure for estimating the dissociation constant K of cbFDH for NAD followed a similar guideline to the one for obtaining the other thermodynamic parameters. However, for this measurement it was necessary to charge the equipment with samples containing different NAD molalities between 0.1 and 100 mmol·kg. The data obtained via nanoDSF were then processed using FoldAffinity [52] (https://spc.embl-hamburg.de/app/foldAffinity (accessed on 3 April 2023)). This software fits the fraction of unfolded molecules versus the initial ligand concentration using an isothermal analysis (near the value for T) to obtain K. This parameter can be defined as per Equation (4)
where stands for the molality of the folded protein, for the molality of the ligand (here NAD), and for the molality of the enzyme–ligand complex.
3. Results
3.1. Kinetics Parameters
The cbFDH reaction follows a bi–bi ordered reaction mechanism consisting of four steps [53], presented in Equation (5) with E = enzyme:
The difficulty of this reaction mechanism, which encompasses a series of intricate steps, is somehow constrained by the irreversible hydride transfer event from the formate ion to the C position of NAD, as previously stated [23,54]. This process yields NADH and CO, as documented in the literature through pre-steady state kinetics investigations [55,56,57]. In the context of practical applications, a greater focus is placed on comprehending how diverse reaction conditions influence the overall kinetic characteristics. Through the data gathered from NADH absorbance, we investigated the kinetic behavior of cbFDH under the influence of various co-solvents, including the aqueous solutions of three osmolytes (betaine (BET), glycerol (GLY), and sorbitol (SOR)) and two DESs (betaine:glycerol(1:2) (DES1) and betaine:sorbitol(1:1) (DES2)) at three different co-solvent concentration in water (5, 10, and 15 wt.%), 25 °C, and atmospheric pressure, as detailed in Section 2. The dynamics of its kinetics, involving the increase in product concentration over time within the specified conditions (characterized by a high concentration of one substrate) [58], could be effectively analyzed using the Michaelis–Menten equation designed for a pseudo-one substrate reaction. Figure S2 illustrates the primary Michaelis–Menten plot curves obtained from this approach as Equation (6) for each system of this work as follows:
where k is the catalytic constant, K is the Michaelis constant, and is the molality of formate. From the observations, it is clear that using a higher concentration of co-solvent leads to an increase in the reaction rate, with the sole exception of the 15 wt. % BET co-solvent. This provides an early indication of the positive stabilization effects of adding co-solvents in the reaction medium. The values for the kinetic parameters (K, k, and K) were obtained using a non-linear fitting with the Levenberg–Marquardt algorithm and are illustrated graphically in Figure 2. A summary of the obtained kinetic parameters is presented in Table S2.
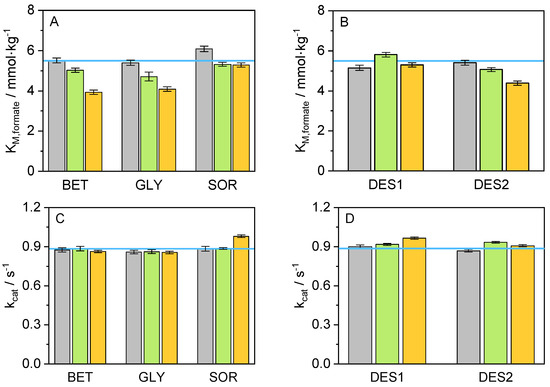
Figure 2.
Values for (A) K in presence of co-solvents, (B) K in presence of DESs, (C) k in presence of co-solvents, and (D) k in presence of DES at (T = 25 °C, p = 1 atm, pH = 8.5), at different concentrations (grey: 5 wt.-%, green: 10 wt.-%, orange: 15 wt.-%, and blue line: neat buffer).
From Figure 2 and Table S1, it is possible to distinguish a decrease in the values for K with increasing co-solvent concentration, compared to the neat buffer environment. This decrease could be interpreted as an improvement in the affinity of cbFDH towards the formate when the co-solvent is present in the cbFDH environment. It is known that osmolytes could interact favorably with water, enriching the hydrogen bond network in the cbFDH vicinity. Nevertheless, SOR represents an exception to this trend since adding it had practically no significant effect on K, regardless of SOR concentration, probably due to solvent structure limitations. Further, the values for k, in contrast to K, show slight negative changes for BET and GLY and a small improvement in the presence of SOR and DES systems. Interestingly, non-monotonic behavior is observed for DES2. Finally, with the help of the relation between k and K, it was possible to determine the catalytic efficiency (K); this measure provides an insight into the overall performance of a cbFDH for a specific reaction and substrate. The effect of the co-solvents used in this work on the catalytic efficiency is shown graphically in Figure 3.
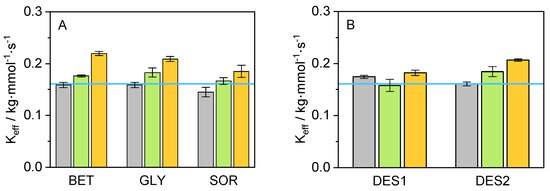
Figure 3.
Values for K obtained from the ratio between k and K in the presence of (A) various co-solvents and (B) in presence of DES on the reaction medium (T = 25 °C, p = 1 atm, pH = 8.5) at different concentrations (grey: 5 wt.-%, green: 10 wt.-%, orange: 15 wt.-%, and blue line: neat buffer).
