Author Contributions
Conceptualization, K.V.B. and J.R.; methodology, K.V.B., J.R., A.Y., I.P., A.P.F. and J.-M.A.; software, K.V.B., J.R., A.P.F. and A.Y.; validation, K.V.B. and J.R.; formal analysis, K.V.B. and J.R.; investigation, K.V.B. and J.R.; resources, I.P. and J.-M.A.; data curation, K.V.B. and J.R.; writing—original draft preparation, K.V.B.; writing—review and editing, K.V.B., A.Y., A.P.F., I.P. and J.-M.A.; visualisation, K.V.B.; supervision, I.P. and J.-M.A.; project administration, J.-M.A.; funding acquisition, I.P. and J.-M.A. All authors have read and agreed to the published version of the manuscript.
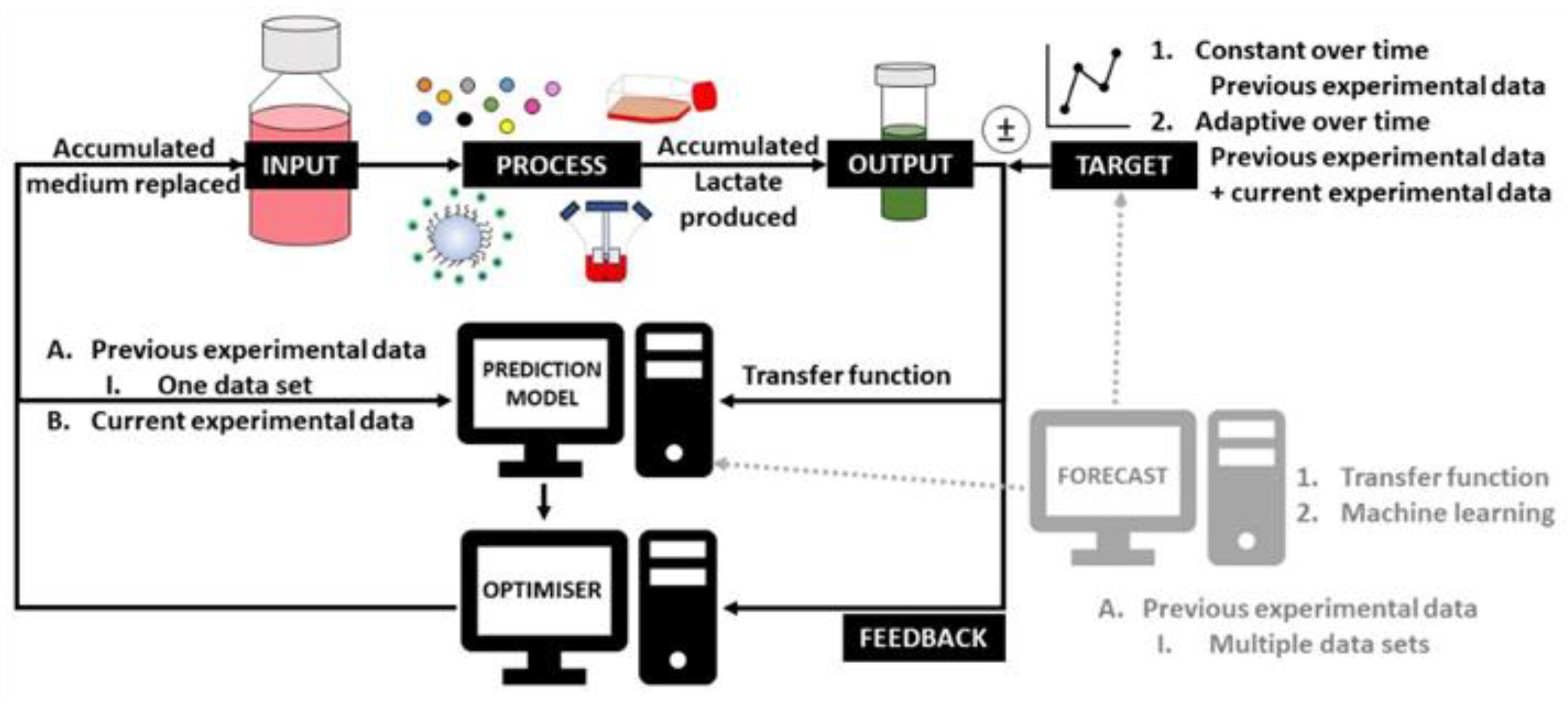
Figure 1.
Different modelling techniques in the model predictive controller (MPC) of this work.
Figure 1.
Different modelling techniques in the model predictive controller (MPC) of this work.
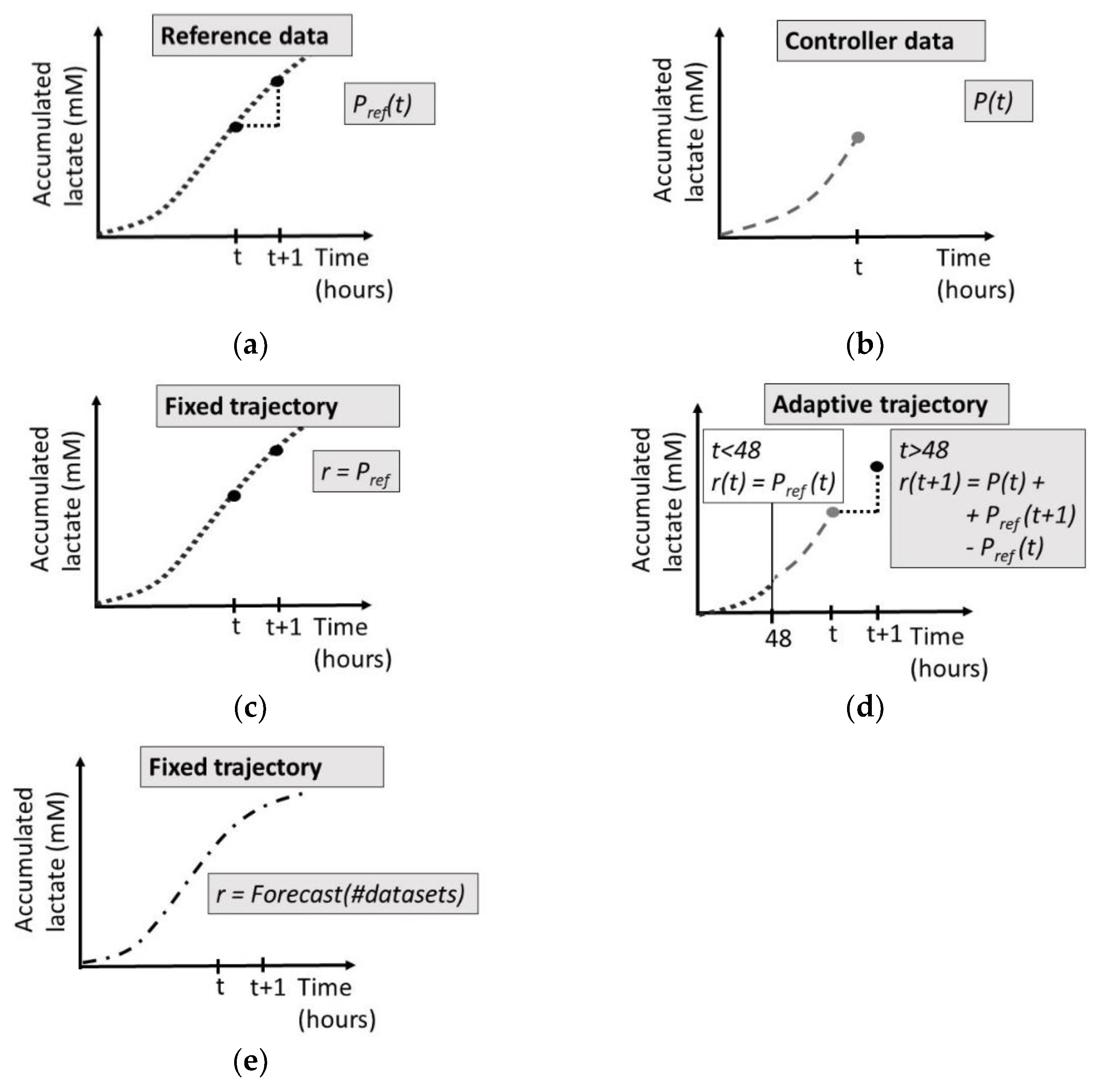
Figure 2.
