Adjusting the Structure of a Peptide Nucleic Acid (PNA) Molecular Beacon and Promoting Its DNA Detection by a Hybrid with Quencher-Modified DNA
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
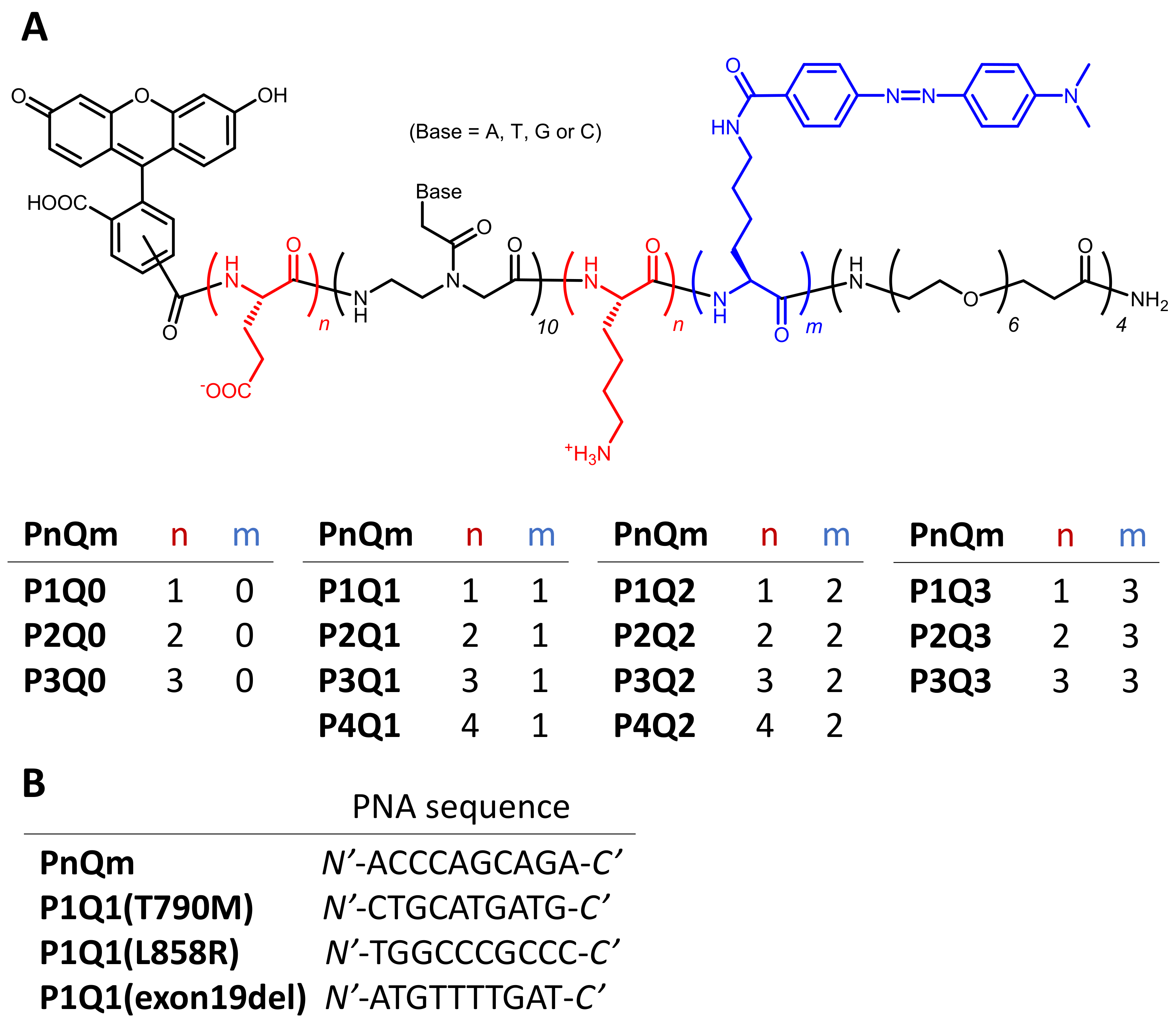
2.2. Peptide Synthesis
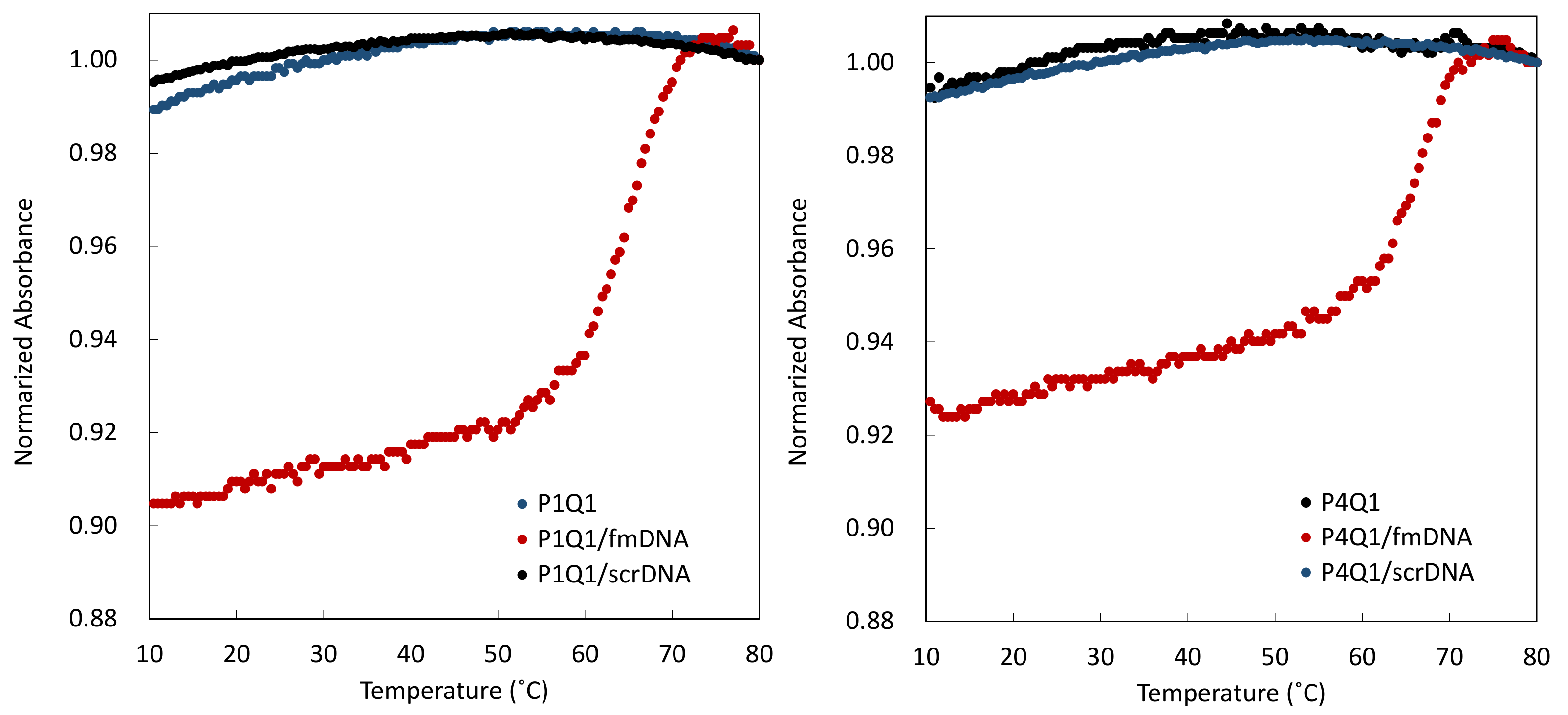
2.3. Melting Curves of Mixtures of PNA Beacon and DNA
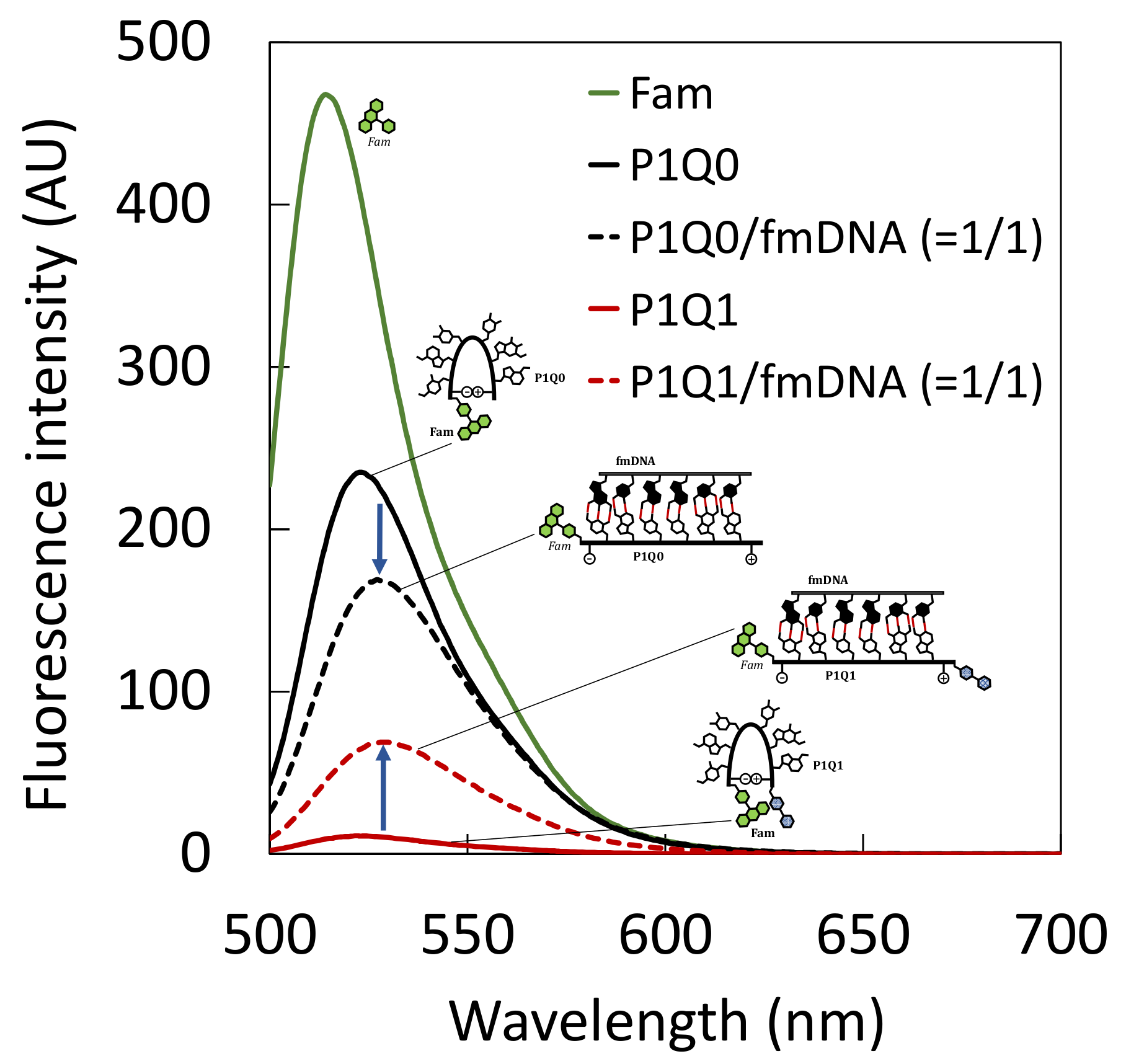
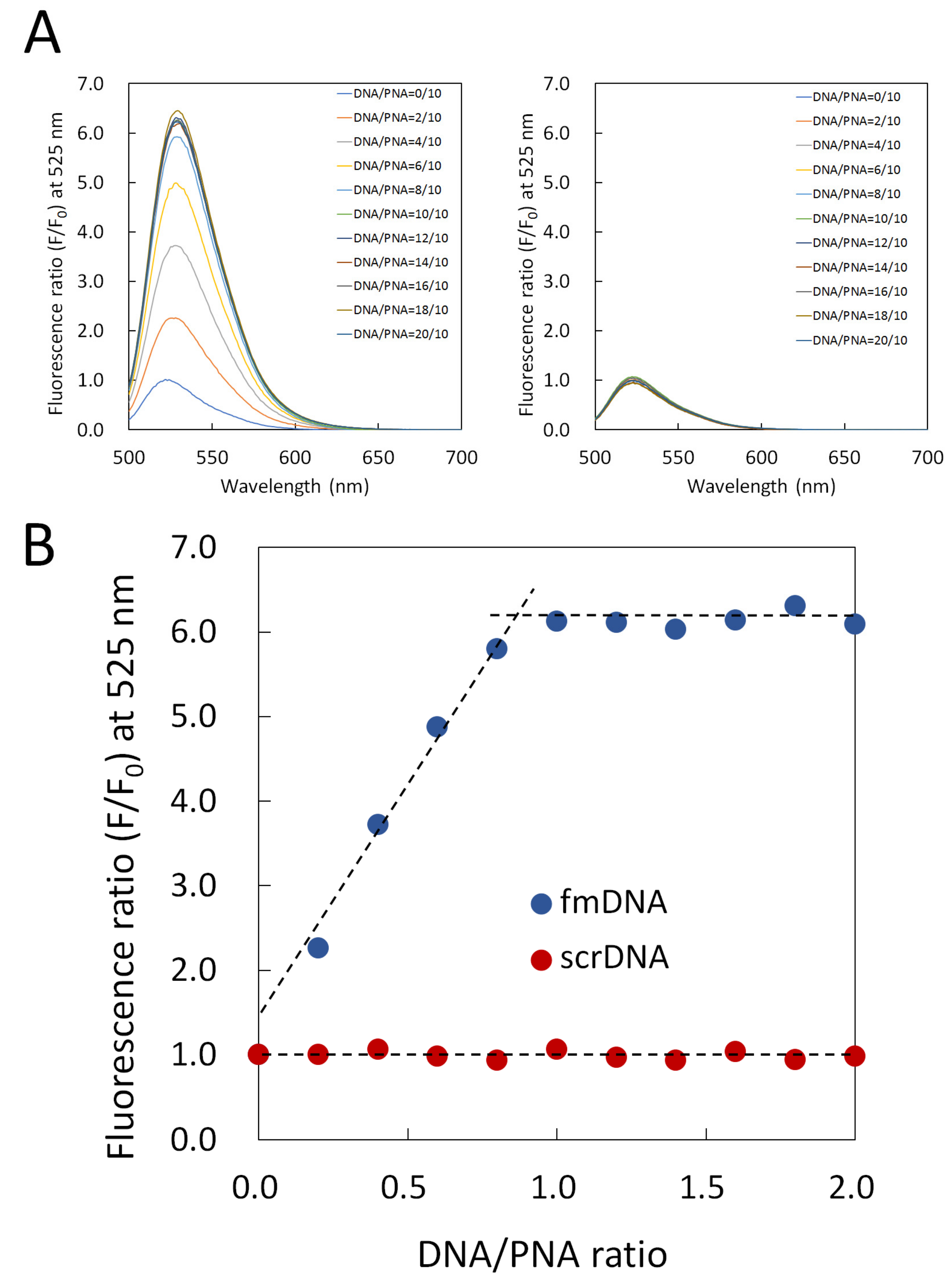
2.4. Fluorescence Titration Curves of PNA Beacon and DNA
2.5. Relative Fluorescence of PNA Beacon and PNA Beacon/Q-DNA Hybrid with Target DNA
3. Results
3.1. Hybrid Formation of DNA with PNA Beacon and Fluorescence Detection of DNA by PNA Beacon
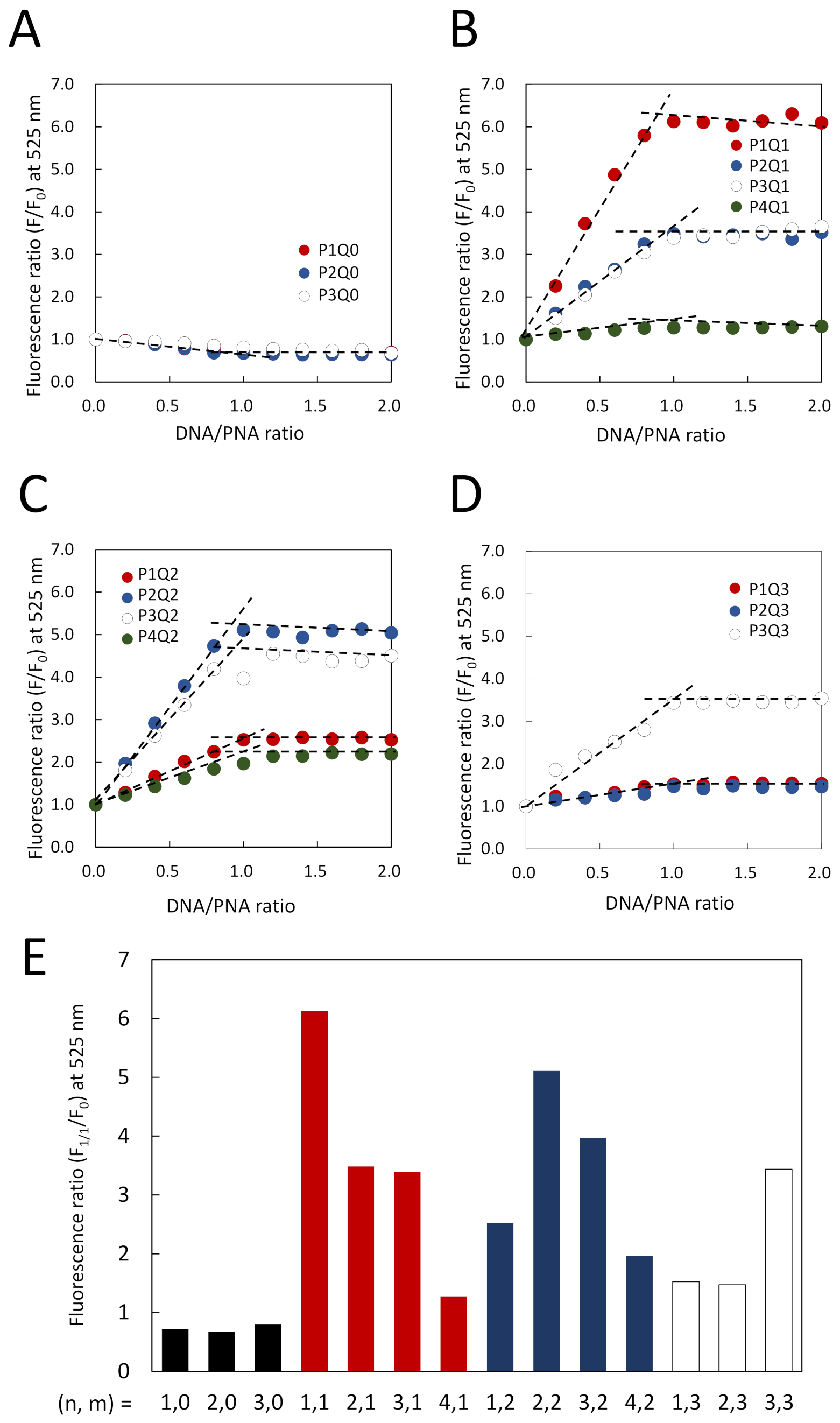
3.2. Elaborate Adjustment of PnQms for Fluorescence Detection of DNA
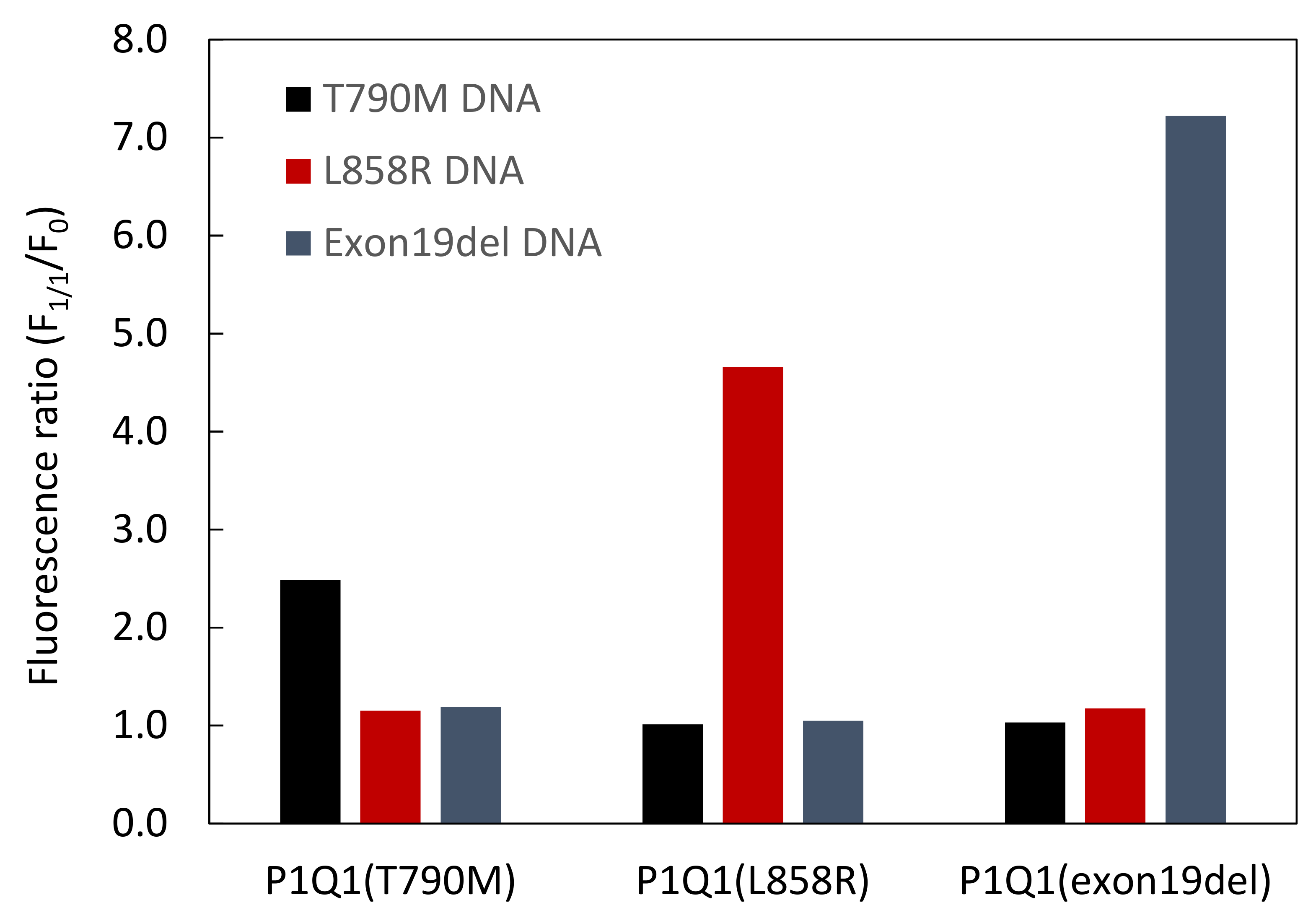
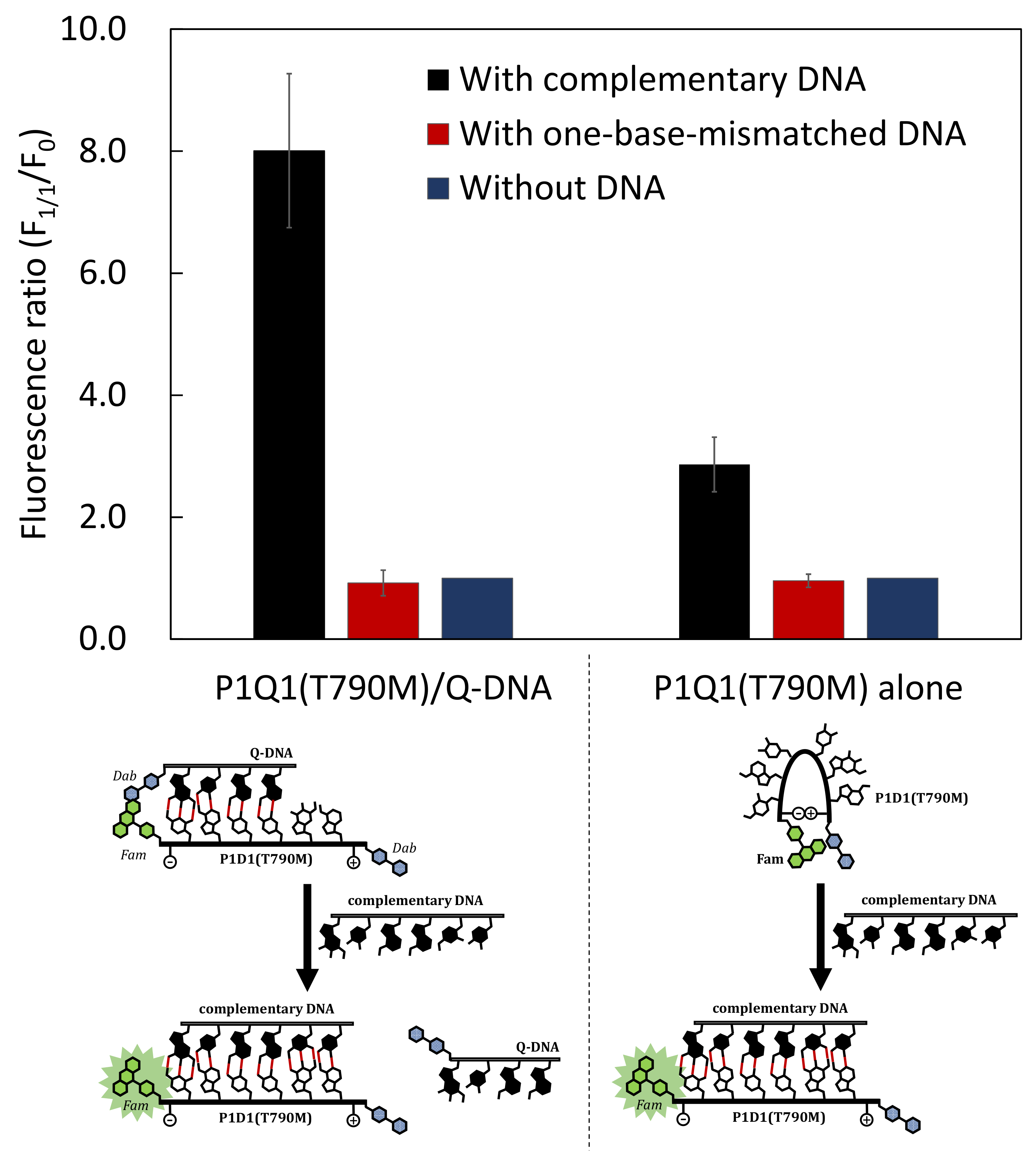
3.3. Comparison of Fluorescence Detection for Target DNA of PNA Beacon and PNA Beacon/Quencher-Modified DNA Hybrid
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Nielsen, P.E.; Egholm, M.; Berg, R.H.; Buchardt, O. Sequence-Selective Recognition of DNA by Strand Displacement with a Thymine-Substituted Polyamide. Science 1991, 254, 1497–1500. [Google Scholar] [CrossRef]