A clear trend can be observed for K: the catalytic efficiency of cbFDH for the reaction considered in this work increases with an increment of the co-solvent concentration. It is also possible to recognize an improvement in K for the 10 wt.% and 15 wt.% compared to the neat buffer. The system containing 15 wt.% BET yielded the lowest value for K (3.933 mmol·kg), followed by 15 wt.% GLY and 15 wt.% DES2 with 4.090 mmol·kg and 4.392 mmol·kg, respectively. Thus, adding BET at 15 wt.% to the reaction medium was responsible for the highest increase in the formate affinity compared to the remaining co-solutes. A possible explanation for this effect may be the preferential exclusion effect of BET from the protein backbone hydration layers [59]. BET molecules can be located mainly in a second solvation layer around 2.7 Å, due to a hydrophobic effect endorsed by the methyl groups. Therefore, a water enhancement occurs on the protein surface [60], while the density of the co-solvent in the second solvation layer increases, promoting favorable non-covalent and hydrogen bond interactions in the system. Concerning the catalytic constant, the highest value for k was achieved in the system containing 15 wt.% SOR (0.980 s), followed closely by 15 wt.% DES1 (0.965 s) and 10 wt.% DES2 (0.933 s). This positive effect of SOR on k can be attributed to its direct interaction with the peptide backbone of the cbFDH, which results in cbFDH structural and hydration changes [61], promoting possible open cbFDH conformations that accelerate the reaction kinetics.
3.2. Thermal Stability
To evaluate cbFDH stability in its native folded state under the studied conditions, we measured the unfolding temperature (T). In the presence of co-solvent, a higher T indicates higher cbFDH thermal stability. Our investigation has revealed a slight elevation in the T upon adding some co-solvents. In Figure S3, the unfolding curves obtained, experimentally expressed as unfolded fractions (f) over the temperature, are shown. These plots provide an insight into the T of the cbFDH (i.e., the inflection point of the sigmoid curve) and how it changes depending on the co-solvent present in the solution. Therefore, it is possible to distinguish a trend for a shift to higher T with the addition of co-solvents (right side), compared to neat buffer, and a further shift as their concentration increases. Figure 4 shows the results for the system studied in this work, allowing for a better comparison of the co-solvent influence on the cbFDH unfolding temperature. From Figure 4, it can be interpreted that the inclusion of co-solvents enhances the enzyme’s stability against thermal stress since, for most of the systems, an improvement in T and hence the thermal stability is observed when compared to the neat buffer condition. BET appears to be the only exception to this behavior as the addition of BET yielded T lower than that of the pure buffer, regardless of its concentration. This is not the case for GLY, DES1, and DES2, where the T values increased with an increment in the co-solvent concentration.
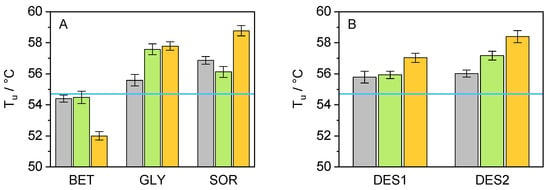
Figure 4.
Graphical representation of the results obtained for the T of cbFDH after thermal assay via nanoDSF in the presence of various co-solvents ((A): BET, GLY, and SOR/(B): DES1, DES2) at different concentrations (grey: 5 wt.-%, green: 10 wt.-%, orange: 15 wt.-%, and blue line: neat buffer) at p = 1 atm and pH= 8.5.
At first sight, this behavior indicates an apparent cbFDH stability improvement against thermal denaturation (except BET). However, ranking the co-solvent effect based exclusively on T may produce misleading results [50]. Therefore, additional thermodynamic parameters were determined to make a supported statement. Further analysis of the data obtained from the fluorescence screening allowed for an evaluation of these properties. A fair comparison of the cbFDH stability could be obtained using the apparent unfolding energy ΔG. To calculate this parameter, the Gibbs–Helmholtz equation was applied; however, to ensure a practical comparison between systems, the T of cbFDH in neat buffer (T = 54.7 °C) was used as a reference, as shown in Equation (7), allowing one to obtain the difference in ΔG between the co-solvent system and the neat buffer. The values for the unfolding enthalpy (ΔH) and apparent heat capacity change in the co-solvent presence (ΔC) were estimated from the unfolding curves. The results are shown in Figure 5.
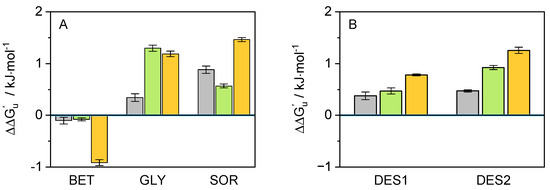
Figure 5.
Graphical representation of the results obtained for the of cbFDH in the presence of various co-solvents ((A): BET, GLY, and SOR/(B): DES1, DES2) using T = 54.7 °C, at different concentrations (grey: 5 wt.-%, green: 10 wt.-%, orange: 15 wt.-%, and value 0: neat buffer) at pH = 8.5 and p = 1 atm.