(a) Reference experimental data, (b) ongoing controller experimental data, (c) fixed target function, defined as the accumulated lactate values generated during one reference experiment, (d) adaptive target function, starting after 48 h, combining the measured accumulated lactate value of the ongoing controller experiment at time t together with the increase in accumulated lactate values of the reference experiment between time t and t + 1, (e) fixed target function, defined by a forecast of the accumulated lactate based on multiple previously gathered datasets.
Figure 2.
(a) Reference experimental data, (b) ongoing controller experimental data, (c) fixed target function, defined as the accumulated lactate values generated during one reference experiment, (d) adaptive target function, starting after 48 h, combining the measured accumulated lactate value of the ongoing controller experiment at time t together with the increase in accumulated lactate values of the reference experiment between time t and t + 1, (e) fixed target function, defined by a forecast of the accumulated lactate based on multiple previously gathered datasets.
Figure 3.
Bright field microscopic images of the hPDCs cultured in tissue flasks (growing on the bottom of the tissue flasks): (a) low cell culture confluency one day after cell seeding, (b) medium cell culture confluency four days after cell seeding, (c) high cell culture confluency seven days after cell seeding.
Figure 3.
Bright field microscopic images of the hPDCs cultured in tissue flasks (growing on the bottom of the tissue flasks): (a) low cell culture confluency one day after cell seeding, (b) medium cell culture confluency four days after cell seeding, (c) high cell culture confluency seven days after cell seeding.
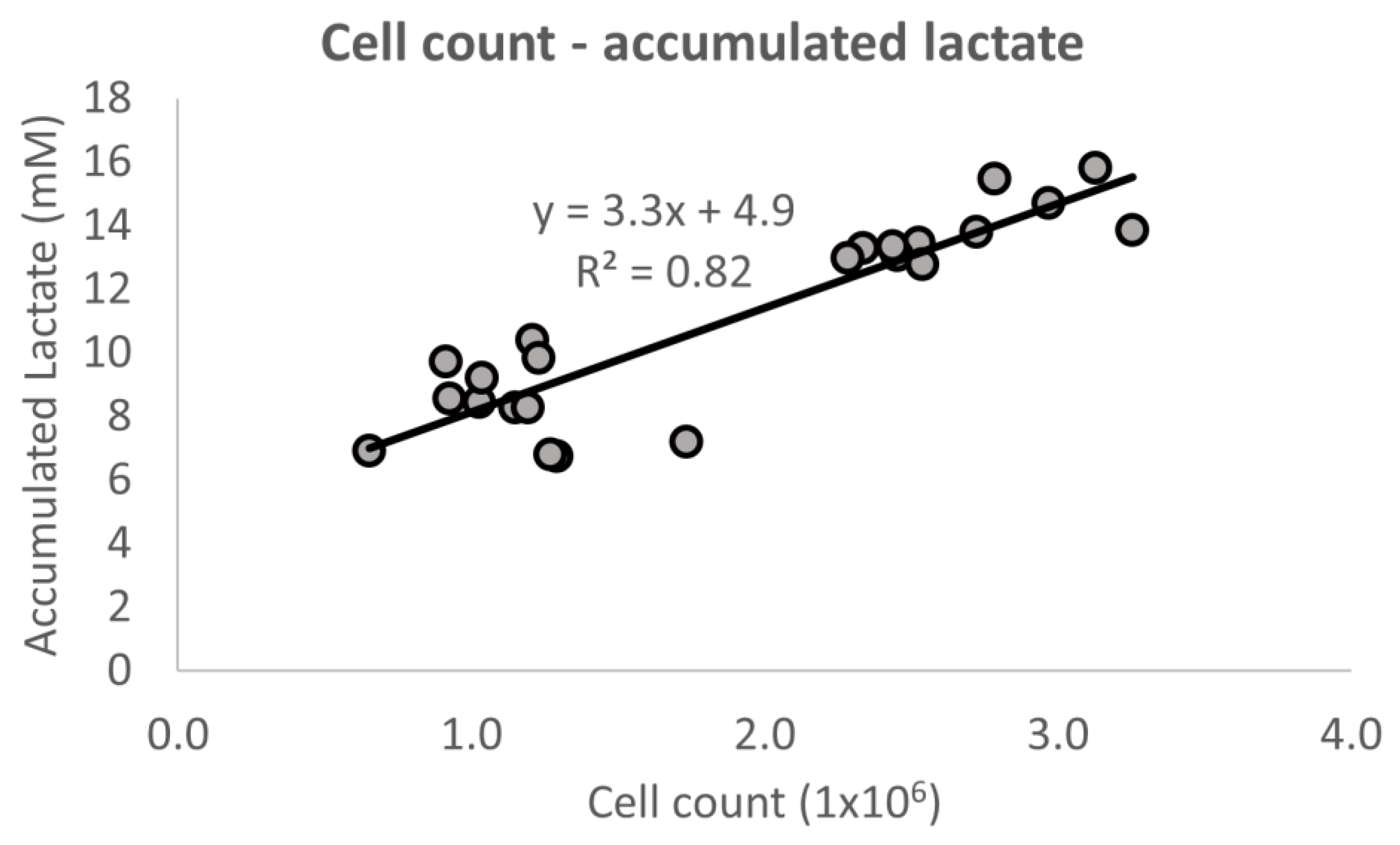
Figure 4.
Regression between the number of cells counted and the total amount of accumulated lactate produced at the end of the cell culture expansion. The data are gathered from both the reference experiment as well as the two controller experiments.
Figure 4.
Regression between the number of cells counted and the total amount of accumulated lactate produced at the end of the cell culture expansion. The data are gathered from both the reference experiment as well as the two controller experiments.