- Egholm, M.; Buchardt, O.; Christensen, L.; Behrens, C.; Freier, S.M.; Driver, D.A.; Berg, R.H.; Kim, S.K.; Norden, B.; Nielsen, P.E. PNA Hybridizes to Complementary Oligonucleotides Obeying the Watson-Crick Hydrogen Bonding Rules. Nature 1993, 365, 566–568. [Google Scholar] [CrossRef]
- Smith, J.O.; Olson, D.A.; Armitage, B.A. Molecular Recognition of PNA-Containing Hybrids: Spontaneous Assembly of Helical Cyanine Dye Aggregates on PNA Templates. J. Am. Chem. Soc. 1999, 121, 2686–2695. [Google Scholar] [CrossRef]
- Datta, B.; Armitage, B.A. Hybridization of PNA to Structured DNA Targets: Quadruplex Invasion and the Overhang Effect. J. Am. Chem. Soc. 2001, 123, 9612–9619. [Google Scholar] [CrossRef]
- Kushon, S.A.; Jordan, J.P.; Seifert, J.L.; Nielsen, H.; Nielsen, P.E.; Armitage, B.A. Effect of Secondary Structure on the Thermodynamics and Kinetics of PNA Hybridization to DNA Hairpins. J. Am. Chem. Soc. 2001, 123, 10805–10813. [Google Scholar] [CrossRef]
- Renikuntla, B.R.; Armitage, B.A. Role Reversal in a Supramolecular Assembly: A Chiral Cyanine Dye Controls the Helicity of a Peptide-Nucleic Acid Duplex. Langmuir 2005, 21, 5362–5366. [Google Scholar] [CrossRef]
- Kanjanawarut, R.; Su, X. Colorimetric Detection of DNA Using Unmodified Metallic Nanoparticles and Peptide Nucleic Acid Probes. Anal. Chem. 2009, 81, 6122–6129. [Google Scholar] [CrossRef]
- Su, X.; Kanjanawarut, R. Control of Metal Nanoparticles Aggregation and Dispersion by PNA and PNA-DNA Complexes, and its Application for Colorimetric DNA Detection. ACS Nano 2009, 3, 2751–2759. [Google Scholar] [CrossRef]