Knowledge of this is essential since it describes how thermally stable the enzyme’s folded state is in the respective co-solvent compared to the buffer. This value could be related with other thermodynamic properties such as B (cf. Figure S4). As shown in Figure 5, the results for correlates with the T results. This confirms the conclusions drawn after the interpretation of Figure 4, i.e., adding the considered co-solvents seems to provide the cbFDH with greater thermal protection compared to neat buffer, and it can be improved with increased concentration. BET was the only co-solvent that resulted in negative values for , regardless of its concentration in the solution. The non-monotonous effect of SOR concentration on thermal stability is worth mentioning as hydrogen bond dynamics between cbFDH/co-solvent/water change dramatically with the amount of water. In contrast, DESs cause linear increases in thermal stability, which are mainly caused by the additive effects of the DESs constituents, i.e., GLY and SOR compensate for the destabilization effect of BET.
The classical thermodynamic model for protein unfolding assumes that the process is completely reversible and that equilibrium is achieved at each stage of a thermal unfolding study. However, in reality, proteins seldom behave this way, and a kinetic explanation for protein unfolding has been put forward using the apparent unfolding constant rate (k) [62]. Values for pk were determined using Equation (8) as follows:
where p is the negative logarithm of the apparent rate constant of unfolding extrapolated to a reference temperature of 298.15 K, E is the activation energy of unfolding, and T is the temperature where k is equal to one, as stated in the two-state theory from native state to unfolded state (N → D) [62]. T and E are sigmoidal unfolding curve explicit characteristics obtained directly from the nanoDSF measurements. Figure 6 shows the results for the systems used in this work. Values higher than the neat buffer denote a decrease in the denaturation rate of cbFDH. This appears to be the case for the systems GLY and SOR, which show an improvement in the values of pk, regardless of their concentration. DES1 and DES2 slightly increased the unfolding rate, while BET induced the same effect. Interestingly, the individual effects of the co-solvents (BET, GLY, and SOR) seem to be additive across all the concentrations when evaluated as DES, where GLY (in DES1) and SOR (in DES2) manage to partially reduce the negative effects of BET in k.
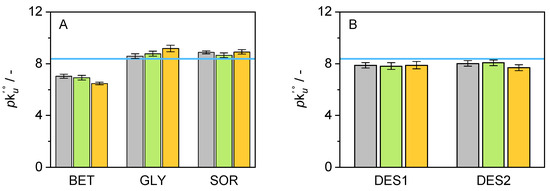
Figure 6.
Graphical representation of the results obtained for pk of cbFDH in the presence of various co-solvents ((A): BET, GLY, and SOR/(B): DES1, DES2) at different concentrations (grey: 5 wt.-%, green: 10 wt.-%, orange: 15 wt.-%, and blue line: neat buffer) at pH = 8.5 and p = 1 atm. Unfolding curves obtained between 30 and 70 °C.
3.3. Modulated Scanning Fluorometry
The potential for unfolding to be reversible is influenced by protein environment factors such as pH, salt, or co-solvent concentration. Thus, it is necessary to evaluate the reversibility of thermal unfolding under the influence of the co-solvents used in this work. In this work, the ability of cbFDH to refold after exposure to thermal stress was assessed using modulated scanning fluorometry (MSF) to allow a swift evaluation of conditions promoting reversible thermal unfolding in proteins [63]. MSF utilizes rapid cycles of heating and cooling across multiple samples concurrently. Each successive heating cycle achieves a slightly elevated temperature [64]. Additionally, MSF assists in identifying the temperature threshold at which irreversible protein unfolding initiates, thereby enabling the measurement, ordering, and contrasting of stability profiles obtained from nanoDSF. Figure 7 shows an example of the unfolding and non-reversibility curves obtained from MSF for the 10 wt.-% concentration of each co-solvent. The remaining co-solvent results are presented in Figures S5 and S6 and summarized in Table S3.
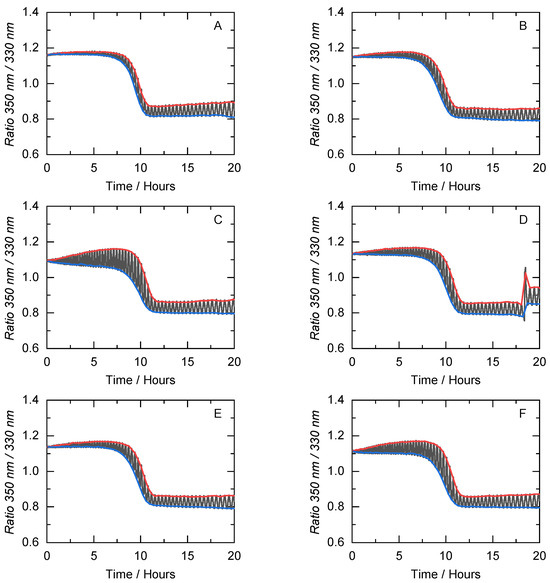
Figure 7.
Unfolding (blue) and non-reversibility (red) curves of cbFDH obtained from the MSF measurements in (A) buffer and under the influence of various co-solvents at 10 wt.-% concentration ((B): BET, (C): GLY, (D): SOR, (E): DES1, and (F): DES2) at pH = 8.5 and p = 1 atm. Here, the fluorescence ratio of 350 nm to 330 nm is plotted against the time. x-axis correlates to the temperature during the measurement (between 20–80 °C).