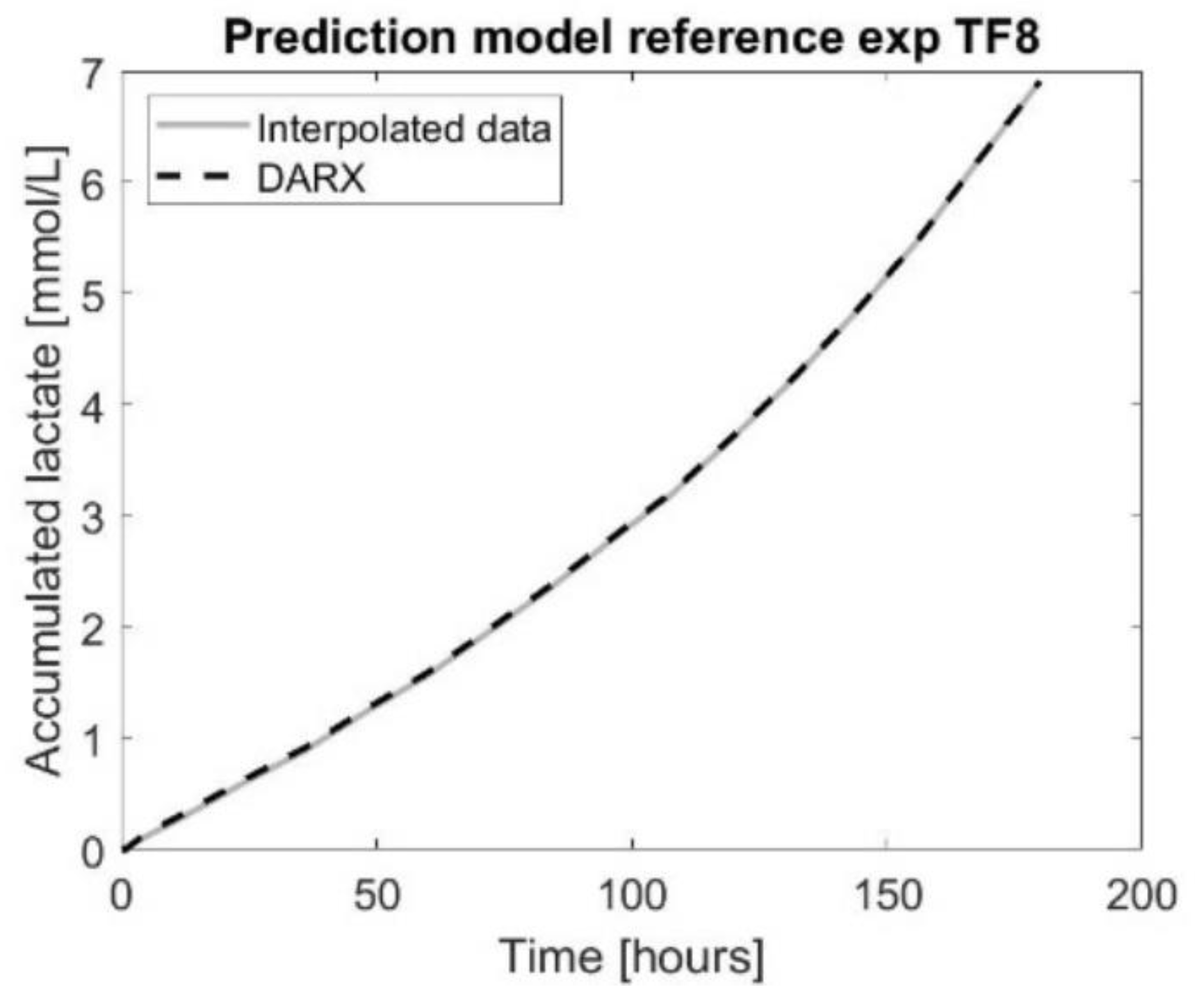
Figure 5.
The prediction model using a dynamic auto-regressive exogenous (DARX) [1 1 1] model for accumulated lactate (mM) compared to the interpolated data for the experimental data of tissue flask eight of the reference experiment.
Figure 5.
The prediction model using a dynamic auto-regressive exogenous (DARX) [1 1 1] model for accumulated lactate (mM) compared to the interpolated data for the experimental data of tissue flask eight of the reference experiment.
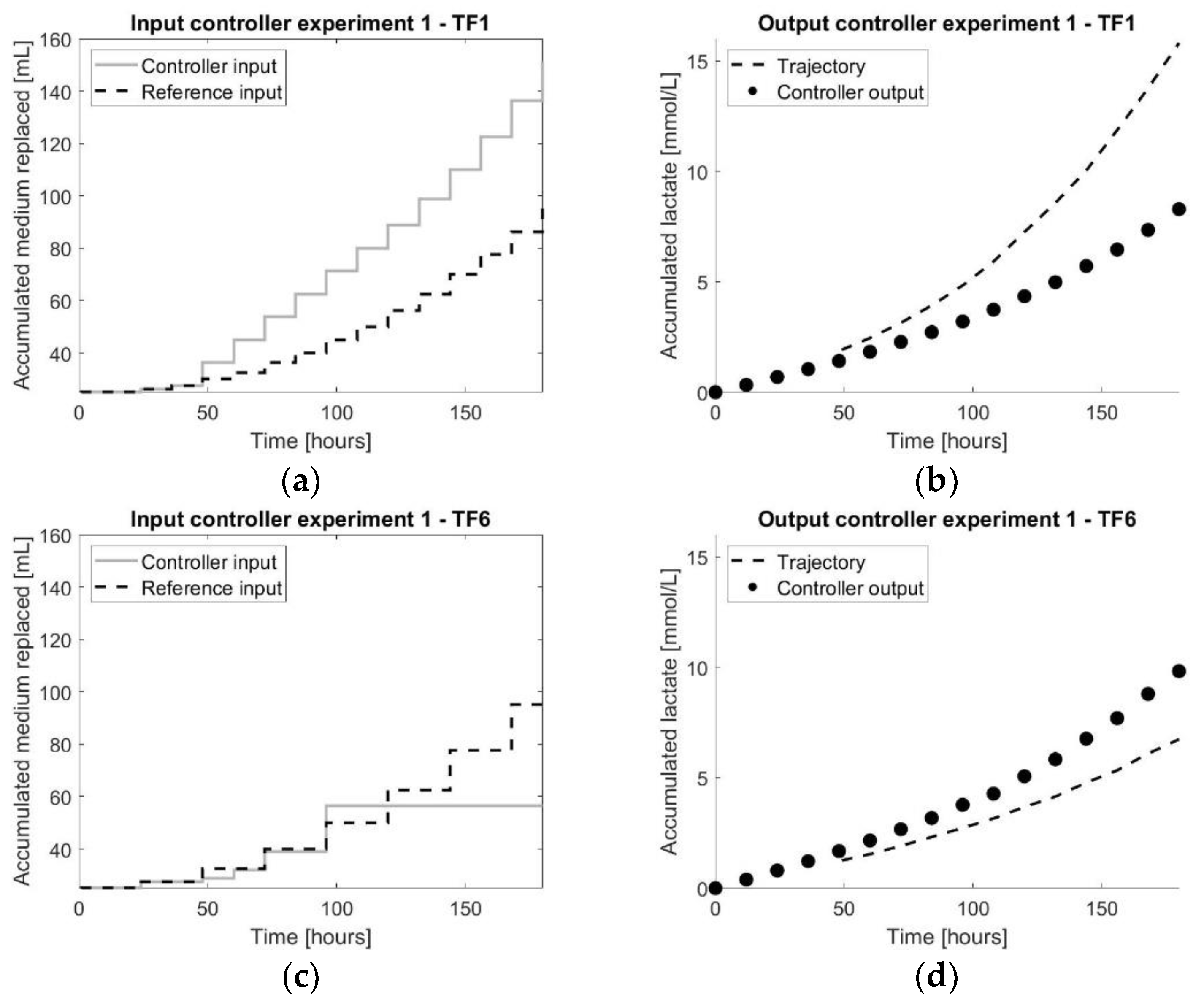
Figure 6.