- Ali, M.M.; Li, Y. Colorimetric Sensing by Using Allosteric-DNAzyme-Coupled Rolling Circle Amplification and a Peptide Nucleic Acid–Organic Dye Probe. Angew. Chem. Int. Ed. 2009, 48, 3512–3515. [Google Scholar] [CrossRef]
- Xu, M.; Xing, S.; Zhao, Y.; Zhao, C. Peptide Nucleic Acid-Assisted Colorimetric Detection of Single-nucleotide Polymorphisms Based on the Intrinsic Peroxidase-like Activity of Hemin-Carbon Nanotube Nanocomposites. Talanta 2021, 232, 122420–122427. [Google Scholar] [CrossRef]
- Wang, J.; Nielsen, P.E.; Jiang, M.; Cai, X.; Fernandes, J.R.; Grant, D.H.; Ozsoz, M.; Beglieter, A.; Mowat, M. Mismatch-Sensitive Hybridization Detection by Peptide Nucleic Acids Immobilized on a Quartz Crystal Microbalance. Anal. Chem. 1997, 69, 5200–5202. [Google Scholar] [CrossRef]
- Ross, P.L.; Lee, K.; Belgrader, P. Discrimination of Single-Nucleotide Polymorphisms in Human DNA Using Peptide Nucleic Acid Probes Detected by MALDI-TOF Mass Spectrometry. Anal. Chem. 1997, 69, 4197–4202. [Google Scholar] [CrossRef]
- Chakrabarti, R.; Klibanov, A.M. Nanocrystals Modified with Peptide Nucleic Acids (PNAs) for Selective Self-Assembly and DNA Detection. J. Am. Chem. Soc. 2003, 125, 12531–12540. [Google Scholar] [CrossRef]