Figure 7 shows that regardless of the co-solvent present in the solution, the inflection point of the non-reversible curve (T) is always higher than the unfolding curve (T). As expected, in a neat buffer solution, the MSF results for T and T are almost equivalent. For the 10 wt.-% co-solvent concentrations, GLY and DES2 cause a greater area between the non-reversibility and unfolding curves than the neat buffer. This indicates the ability of the cbFDH in a crowded environment to recover its native state after being subjected to thermal stress and structural perturbations in a crowded environment. Thus, the exposure of cbFDH to temperatures below T does not lead to substantial secondary and tertiary structural changes. These small reversible structural changes can lead to open cbFDH conformations that could be one of the factors contributing to the increase in kinetic efficiency. It is important to note that T does not correlate with other biophysical variables of the unfolding mechanism, as previously demonstrated [51]. To varying degrees, the co-solvents in this work increase the resistance of cbFDH to temperature-induced degradation concerning the neat buffer. In addition, some co-solvents apparently caused protein aggregation at high temperatures (i.e., 10 wt.-% SOR), which was expected due to the crowded hydrogen bond network in the DES systems.
3.4. Binding Affinity
The influence of substrate binding (NAD) on the thermal stability of cbFDH was also studied within the framework of this work, including the effect of the co-solvents. For each of the systems, the unfolding curves were obtained in different environments for several NAD concentrations (cf. Figure S7) for the neat buffer. It is possible to recognize a shift of the unfolding curves to the right side of the graph, i.e., higher unfolding temperature, with increasing NAD concentration. This result suggests that substrate also contributes to cbFDH stability, probably through conformational changes in the binding site. An isothermal fit of the unfolded fraction of cbFDH was performed at several temperatures in the range of T ± 10 °C, as a function of the NAD concentration (cf. Figure S8). Regardless of the temperatures chosen for the isothermal analysis, a decrease in the unfolded fraction was observed with increased substrate molality. From the isothermal fit, obtaining the fractions of the bound and free substrate and, therefore, the dissociation constant is possible. The values for K for the considered systems of this work are presented in Figure 8.
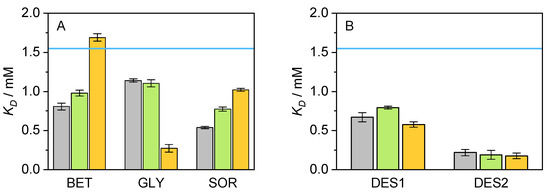
Figure 8.
Graphical representation of the results obtained for the K of cbFDH in the presence of various co-solvents ((A): BET, GLY, and SOR/(B): DES1, DES2) at different concentrations (grey: 5 wt.-%, green: 10 wt.-%, orange: 15 wt.-%, and blue line: neat buffer) at pH = 8.5 and p = 1 atm. After thermal assay via nanoDSF, the obtained data were analyzed with FoldAffinity to estimate K.
Figure 8 shows a particular positive effect of DES2 on K, the co-solvent which causes the highest negative deviation from buffer K value. The lower the values for K, the higher the cbFDH affinity for the substrate. Moderate improvements can be observed for the remaining co-solvents, but their different concentrations do not follow any particular trend. BET is again the only exception, in this case for its 15 wt.-% concentration, since it appears to be the only system unable to improve the value for K compared to a system with an absence of co-solvents. Interestingly, these changes in the binding affinity with NAD do not correlate with the results obtained in the kinetic parameters (cf. Figure 2), i.e., the improvement in the affinity substrate/cbFDH on DES systems is not translated into improvements on K or k above the contribution of the individual co-solvents especially BET and GLY. This validates using a pseudo-one substrate approach for the kinetic curves since the NAD binding is a favored fast process and the cbFDH reaction rate is limited by the hydride transfer event from the formate ion on the active site, and therefore K is rather affected by the formate affinity improvement due to co-solvent/formate interactions.
4. Conclusions
The influence of single co-solvents and co-solvent mixtures on the oxidation reaction of formate to carbon dioxide catalyzed by cbFDH was studied and evaluated experimentally. To efficiently characterize these effects, a complete description of the cbFDH reaction kinetics and the thermal stress stability was performed. The main goal was to evaluate the overall performance of the co-solvent mixtures as boosters of the mentioned enzymatic properties, compared to the effectiveness of the considered single co-solvents effects. The study of cbFDH’s kinetics revealed that adding co-solvents and co-solvent mixtures to the aqueous reaction medium effectively increased the catalytic efficiency cbFDH. All of the considered co-solvents outperformed the neat buffer at their highest concentration (15 wt.-%). K values were remarkably improved in the presence of BET, GLY, and DES2 (BET:SOR 1:1), due mainly to the improvement of the formate affinity on the co-solvent environment. BET tends to promote water enhancement on the protein surface through preferential exclusion from the cbFDH hydration layer, which allows effectively non-covalent interactions such as hydrogen bonds, thus showing the most significant decrease in K. However, a substantial improvement to k was not observed for any co-solvent, suggesting that the cbFDH secondary structure remains stable. BET was shown to negatively affect the stability of cbFDH’s through ΔG. At the same time, GLY, SOR, and DESs promote overall higher protection against thermal stress, improving T. These co-solvents influence the enzyme surface through molecular crowding, excluding water molecules and locating themselves near cbFDH surface. Combined in a mixture as DESs, the formed network of hydrogen bonds acts as a biochemical chaperone, increasing the unfolding reversibility in temperatures above T and improving the affinity towards NAD. Overall, the studied DESs appear to be an efficient alternative for enhancing the biocatalytic process from conventional aqueous systems as it is possible to tailor the constituents and take advantage of the individual contributions to the cbFDH stability. Further, a settlement between the enhancement of the kinetic properties (i.e., BET systems) and the stability of the enzyme (e.g., through GLY or SOR) can be achieved in mixtures without hindering one or the other.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/pr11102815/s1, Figure S1: SDS-Page of purified cbFDH, Figure S2: Michaelis-Menten primary plots, Figure S3: Sigmoidal unfolding curves, Figure S4: , Figure S5: MSF at 5 wt.-%, Figure S6: MSF at 15 wt.-%, Figure S7: Unfolding sigmoidal curves in binding experiments, Figure S8: Isothermal binding fit; Table S1: Chemicals used in this Work, Table S2: Kinetic parameters, Table S3: Thermal unfolding parameters.