The results of the first controller experiment: (a,c) representing both the input values of the controller experiment as well as the reference experiment for TF1 and TF6, respectively; (b,d) representing both the output measurements of the controller experiment as well as the reference experiment for TF1 and TF6, respectively.
Figure 6.
The results of the first controller experiment: (a,c) representing both the input values of the controller experiment as well as the reference experiment for TF1 and TF6, respectively; (b,d) representing both the output measurements of the controller experiment as well as the reference experiment for TF1 and TF6, respectively.
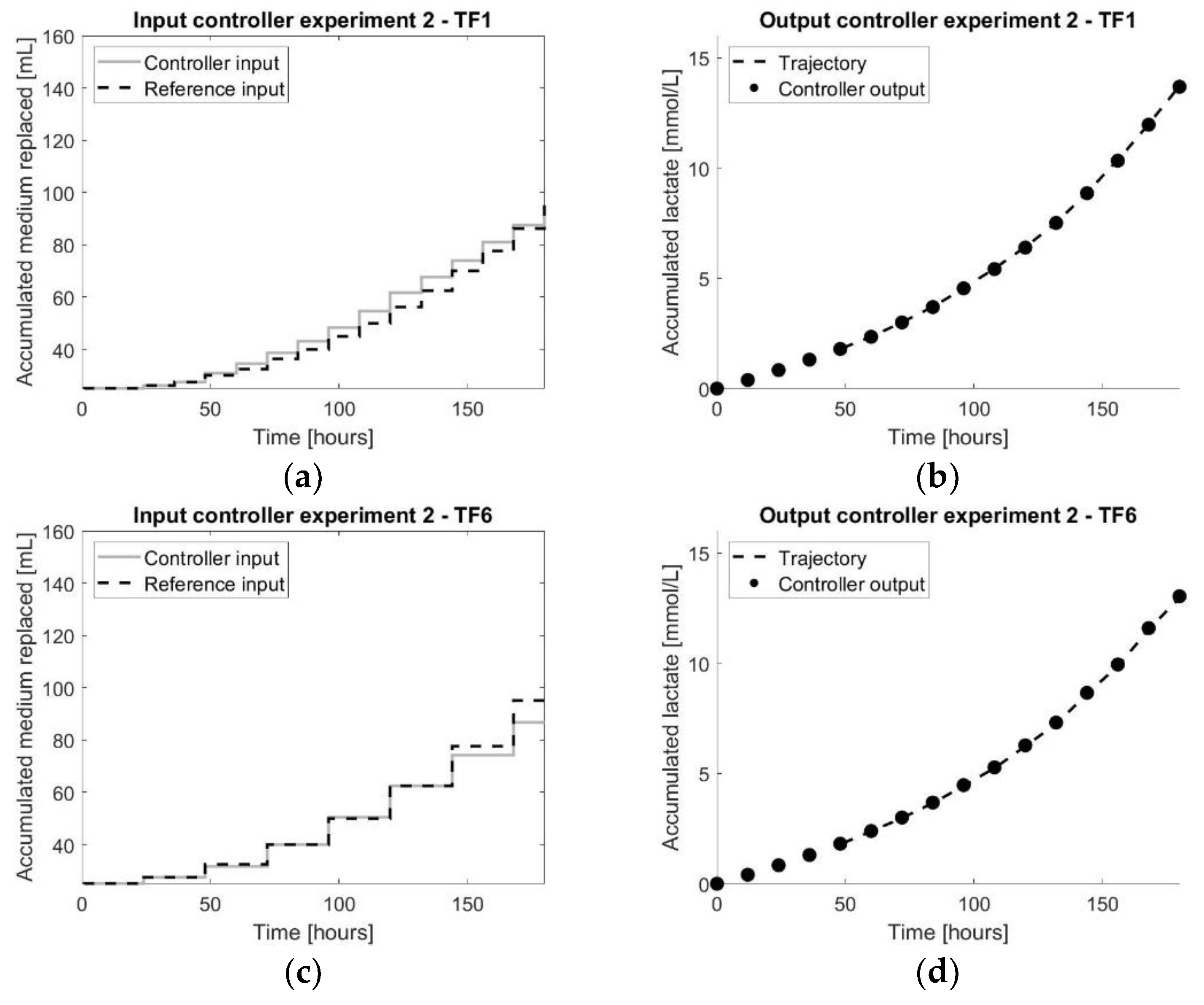
Figure 7.
The results of the second controller experiment: (a,c) representing both the input values of the controller experiment as well as the reference experiment for TF1 and TF6, respectively; (b,d) representing both the output measurements of the controller experiment as well as the reference experiment for TF1 and TF6, respectively.
Figure 7.
The results of the second controller experiment: (a,c) representing both the input values of the controller experiment as well as the reference experiment for TF1 and TF6, respectively; (b,d) representing both the output measurements of the controller experiment as well as the reference experiment for TF1 and TF6, respectively.
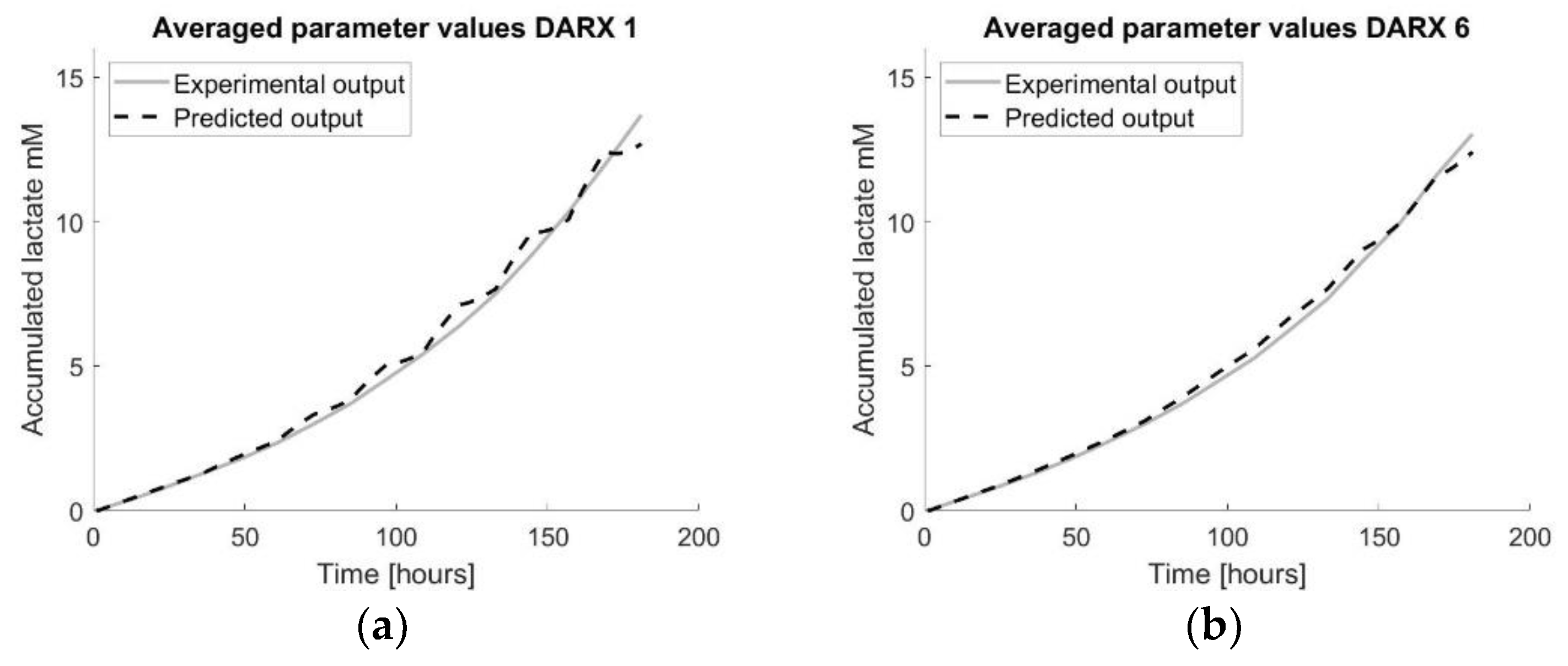
Figure 8.