- Wang, J.; Palecek, E.; Nielsen, P.E.; Rivas, G.; Cai, X.; Shiraishi, H.; Dontha, N.; Luo, D.; Farias, P.A.M. Peptide Nucleic Acid Probes for Sequence-Specific DNA Biosensors. J. Am. Chem. Soc. 1996, 118, 7667–7670. [Google Scholar] [CrossRef]
- Cai, B.; Wang, S.; Huang, L.; Ning, Y.; Zhang, Z.; Zhang, G.-J. Ultrasensitive Label-Free Detection of PNA-DNA Hybridization by Reduced Graphene Oxide Field-Effect Transistor Biosensor. ACS Nano 2014, 8, 2632–2638. [Google Scholar] [CrossRef]
- Ali, M.; Neumann, R.; Ensinger, W. Sequence-Specific Recognition of DNA Oligomer Using Peptide Nucleic Acid (PNA)-Modified Synthetic Ion Channels: PNA/DNA Hybridization in Nanoconfined Environment. ACS Nano 2010, 4, 7267–7274. [Google Scholar] [CrossRef]
- Xuan, F.; Fan, T.W.; Hsing, I.-M. Electrochemical Interrogation of Kinetically-Controlled Dendritic DNA/PNA Assembly for Immobilization-Free and Enzyme-Free Nucleic Acids Sensing. ACS Nano 2015, 9, 5027–5033. [Google Scholar] [CrossRef]
- Brandt, O.; Hoheisel, J.D. Peptide Nucleic Acids on Microarrays and Other Biosensors. Trends Biotechnol. 2004, 22, 617–622. [Google Scholar] [CrossRef]
- Lai, Q.; Chen, W.; Zhang, Y.; Liu, Z. Application Strategies of Peptide Nucleic Acids toward Electrochemical Nucleic Acid Sensors. Analyst 2021, 146, 5822–5835. [Google Scholar] [CrossRef]