Author Contributions
Conceptualization, investigation, and writing—original draft preparation: N.F.G.-P.; methodology, data curation, and formal analysis: A.F.A.-A., A.V. and G.R.; and supervision, project administration, funding acquisition, and writing—review and editing: R.I.C., T.H., G.S. and C.H. All authors have read and agreed to the published version of the manuscript.
Funding
The authors thank the funding from the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) under Germany’s Excellence Strategy–EXC 2033—390677874— RESOLV. Nicolás Gajardo’s work was supported by the German Academic Exchange Service (DAAD) under the Graduate School Scholarship Programme, 2020 (57516591).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data from the research are contained within the manuscript and Supplementary Materials; further inquiries can be directed to the corresponding author.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Gao, M.; Held, C.; Patra, S.; Arns, L.; Sadowski, G.; Winter, R. Crowders and Cosolvents-Major Contributors to the Cellular Milieu and Efficient Means to Counteract Environmental Stresses. Chemphyschem Eur. J. Chem. Phys. Phys. Chem. 2017, 18, 2951–2972. [Google Scholar] [CrossRef] [PubMed]
- Zaks, A.; Klibanov, A.M. Enzymatic catalysis in nonaqueous solvents. J. Biol. Chem. 1988, 263, 3194–3201. [Google Scholar] [CrossRef] [PubMed]
- Robertson, D.E.; Steer, B.A. Recent progress in biocatalyst discovery and optimization. Curr. Opin. Chem. Biol. 2004, 8, 141–149. [Google Scholar] [CrossRef] [PubMed]
- Timson, D.J. Four Challenges for Better Biocatalysts. Fermentation 2019, 5, 39. [Google Scholar] [CrossRef]
- Woodley, J.M. Accelerating the implementation of biocatalysis in industry. Appl. Microbiol. Biotechnol. 2019, 103, 4733–4739. [Google Scholar] [CrossRef] [PubMed]
- Fernandes, P. Enzymes in food processing: A condensed overview on strategies for better biocatalysts. Enzym. Res. 2010, 2010, 862537. [Google Scholar] [CrossRef]
- Huisman, G.W.; Collier, S.J. On the development of new biocatalytic processes for practical pharmaceutical synthesis. Curr. Opin. Chem. Biol. 2013, 17, 284–292. [Google Scholar] [CrossRef]
- Turner, N.J. Directed evolution of enzymes for applied biocatalysis. Trends Biotechnol. 2003, 21, 474–478. [Google Scholar] [CrossRef]
- Queiroga, A.C.; Pintado, M.M.; Malcata, F.X. Novel microbial-mediated modifications of wool. Enzym. Microb. Technol. 2007, 40, 1491–1495. [Google Scholar] [CrossRef]
- Maijala, P.; Kleen, M.; Westin, C.; Poppius-Levlin, K.; Herranen, K.; Lehto, J.H.; Reponen, P.; Mäentausta, O.; Mettälä, A.; Hatakka, A. Biomechanical pulping of softwood with enzymes and white-rot fungus Physisporinus rivulosus. Enzym. Microb. Technol. 2008, 43, 169–177. [Google Scholar] [CrossRef]
- Bisswanger, H. Enzyme Kinetics; Wiley: Hoboken, NJ, USA, 2008. [Google Scholar] [CrossRef]
- Jaeger, K.E.; Eggert, T. Enantioselective biocatalysis optimized by directed evolution. Curr. Opin. Biotechnol. 2004, 15, 305–313. [Google Scholar] [CrossRef] [PubMed]
- Karabec, M.; Łyskowski, A.; Tauber, K.C.; Steinkellner, G.; Kroutil, W.; Grogan, G.; Gruber, K. Structural insights into substrate specificity and solvent tolerance in alcohol dehydrogenase ADH-‘A’ from Rhodococcus ruber DSM 44541. Chem. Commun. 2010, 46, 6314–6316. [Google Scholar] [CrossRef] [PubMed]
- Nishigaki, J.I.; Ishida, T.; Honma, T.; Haruta, M. Oxidation of β-Nicotinamide Adenine Dinucleotide (NADH) by Au Cluster and Nanoparticle Catalysts Aiming for Coenzyme Regeneration in Enzymatic Glucose Oxidation. ACS Sustain. Chem. Eng. 2020, 8, 10413–10422. [Google Scholar] [CrossRef]
- Bolivar, J.M.; Wilson, L.; Ferrarotti, S.A.; Fernandez-Lafuente, R.; Guisan, J.M.; Mateo, C. Evaluation of different immobilization strategies to prepare an industrial biocatalyst of formate dehydrogenase from Candida boidinii. Enzym. Microb. Technol. 2007, 40, 540–546. [Google Scholar] [CrossRef]