Visualisation of the prediction models using multiple datasets by applying a transfer function model for all datasets and averaging the model parameters a and b into one average transfer function model. The number in the title refers to the number of the tissue flask used as the test set. (a) visualizes the prediction model developed using the average parameter values obtained by applying DARX transfer function models on each training set using TF1 as the test set. (b) visualizes the prediction model developed using the average parameter values obtained by applying DARX transfer function models on each training set using TF6 as the test set.
Figure 8.
Visualisation of the prediction models using multiple datasets by applying a transfer function model for all datasets and averaging the model parameters a and b into one average transfer function model. The number in the title refers to the number of the tissue flask used as the test set. (a) visualizes the prediction model developed using the average parameter values obtained by applying DARX transfer function models on each training set using TF1 as the test set. (b) visualizes the prediction model developed using the average parameter values obtained by applying DARX transfer function models on each training set using TF6 as the test set.
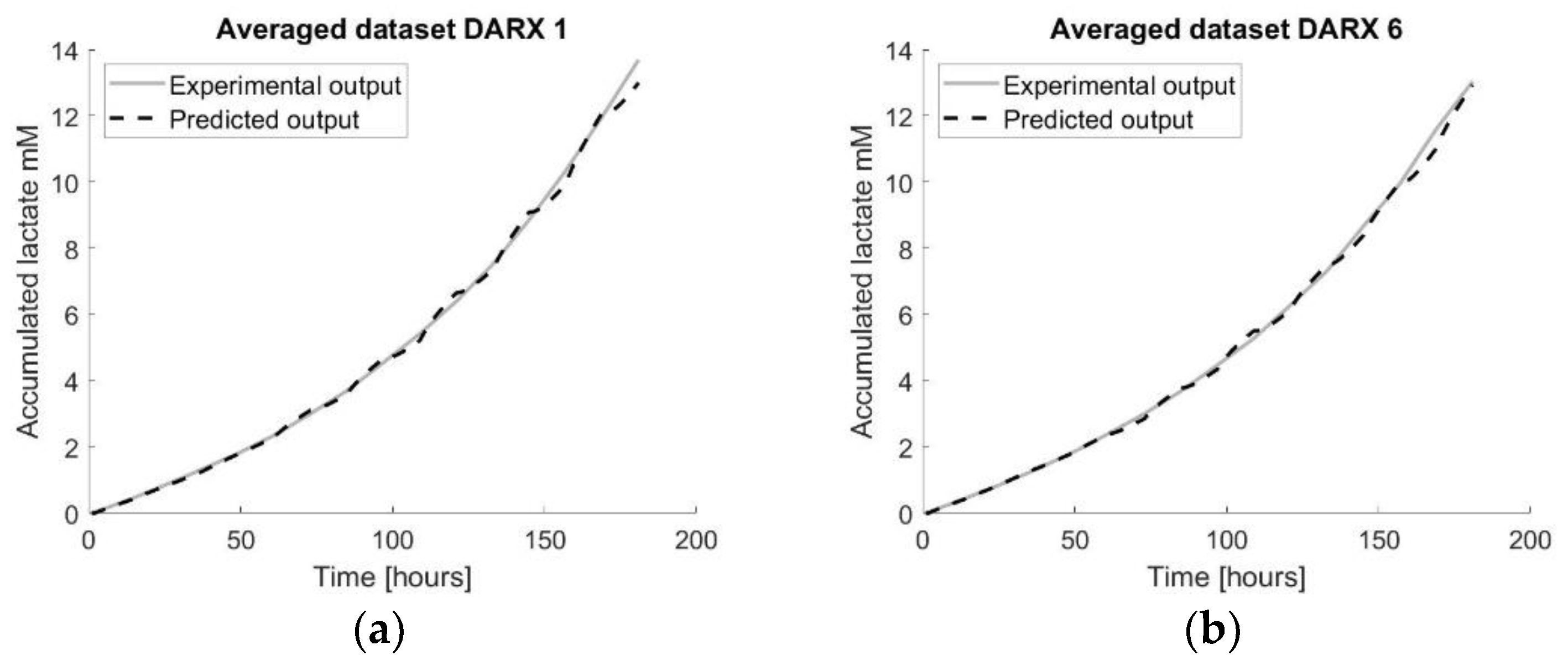
Figure 9.
Visualisation of the forecast models using multiple datasets by applying a transfer function on the averaged dataset. The number in the title refers to the number of the tissue flask used as the test set. (a) visualizes the prediction model developed using the DARX transfer function model on the average of the whole training set with TF1 as the test set. (b) visualizes the prediction model developed using the DARX transfer function model on the average of the whole training set with TF6 as the test set.
Figure 9.
Visualisation of the forecast models using multiple datasets by applying a transfer function on the averaged dataset. The number in the title refers to the number of the tissue flask used as the test set. (a) visualizes the prediction model developed using the DARX transfer function model on the average of the whole training set with TF1 as the test set. (b) visualizes the prediction model developed using the DARX transfer function model on the average of the whole training set with TF6 as the test set.