- Moccia, M.; Antonacci, A.; Saviano, M.; Caratelli, V.; Arduini, F.; Scognamiglio, V. Emerging Technologies in the Design of Peptide Nucleic Acids (PNAs) Based Biosensors. Trends Analyt. Chem. 2020, 132, 116062–116076. [Google Scholar] [CrossRef]
- Endo, T.; Kerman, K.; Nagatani, N.; Takamura, Y.; Tamiya, E. Label-Free Detection of Peptide Nucleic Acid-DNA Hybridization Using Localized Surface Plasmon Resonance Based Optical Biosensor. Anal. Chem. 2005, 77, 6976–6984. [Google Scholar] [CrossRef] [PubMed]
- Vilaivan, T. Fluorogenic PNA Probes. Beilstein J. Org. Chem. 2018, 14, 253–281. [Google Scholar] [CrossRef] [PubMed]
- Kuhn, H.; Demidov, V.V.; Coull, J.M.; Fiandaca, M.J.; Gildea, B.D.; Frank-Kamenetskii, M.D. Hybridization of DNA and PNA Molecular Beacons to Single-Stranded and Double-Stranded DNA Targets. J. Am. Chem. Soc. 2002, 124, 1097–1103. [Google Scholar] [CrossRef] [PubMed]
- Xi, C.; Balberg, M.; Boppart, S.A.; Raskin, L. Use of DNA and Peptide Nucleic Acid Molecular Beacons for Detection and Quantification of rRNA in Solution and in Whole Cells. Appl. Environ. Microbiol. 2003, 69, 5673–5678. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Smolina, I.V.; Demidov, V.V.; Soldatenkov, V.A.; Chasovskikh, S.G.; Frank-Kamenetskii, M.D. End Invasion of Peptide Nucleic Acids (PNAs) with Mixed-Base Composition into Linear DNA Duplexes. Nucleic Acids Res. 2005, 33, e146. [Google Scholar] [CrossRef]