- Woodley, J.M. New frontiers in biocatalysis for sustainable synthesis. Curr. Opin. Green Sustain. Chem. 2020, 21, 22–26. [Google Scholar] [CrossRef]
- Bandaria, J.N.; Dutta, S.; Hill, S.E.; Kohen, A.; Cheatum, C.M. Fast Enzyme Dynamics at the Active Site of Formate Dehydrogenase. J. Am. Chem. Soc. 2008, 130, 22–23. [Google Scholar] [CrossRef] [PubMed]
- Schirwitz, K.; Schmidt, A.; Lamzin, V.S. High-resolution structures of formate dehydrogenase from Candida boidinii. Protein Sci. 2007, 16, 1146–1156. [Google Scholar] [CrossRef] [PubMed]
- Yadav, R.K.; Baeg, J.O.; Oh, G.H.; Park, N.J.; Kong, K.j.; Kim, J.; Hwang, D.W.; Biswas, S.K. A Photocatalyst–Enzyme Coupled Artificial Photosynthesis System for Solar Energy in Production of Formic Acid from CO2. J. Am. Chem. Soc. 2012, 134, 11455–11461. [Google Scholar] [CrossRef]
- Yang, J.Y.; Kerr, T.A.; Wang, X.S.; Barlow, J.M. Reducing CO2 to HCO2– at Mild Potentials: Lessons from Formate Dehydrogenase. J. Am. Chem. Soc. 2020, 142, 19438–19445. [Google Scholar] [CrossRef]
- Tishkov, V.I.; Popov, V.O. Catalytic mechanism and application of formate dehydrogenase. Biochem. Biokhimiia 2004, 69, 1252–1267. [Google Scholar] [CrossRef]
- Ordu, E.B.; Cameron, G.; Clarke, A.R.; Karagüler, N.G. Kinetic and thermodynamic properties of the folding and assembly of formate dehydrogenase. FEBS Lett. 2009, 583, 2887–2892. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Jaworek, M.W.; Gajardo-Parra, N.F.; Sadowski, G.; Winter, R.; Held, C. Boosting the kinetic efficiency of formate dehydrogenase by combining the effects of temperature, high pressure and co-solvent mixtures. Colloids Surf. Biointerfaces 2021, 208, 112127. [Google Scholar] [CrossRef] [PubMed]
- Castillo, R.; Oliva, M.; Martí, S.; Moliner, V. A Theoretical Study of the Catalytic Mechanism of Formate Dehydrogenase. J. Phys. Chem. B 2008, 112, 10012–10022. [Google Scholar] [CrossRef] [PubMed]
- Zheng, J.; Yang, T.; Zhou, J.; Xu, M.; Zhang, X.; Rao, Z. Elimination of a Free Cysteine by Creation of a Disulfide Bond Increases the Activity and Stability of Candida boidinii Formate Dehydrogenase. Appl. Environ. Microbiol. 2017, 83, e02624-16. [Google Scholar] [CrossRef] [PubMed]
- Magri, A.; Pecorari, T.; Pereira, M.M.; Cilli, E.M.; Greaves, T.L.; Pereira, J.F.B. Enhancing the Biocatalytic Activity of l -Asparaginase Using Aqueous Solutions of Cholinium-Based Ionic Liquids. ACS Sustain. Chem. Eng. 2019, 7, 19720–19731. [Google Scholar] [CrossRef]
- Xu, P.; Zheng, G.W.; Zong, M.H.; Li, N.; Lou, W.Y. Recent progress on deep eutectic solvents in biocatalysis. Bioresour. Bioprocess. 2017, 4, 1–18. [Google Scholar] [CrossRef]
- Bittner, J.P.; Zhang, N.; Huang, L.; Domínguez de María, P.; Jakobtorweihen, S.; Kara, S. Impact of deep eutectic solvents (DESs) and individual DES components on alcohol dehydrogenase catalysis: Connecting experimental data and molecular dynamics simulations. Green Chem. 2022, 24, 1120–1131. [Google Scholar] [CrossRef]
- Pätzold, M.; Siebenhaller, S.; Kara, S.; Liese, A.; Syldatk, C.; Holtmann, D. Deep eutectic solvents as efficient solvents in biocatalysis. Trends Biotechnol. 2019, 37, 943–959. [Google Scholar] [CrossRef]
- Hansen, B.B.; Spittle, S.; Chen, B.; Poe, D.; Zhang, Y.; Klein, J.M.; Horton, A.; Adhikari, L.; Zelovich, T.; Doherty, B.W.; et al. Deep eutectic solvents: A review of fundamentals and applications. Chem. Rev. 2020, 121, 1232–1285. [Google Scholar] [CrossRef]
- Abbott, A.P.; Capper, G.; Davies, D.L.; Rasheed, R.K.; Tambyrajah, V. Novel solvent properties of choline chloride/urea mixtures. Chem. Commun. 2003, 70–71. [Google Scholar] [CrossRef]
- Palmelund, H.; Andersson, M.P.; Asgreen, C.J.; Boyd, B.J.; Rantanen, J.; Löbmann, K. Tailor-made solvents for pharmaceutical use? Experimental and computational approach for determining solubility in deep eutectic solvents (DES). Int. J. Pharm. X 2019, 1, 100034. [Google Scholar] [CrossRef] [PubMed]