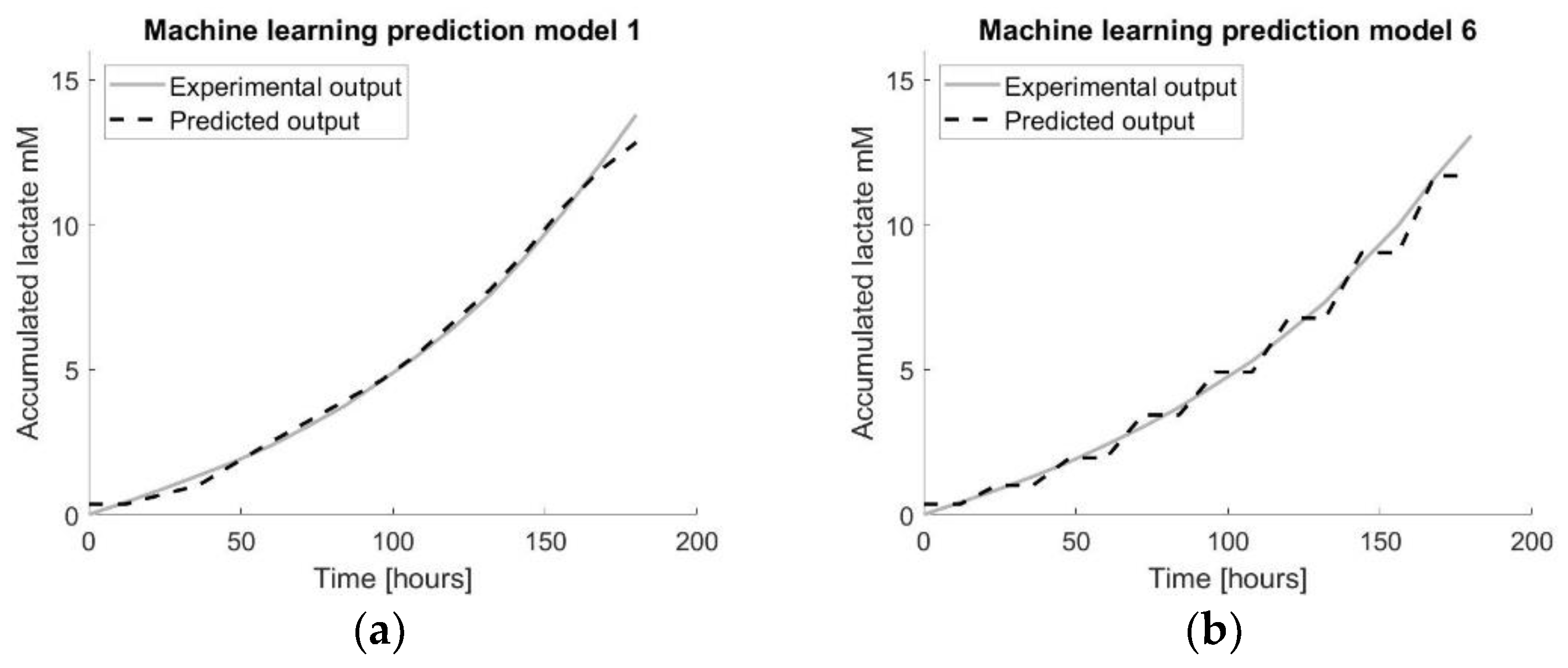
Figure 10.
Visualisation of the prediction models using multiple datasets by applying machine learning on all datasets. The number in the title refers to the number of the tissue flask used as the test set. (a) visualizes the prediction model developed using RBF LS-SVM with TF1 (from the dataset) as the test set. (b) visualizes the prediction model developed using RBF LS-SVM with TF6 (from the dataset) as the test set.
Figure 10.
Visualisation of the prediction models using multiple datasets by applying machine learning on all datasets. The number in the title refers to the number of the tissue flask used as the test set. (a) visualizes the prediction model developed using RBF LS-SVM with TF1 (from the dataset) as the test set. (b) visualizes the prediction model developed using RBF LS-SVM with TF6 (from the dataset) as the test set.
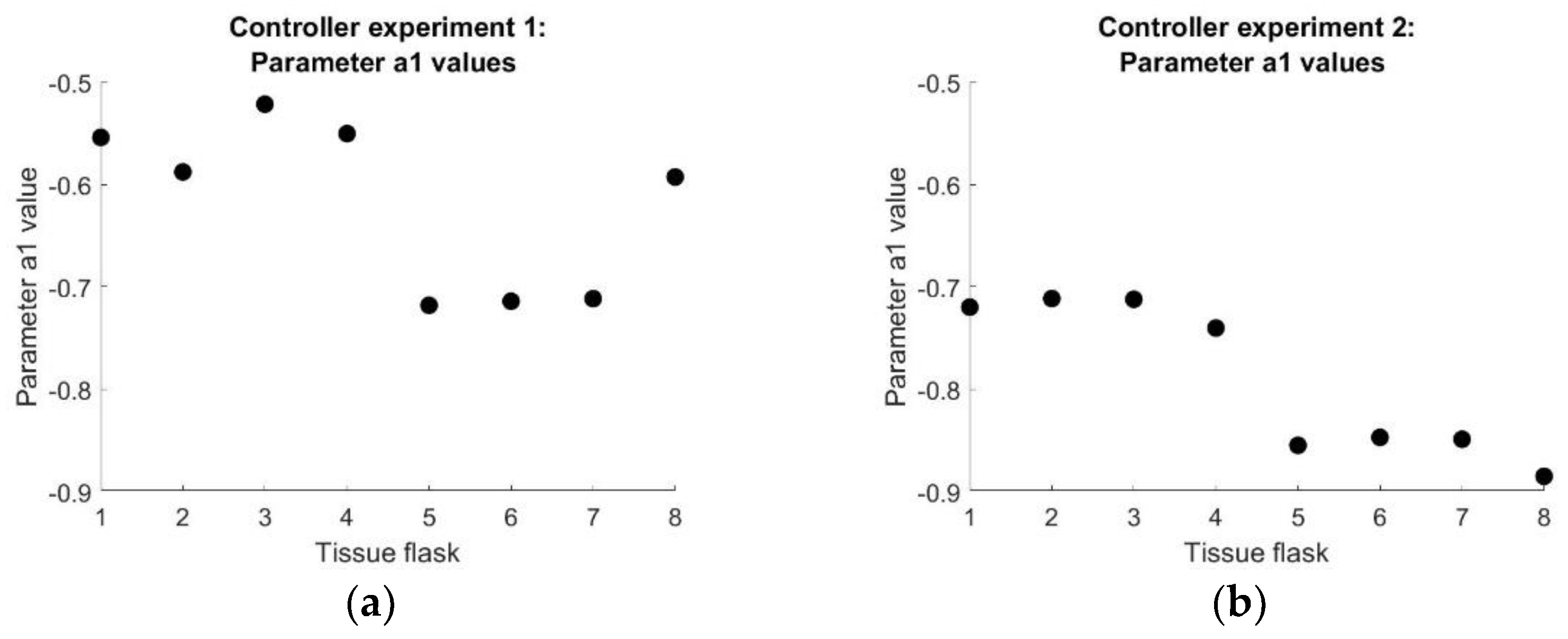
Figure 11.
(a) The fixed parameter of the resulting DARX model for all tissue flasks of controller experiment 1. (b) The fixed parameter of the resulting DARX model for all tissue flasks of controller experiment 2.
Figure 11.
(a) The fixed parameter of the resulting DARX model for all tissue flasks of controller experiment 1. (b) The fixed parameter of the resulting DARX model for all tissue flasks of controller experiment 2.