- Xi, C.; Raskin, L.; Boppart, S.A. Evaluation of Microfluidic Biosensor Development Using Microscopic Analysis of Molecular Beacon Hybridization Kinetics. Biomed. Microdevices 2005, 7, 7–12. [Google Scholar] [CrossRef][Green Version]
- Ye, S.; Miyajima, Y.; Ohnishi, T.; Yamamoto, Y.; Komiyama, M. Combination of Peptide Nucleic Acid Beacon and Nuclease S1 for Clear-Cut Genotyping of Single Nucleotide Polymorphisms. Anal. Biochem. 2007, 363, 300–302. [Google Scholar] [CrossRef]
- Smolina, I.V.; Kuhn, H.; Lee, C.; Frank-Kamenetskii, M.D. Fluorescence-Based Detection of Short DNA Sequences under Non-Denaturing Conditions. Bioorg. Med. Chem. 2008, 16, 84–93. [Google Scholar] [CrossRef]
- Li, X.; Morgenroth, E.; Raskin, L. Quantitative rRNA-Targeted Solution-Based Hybridization Assay Using Peptide Nucleic Acid Molecular Beacons. Appl. Environ. Microbiol. 2008, 74, 7297–7305. [Google Scholar] [CrossRef]
- Totsingan, F.; Rossi, S.; Corradini, R.; Tedeschi, T.; Sforza, S.; Juris, A.; Scaravelli, E.; Marchelli, R. Label-Free Selective DNA Detection with High Mismatch Recognition by PNA Beacons and Ion Exchange HPLC. Org. Biomol. Chem. 2008, 6, 1232–1237. [Google Scholar] [CrossRef]
- Totsingan, F.; Tedeschi, T.; Sforza, S.; Corradini, R.; Marchelli, R. Highly Selective Single Nucleotide Polymorphism Recognition by a Chiral (5S) PNA Beacon. Chirality 2009, 21, 245–253. [Google Scholar] [CrossRef] [PubMed]