- Meyer, L.E.; Andersen, M.B.; Kara, S. A deep eutectic solvent thermomorphic multiphasic system for biocatalytic applications. Angew. Chem. Int. Ed. 2022, 61, e202203823. [Google Scholar] [CrossRef] [PubMed]
- Domínguez de María, P.; Maugeri, Z. Ionic liquids in biotransformations: From proof-of-concept to emerging deep-eutectic-solvents. Curr. Opin. Chem. Biol. 2011, 15, 220–225. [Google Scholar] [CrossRef]
- Abbasi, N.M.; Farooq, M.Q.; Anderson, J.L. Investigating the Variation in Solvation Interactions of Choline Chloride-Based Deep Eutectic Solvents Formed Using Different Hydrogen Bond Donors. ACS Sustain. Chem. Eng. 2021, 9, 11970–11980. [Google Scholar] [CrossRef]
- Chanquia, S.N.; Huang, L.; García Liñares, G.; Domínguez de María, P.; Kara, S. Deep eutectic solvents as smart cosubstrate in alcohol dehydrogenase-catalyzed reductions. Catalysts 2020, 10, 1013. [Google Scholar] [CrossRef]
- Yadav, N.; Venkatesu, P. Current understanding and insights towards protein stabilization and activation in deep eutectic solvents as sustainable solvent media. Phys. Chem. Chem. Phys. PCCP 2022, 24, 13474–13509. [Google Scholar] [CrossRef]
- Zhao, H.; Baker, G.A.; Holmes, S. Protease activation in glycerol-based deep eutectic solvents. J. Mol. Catal. B Enzym. 2011, 72, 163–167. [Google Scholar] [CrossRef]
- Gajardo-Parra, N.F.; Meneses, L.; Duarte, A.R.C.; Paiva, A.; Held, C. Assessing the Influence of Betaine-Based Natural Deep Eutectic Systems on Horseradish Peroxidase. ACS Sustain. Chem. Eng. 2022, 10, 12873–12881. [Google Scholar] [CrossRef]
- Meneses, L.; Gajardo-Parra, N.F.; Cea-Klapp, E.; Garrido, J.M.; Held, C.; Duarte, A.R.; Paiva, A. Improving the activity of horseradish peroxidase in betaine-based natural deep eutectic systems. RSC Sustain. 2023, 1, 886–897. [Google Scholar] [CrossRef]
- Delorme, A.E.; Andanson, J.M.; Verney, V. Improving laccase thermostability with aqueous natural deep eutectic solvents. Int. J. Biol. Macromol. 2020, 163, 919–926. [Google Scholar] [CrossRef]
- Toledo, M.L.; Pereira, M.M.; Freire, M.G.; Silva, J.P.; Coutinho, J.A.; Tavares, A.P. Laccase activation in deep eutectic solvents. ACS Sustain. Chem. Eng. 2019, 7, 11806–11814. [Google Scholar] [CrossRef]
- Sanchez-Fernandez, A.; Basic, M.; Xiang, J.; Prevost, S.; Jackson, A.J.; Dicko, C. Hydration in Deep Eutectic Solvents Induces Non-monotonic Changes in the Conformation and Stability of Proteins. J. Am. Chem. Soc. 2022, 144, 23657–23667. [Google Scholar] [CrossRef] [PubMed]
- Esquembre, R.; Sanz, J.M.; Wall, J.G.; del Monte, F.; Mateo, C.R.; Ferrer, M.L. Thermal unfolding and refolding of lysozyme in deep eutectic solvents and their aqueous dilutions. Phys. Chem. Chem. Phys. 2013, 15, 11248–11256. [Google Scholar] [CrossRef]
- Panić, M.; Cvjetko Bubalo, M.; Radojčić Redovniković, I. Designing a biocatalytic process involving deep eutectic solvents. J. Chem. Technol. Biotechnol. 2021, 96, 14–30. [Google Scholar] [CrossRef]
- Ahmad, I.; Syakfanaya, A.M.; Azminah, A.; Saputri, F.C.; Mun’im, A. Optimization of betaine-sorbitol natural deep eutectic solvent-based ultrasound-assisted extraction and pancreatic lipase inhibitory activity of chlorogenic acid and caffeine content from robusta green coffee beans. Heliyon 2021, 7, e07702. [Google Scholar] [CrossRef]
- Abranches, D.O.; Silva, L.P.; Martins, M.A.; Pinho, S.P.; Coutinho, J.A. Understanding the formation of deep eutectic solvents: Betaine as a universal hydrogen bond acceptor. ChemSusChem 2020, 13, 4916–4921. [Google Scholar] [CrossRef]
- Khodaverdian, S.; Dabirmanesh, B.; Heydari, A.; Dashtban-Moghadam, E.; Khajeh, K.; Ghazi, F. Activity, stability and structure of laccase in betaine based natural deep eutectic solvents. Int. J. Biol. Macromol. 2018, 107, 2574–2579. [Google Scholar] [CrossRef]