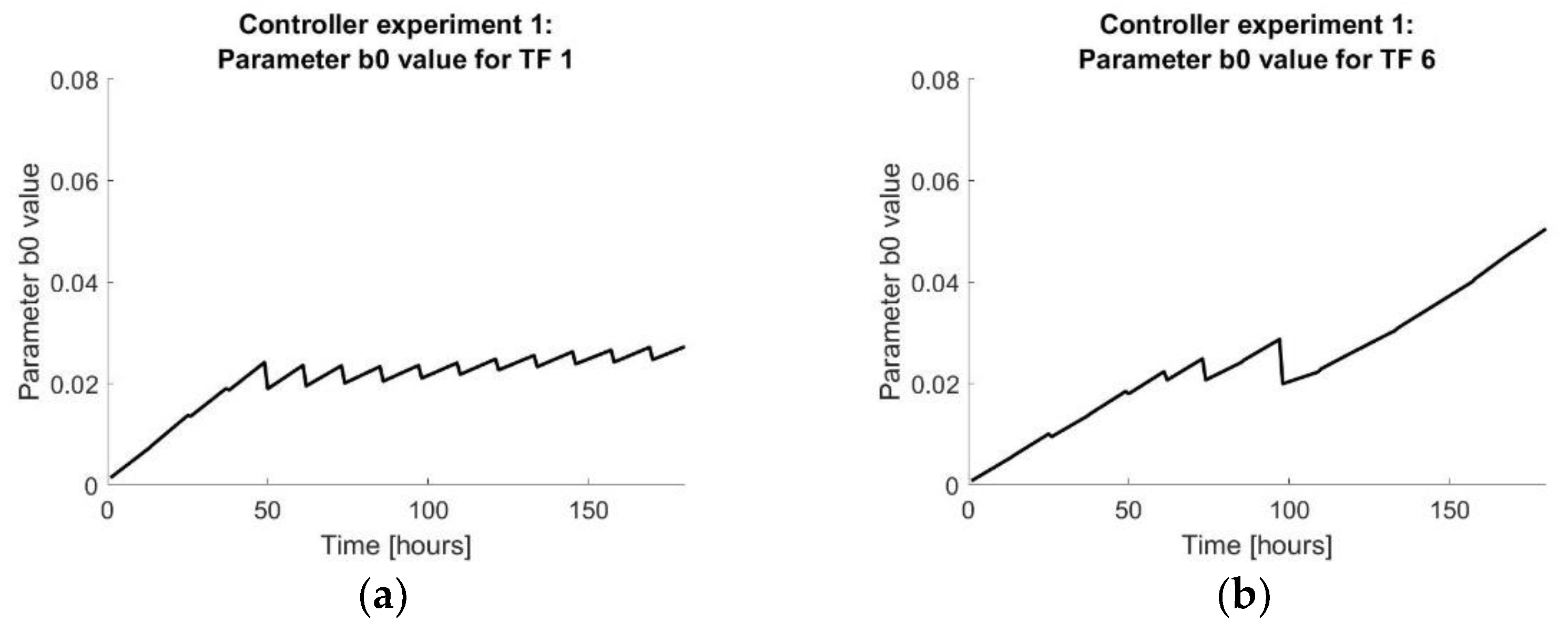
Figure 12.
(a) The value over time of the resulting DARX model for tissue flasks 1 of controller experiment 1. (b) The value over time of the resulting DARX model for tissue flasks 6 of controller experiment 1. (c) The value over time of the resulting DARX model for tissue flasks 1 of controller experiment 2. (d) The value over time of the resulting DARX model for tissue flasks 6 of controller experiment 2.
Figure 12.
(a) The value over time of the resulting DARX model for tissue flasks 1 of controller experiment 1. (b) The value over time of the resulting DARX model for tissue flasks 6 of controller experiment 1. (c) The value over time of the resulting DARX model for tissue flasks 1 of controller experiment 2. (d) The value over time of the resulting DARX model for tissue flasks 6 of controller experiment 2.
Table 1.
Medium replacement scheme of the reference experiment indicating the percentage and volume of medium replaced at each time point, where a 100% medium replacement is equal to 25 mL.
Table 1.
Medium replacement scheme of the reference experiment indicating the percentage and volume of medium replaced at each time point, where a 100% medium replacement is equal to 25 mL.
| | | A (TF 1, 2) | B (TF 3, 4) | C (TF 5, 6) | D (TF 7, 8) |
|---|
| | Hours | % | mL | % | mL | % | mL | % | mL |
|---|
| Day 1 | 24 | 5 | 1.25 | 5 | 1.25 | 10 | 2.5 | 10 | 2.5 |
| | 36 | 5 | 1.25 | 5 | 1.25 | - | | - | |
| Day 2 | 48 | 10 | 2.5 | 7.5 | 1.875 | 20 | 5 | 15 | 3.75 |
| | 60 | 10 | 2.5 | 7.5 | 1.875 | - | | - | |
| Day 3 | 72 | 15 | 3.75 | 10 | 2.5 | 30 | 7.5 | 20 | 5 |
| | 84 | 15 | 3.75 | 10 | 2.5 | - | | - | |
| Day 4 | 96 | 20 | 5 | 12.5 | 3.125 | 40 | 10 | 25 | 6.25 |
| | 108 | 20 | 5 | 12.5 | 3.125 | - | | - | |
| Day 5 | 120 | 25 | 6.25 | 15 | 3.75 | 50 | 12.5 | 30 | 7.5 |
| | 132 | 25 | 6.25 | 15 | 3.75 | - | | - | |
| Day 6 | 144 | 30 | 7.5 | 17.5 | 4.375 | 60 | 15 | 35 | 8.75 |
| | 156 | 30 | 7.5 | 17.5 | 4.375 | - | | - | |
| Day 7 | 168 | 35 | 8.75 | 20 | 5 | 70 | 17.5 | 40 | 10 |
| | 180 | 35 | 8.75 | 20 | 5 | - | | - | |
Table 2.
Scheme of the frequency and amount of medium replacements of the different tissue flasks of the reference experiment. The total amount is the sum of the initial 25 mL and all replacements as described in
Table 1.
Table 2.
Scheme of the frequency and amount of medium replacements of the different tissue flasks of the reference experiment. The total amount is the sum of the initial 25 mL and all replacements as described in
Table 1.
| Tissue Flasks | Frequency | Total Amount |
|---|
| TF 1 & 2 | Every 12 h | 95 mL |
| TF 3 & 4 | Every 12 h | 68.75 mL |
| TF 5 & 6 | Every 24 h | 95 mL |
| TF 7 & 8 | Every 24 h | 68.75 mL |
Table 3.
Scheme to indicate which reference data were used in the target function for the two controller experiments.
Table 3.
Scheme to indicate which reference data were used in the target function for the two controller experiments.
| Controller Experiment 1 | Controller Experiment 2 |
|---|
| Target Function | TF Controller | Target Function | TF Controller |
|---|
| TF1REF | TF1 | TF1REF + TF1 | TF1 |
| TF2REF | TF2 | TF1REF + TF2 | TF2 |
| TF3REF | TF3 | TF1REF + TF3 | TF3 |
| TF4REF | TF4 | Control: TF1REF | TF4 |
| TF5REF | TF5 | TF6REF + TF5 | TF5 |
| TF6REF | TF6 | TF6REF + TF6 | TF6 |
| TF7REF | TF7 | TF6REF + TF7 | TF7 |
| TF8REF | TF8 | Control: TF6REF | TF8 |
Table 4.
Cell expansion results from the reference experiment. Number of cells counted, total amount of accumulated lactate produced (mM), total amount of medium supplied (mL) and the overall medium efficiency (cells/mL).
Table 4.
Cell expansion results from the reference experiment. Number of cells counted, total amount of accumulated lactate produced (mM), total amount of medium supplied (mL) and the overall medium efficiency (cells/mL).
| | TF1 | TF2 | TF3 | TF4 | TF5 | TF6 | TF7 | TF8 |
|---|
| Cells counted (1.0 × 106) | 3.13 | 2.79 | 3.26 | 2.97 | 1.74 | 1.29 | 1.27 | 0.65 |
| Accumulated lactate (mM) | 15.81 | 15.48 | 13.86 | 14.70 | 7.21 | 6.74 | 6.79 | 6.91 |
| Accumulated medium (mL) | 95.00 | 95.00 | 68.75 | 68.75 | 95.00 | 95.00 | 68.75 | 68.75 |
| Medium efficiency (1.0 × 104 cells/mL) | 3.29 | 2.93 | 4.73 | 4.32 | 1.83 | 1.36 | 1.85 | 0.95 |
Table 5.