- Zanoli, L.M.; Licciardello, M.; D’Agata, R.; Lantano, C.; Calabretta, A.; Corradini, R.; Marchelli, R.; Spoto, G. Peptide Nucleic Acid Molecular Beacons for the Detection of PCR Amplicons in Droplet-Based Microfluidic Devices. Anal. Bioanal. Chem. 2013, 405, 615–624. [Google Scholar] [CrossRef] [PubMed]
- Matsumoto, K.; Nakata, E.; Tamura, T.; Saito, I.; Aizawa, Y.; Morii, T. A Peptide Nucleic Acid (PNA) Heteroduplex Probe Containing an Inosine-Cytosine Base Pair Discriminates a Single-Nucleotide Difference in RNA. Chem. Eur. J. 2013, 19, 5034–5040. [Google Scholar] [CrossRef]
- Mach, K.E.; Kaushik, A.M.; Hsieh, K.; Wong, P.K.; Wang, T.-H.; Liao, J.C. Optimizing Peptide Nucleic Acid Probes for Hybridization-Based Detection and Identification of Bacterial Pathogens. Analyst 2019, 144, 1565–1574. [Google Scholar] [CrossRef]
- Shigeto, H.; Ohtsuki, T.; Iizuka, A.; Akiyama, Y.; Yamamura, S. Imaging Analysis of EGFR Mutated Cancer Cells Using Peptide Nucleic Acid (PNA)–DNA Probes. Analyst 2019, 144, 4613–4621. [Google Scholar] [CrossRef]
- Shigeto, H.; Yamada, E.; Kitamatsu, M.; Ohtsuki, T.; Iizuka, A.; Akiyama, Y.; Yamamura, S. Analysis of Single Nucleotide-Mutated Single Cancer Cells Using the Combined Technologies of Single-Cell Microarray Chips and Peptide Nucleic Acid-DNA Probes. Micromachines 2020, 11, 628. [Google Scholar] [CrossRef]