- Varriale, S.; Delorme, A.E.; Andanson, J.M.; Devemy, J.; Malfreyt, P.; Verney, V.; Pezzella, C. Enhancing the thermostability of engineered laccases in aqueous betaine-based natural deep eutectic solvents. ACS Sustain. Chem. Eng. 2021, 10, 572–581. [Google Scholar] [CrossRef]
- Kotov, V.; Mlynek, G.; Vesper, O.; Pletzer, M.; Wald, J.; Teixeira-Duarte, C.M.; Celia, H.; Garcia-Alai, M.; Nussberger, S.; Buchanan, S.K.; et al. In-depth interrogation of protein thermal unfolding data with MoltenProt. Protein Sci. Publ. Protein Soc. 2021, 30, 201–217. [Google Scholar] [CrossRef]
- Svilenov, H.L.; Menzen, T.; Richter, K.; Winter, G. Modulated Scanning Fluorimetry Can Quickly Assess Thermal Protein Unfolding Reversibility in Microvolume Samples. Mol. Pharm. 2020, 17, 2638–2647. [Google Scholar] [CrossRef]
- Niebling, S.; Burastero, O.; Bürgi, J.; Günther, C.; Defelipe, L.A.; Sander, S.; Gattkowski, E.; Anjanappa, R.; Wilmanns, M.; Springer, S.; et al. FoldAffinity: Binding affinities from nDSF experiments. Sci. Rep. 2021, 11, 9572. [Google Scholar] [CrossRef]
- Schmidt, T.; Michalik, C.; Zavrel, M.; Spiess, A.; Marquardt, W.; Ansorge-Schumacher, M. Mechanistic model for prediction of formate dehydrogenase kinetics under industrially relevant conditions. Biotechnol. Prog. 2010, 26, 73–78. [Google Scholar] [CrossRef] [PubMed]
- Demchenko, A.P.; Rusyn, O.I.; Egorov, A.M.; Tishkov, V.I. The solvent effects on the kinetics of bacterial formate dehydrogenase reaction. Biochim. Biophys. Acta-(Bba)-Protein Struct. Mol. Enzymol. 1990, 1039, 290–296. [Google Scholar] [CrossRef]
- Ranasinghe, C.; Guo, Q.; Sapienza, P.J.; Lee, A.L.; Quinn, D.M.; Cheatum, C.M.; Kohen, A. Protein mass effects on formate dehydrogenase. J. Am. Chem. Soc. 2017, 139, 17405–17413. [Google Scholar] [CrossRef]
- Tishkov, V.I.; Popov, V.O. Protein engineering of formate dehydrogenase. Biomol. Eng. 2006, 23, 89–110. [Google Scholar] [CrossRef] [PubMed]
- Tishkov, V.I.; Galkin, A.G.; Egorov, A.M. Kinetic isotope effect and the presteady-state kinetics of the reaction catalyzed by the bacterial formate dehydrogenase. Biochimie 1989, 71, 551–557. [Google Scholar] [CrossRef] [PubMed]
- Song, J.; Saeman, M.R.; Baer, L.A.; Cai, A.R.; Wade, C.E.; Wolf, S.E. Exercise altered the skeletal muscle microRNAs and gene expression profiles in burn rats with hindlimb unloading. J. Burn. Care Res. 2017, 38, 11–19. [Google Scholar] [CrossRef]
- Harries, D.; Rösgen, J. A practical guide on how osmolytes modulate macromolecular properties. Methods Cell Biol. 2008, 84, 679–735. [Google Scholar] [CrossRef] [PubMed]
- Gajardo-Parra, N.F.; Akrofi-Mantey, H.; Ascani, M.; Cea-Klapp, E.; Garrido, J.M.; Sadowski, G.; Held, C. Osmolyte effect on enzymatic stability and reaction equilibrium of formate dehydrogenase. Phys. Chem. Chem. Phys. PCCP 2022, 24, 27930–27939. [Google Scholar] [CrossRef]
- Sanchez-Fernandez, A.; Edler, K.J.; Arnold, T.; Alba Venero, D.; Jackson, A.J. Protein conformation in pure and hydrated deep eutectic solvents. Phys. Chem. Chem. Phys. PCCP 2017, 19, 8667–8670. [Google Scholar] [CrossRef]
- Mazurenko, S.; Kunka, A.; Beerens, K.; Johnson, C.M.; Damborsky, J.; Prokop, Z. Exploration of Protein Unfolding by Modelling Calorimetry Data from Reheating. Sci. Rep. 2017, 7, 16321. [Google Scholar] [CrossRef] [PubMed]
- Berner, C.; Menzen, T.; Winter, G.; Svilenov, H.L. Combining Unfolding Reversibility Studies and Molecular Dynamics Simulations to Select Aggregation-Resistant Antibodies. Mol. Pharm. 2021, 18, 2242–2253. [Google Scholar] [CrossRef] [PubMed]
- Svilenov, H.L.; Arosio, P.; Menzen, T.; Tessier, P.; Sormanni, P. Approaches to expand the conventional toolbox for discovery and selection of antibodies with drug-like physicochemical properties. mAbs 2023, 15, 2164459. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).