Cell expansion results from the first controller experiment. Number of cells counted, total amount of accumulated lactate produced (mM), total amount of medium supplied (mL) and the overall medium efficiency (cells/mL).
Table 5.
Cell expansion results from the first controller experiment. Number of cells counted, total amount of accumulated lactate produced (mM), total amount of medium supplied (mL) and the overall medium efficiency (cells/mL).
| | TF1 | TF2 | TF3 | TF4 | TF5 | TF6 | TF7 | TF8 |
|---|
| Cells counted (1.0 × 106) | 1.15 | 1.20 | 1.03 | 0.93 | 1.21 | 1.23 | 1.04 | 0.92 |
| Accumulated lactate (mM) | 8.29 | 8.27 | 8.45 | 8.55 | 10.40 | 9.83 | 9.22 | 9.73 |
| Accumulated medium (mL) | 151.25 | 96.25 | 96.88 | 64.38 | 79.07 | 56.45 | 73.01 | 53.20 |
| Medium efficiency (1.0 × 104 cells/mL) | 0.76 | 1.24 | 1.06 | 1.44 | 1.53 | 2.18 | 1.42 | 1.72 |
Table 6.
Cell expansion results from the second controller experiment. Number of cells counted, total amount of accumulated lactate produced (mM), total amount of medium supplied (mL) and the overall medium efficiency (cells/mL).
Table 6.
Cell expansion results from the second controller experiment. Number of cells counted, total amount of accumulated lactate produced (mM), total amount of medium supplied (mL) and the overall medium efficiency (cells/mL).
| | TF1 | TF2 | TF3 | TF4 | TF5 | TF6 | TF7 | TF8 |
|---|
| Cells counted (1.0 × 106) | 2.72 | 2.49 | 2.34 | 2.28 | 2.53 | 2.45 | 2.54 | 2.44 |
| Accumulated lactate (mM) | 13.80 | 13.15 | 13.29 | 12.98 | 13.47 | 13.07 | 12.78 | 13.34 |
| Accumulated medium (mL) | 93.79 | 95.71 | 95.53 | 95.00 | 84.71 | 86.83 | 87.73 | 95.00 |
| Medium efficiency (1.0 × 104 cells/mL) | 2.90 | 2.60 | 2.45 | 2.39 | 2.98 | 2.82 | 2.89 | 2.59 |
Table 7.
NRMSE, model evaluation of the DARX [1 1 1] model for the data of all eight tissue flasks for all three experiments. A NRMSE equal to zero represents a perfect fit of the DARX model when compared to the experimental data.
Table 7.
NRMSE, model evaluation of the DARX [1 1 1] model for the data of all eight tissue flasks for all three experiments. A NRMSE equal to zero represents a perfect fit of the DARX model when compared to the experimental data.
| | NRMSE |
|---|
| | Reference Exp | Controller Exp 1 | Controller Exp 2 |
|---|
| TF1 | 0.0115 | 0.0148 | 0.0088 |
| TF2 | 0.0016 | 0.0048 | 0.0122 |
| TF3 | 0.0074 | 0.0140 | 0.0010 |
| TF4 | 0.0077 | 0.0166 | 0.0021 |
| TF5 | 0.0155 | 0.0017 | 0.0130 |
| TF6 | 0.0021 | 0.0033 | 0.0038 |
| TF7 | 0.0027 | 0.0010 | 0.0142 |
| TF8 | 0.0093 | 0.0010 | 0.0023 |
Table 8.
NRMSE values comparing the accumulated lactate output values of the controller experiment one with the trajectory values using the goodnessOfFit function in MATLAB. The NRMSE closer to zero represents the better fit.
Table 8.
NRMSE values comparing the accumulated lactate output values of the controller experiment one with the trajectory values using the goodnessOfFit function in MATLAB. The NRMSE closer to zero represents the better fit.
| | TF1 | TF2 | TF3 | TF4 | TF5 | TF6 | TF7 | TF8 |
|---|
| NRMSE | 0.9107 | 0.9091 | 0.8739 | 0.8485 | 0.9125 | 1.0486 | 0.7550 | 0.9637 |
Table 9.
NRMSE values comparing the accumulated lactate output values of the controller experiment two with the trajectory values using the goodnessOfFit function in MATLAB. The NRMSE closer to zero represents the better fit.
Table 9.
NRMSE values comparing the accumulated lactate output values of the controller experiment two with the trajectory values using the goodnessOfFit function in MATLAB. The NRMSE closer to zero represents the better fit.
| | TF1 | TF2 | TF3 | TF4 | TF5 | TF6 | TF7 | TF8 |
|---|
| NRMSE | 0.0054 | 0.0071 | 0.0068 | 0.0073 | 0.0149 | 0.0145 | 0.0143 | 0.0147 |
Table 10.
NRMSE comparing the accumulated lactate output values of the forecast model using multiple datasets by applying DARX transfer function models on the different datasets and averaging the parameters using the goodnessOfFit function in MATLAB. The NRMSE closer to zero represents the better fit. The number of the tissue flasks referrers to the tissue flask used as the test data set.
Table 10.
NRMSE comparing the accumulated lactate output values of the forecast model using multiple datasets by applying DARX transfer function models on the different datasets and averaging the parameters using the goodnessOfFit function in MATLAB. The NRMSE closer to zero represents the better fit. The number of the tissue flasks referrers to the tissue flask used as the test data set.
| | TF1 | TF2 | TF3 | TF4 | TF5 | TF6 | TF7 | TF8 |
|---|
| NRMSE | 0.0846 | 0.1441 | 0.1319 | 0.0647 | 0.0752 | 0.0647 | 0.0927 | 0.0023 |
Table 11.
NRMSE comparing the accumulated lactate output values of the forecast model using multiple datasets by applying DARX transfer function models on the averaged dataset using the goodnessOfFit function in MATLAB. The NRMSE closer to zero represents the better fit.
Table 11.
NRMSE comparing the accumulated lactate output values of the forecast model using multiple datasets by applying DARX transfer function models on the averaged dataset using the goodnessOfFit function in MATLAB. The NRMSE closer to zero represents the better fit.
| | TF1 | TF2 | TF3 | TF4 | TF5 | TF6 | TF7 | TF8 |
|---|
| NRMSE | 0.0439 | 0.0932 | 0.0876 | 0.0443 | 0.1037 | 0.0431 | 0.0682 | 0.0849 |
Table 12.
NRMSE comparing the accumulated lactate output values of the forecast model using multiple datasets by applying machine learning using the goodnessOfFit function in MATLAB. The NRMSE closer to zero represents the better fit. The number of the tissue flasks referrers to the tissue flask used as the test data set.
Table 12.
NRMSE comparing the accumulated lactate output values of the forecast model using multiple datasets by applying machine learning using the goodnessOfFit function in MATLAB. The NRMSE closer to zero represents the better fit. The number of the tissue flasks referrers to the tissue flask used as the test data set.
| | TF1 | TF2 | TF3 | TF4 | TF5 | TF6 | TF7 | TF8 |
|---|
| NRMSE | 0.0917 | 0.1292 | 0.1094 | 0.0644 | 0.1944 | 0.1300 | 0.1145 | 0.1740 |