- Tabara, K.; Watanabe, K.; Shigeto, H.; Yamamura, S.; Kishi, T.; Kitamatsu, M.; Ohtsuki, T. Fluorophore–PNA–Quencher/Quencher–DNA Probe for miRNA Detection. Bioorg. Med. Chem. Lett. 2021, 51, 128359–128363. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shigeto, H.; Kishi, T.; Ishii, K.; Ohtsuki, T.; Yamamura, S.; Kitamatsu, M. Adjusting the Structure of a Peptide Nucleic Acid (PNA) Molecular Beacon and Promoting Its DNA Detection by a Hybrid with Quencher-Modified DNA. Processes 2022, 10, 722. https://doi.org/10.3390/pr10040722
Shigeto H, Kishi T, Ishii K, Ohtsuki T, Yamamura S, Kitamatsu M. Adjusting the Structure of a Peptide Nucleic Acid (PNA) Molecular Beacon and Promoting Its DNA Detection by a Hybrid with Quencher-Modified DNA. Processes. 2022; 10(4):722. https://doi.org/10.3390/pr10040722
Chicago/Turabian StyleShigeto, Hajime, Takamasa Kishi, Koki Ishii, Takashi Ohtsuki, Shohei Yamamura, and Mizuki Kitamatsu. 2022. "Adjusting the Structure of a Peptide Nucleic Acid (PNA) Molecular Beacon and Promoting Its DNA Detection by a Hybrid with Quencher-Modified DNA" Processes 10, no. 4: 722. https://doi.org/10.3390/pr10040722
APA StyleShigeto, H., Kishi, T., Ishii, K., Ohtsuki, T., Yamamura, S., & Kitamatsu, M. (2022). Adjusting the Structure of a Peptide Nucleic Acid (PNA) Molecular Beacon and Promoting Its DNA Detection by a Hybrid with Quencher-Modified DNA. Processes, 10(4), 722. https://doi.org/10.3390/pr10040722






