A High-Throughput Screening Procedure (Py-Fe3+) for Enhancing Ethanol Production by Saccharomyces cerevisiae Using ARTP Random Mutagenesis
Abstract
1. Introduction
2. Materials and Methods
2.1. Strain and Medium
2.2. ARTP-Based Random Mutagenesis and Adaptive Evolution
2.3. TTC-Based Screening
2.4. Py-Fe3+-Based Screening
2.5. Shake-Flask Fermentation
2.6. GC Analysis and Cell Growth Assay
2.7. Statistical Analysis
3. Results and Discussion
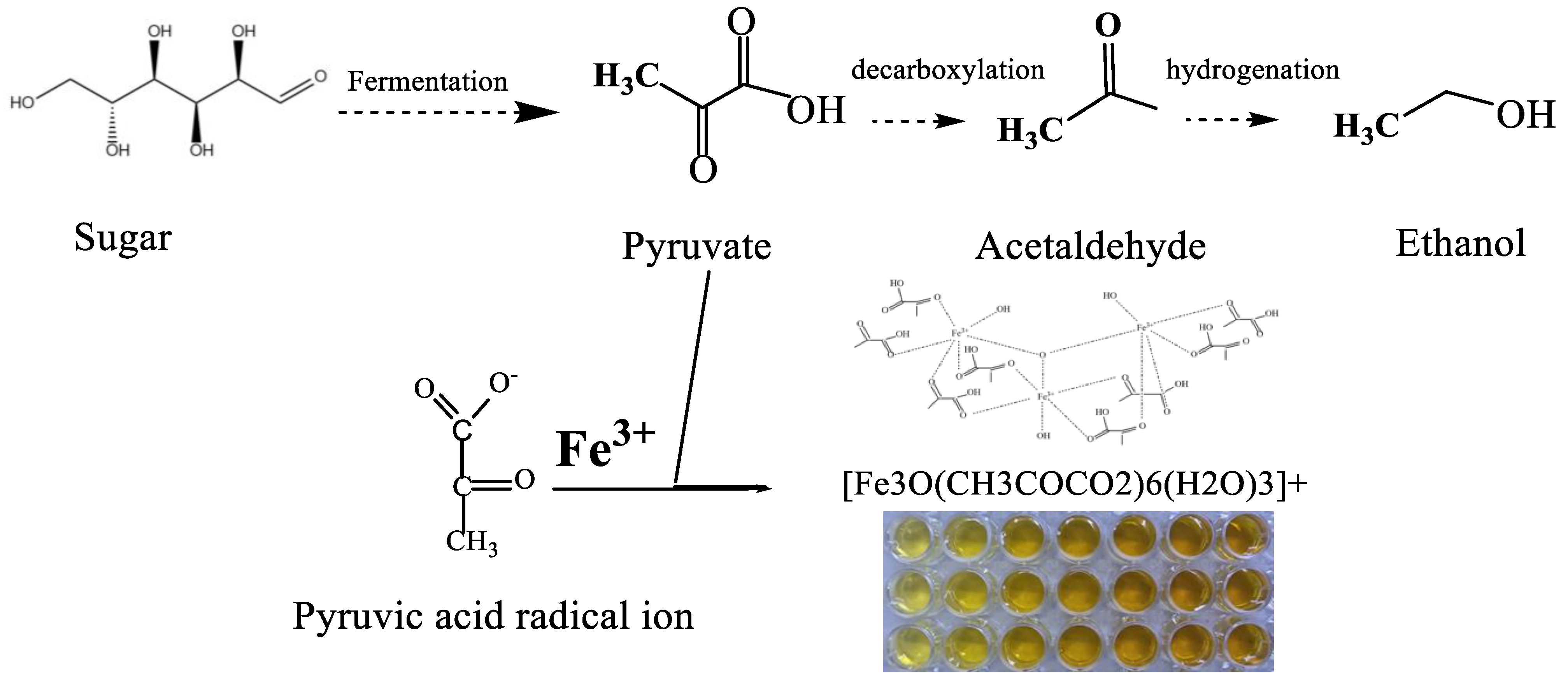
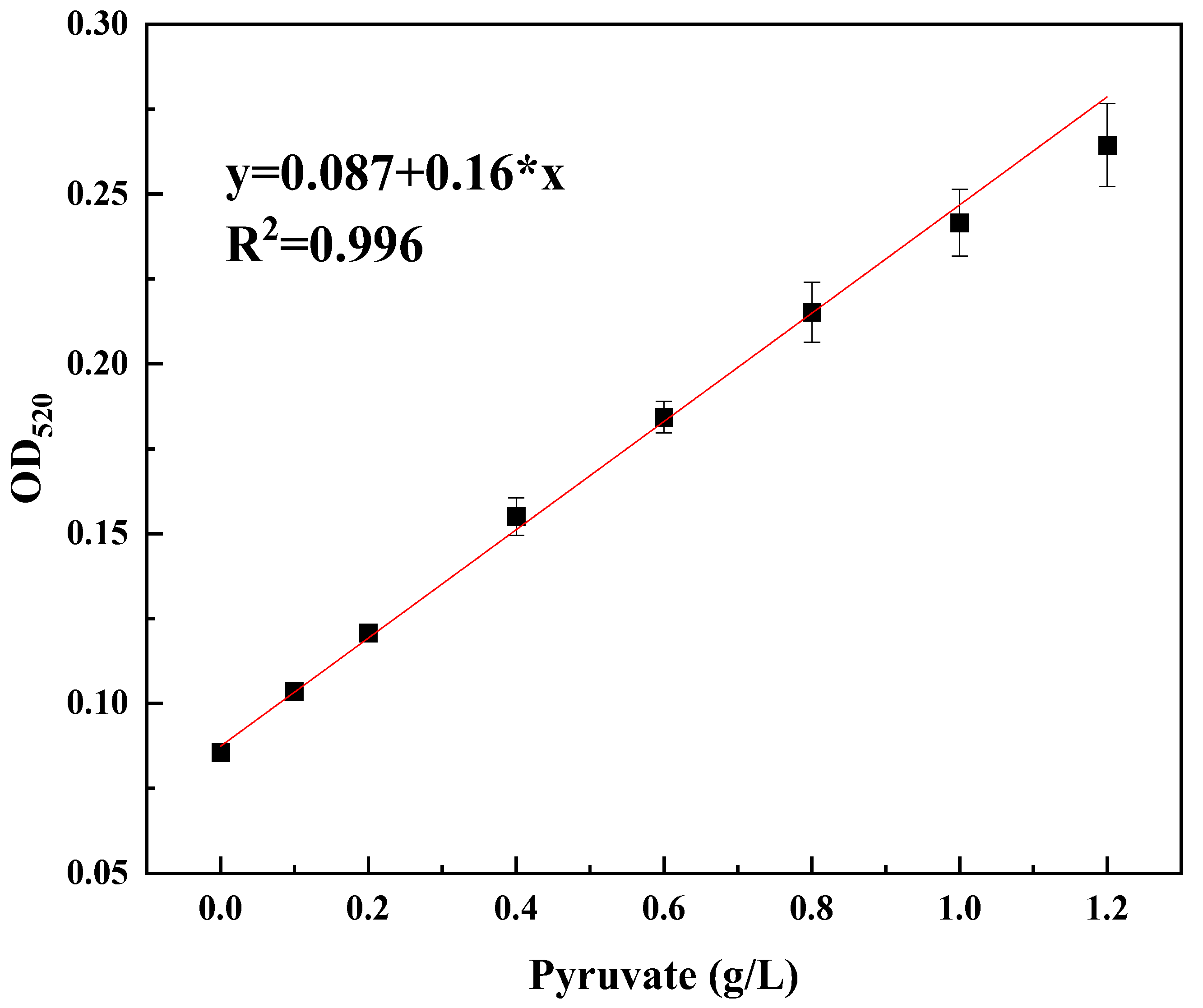
3.1. The Feasibility of Using Py-Fe3+ to Identify High-Titer Ethanol Production
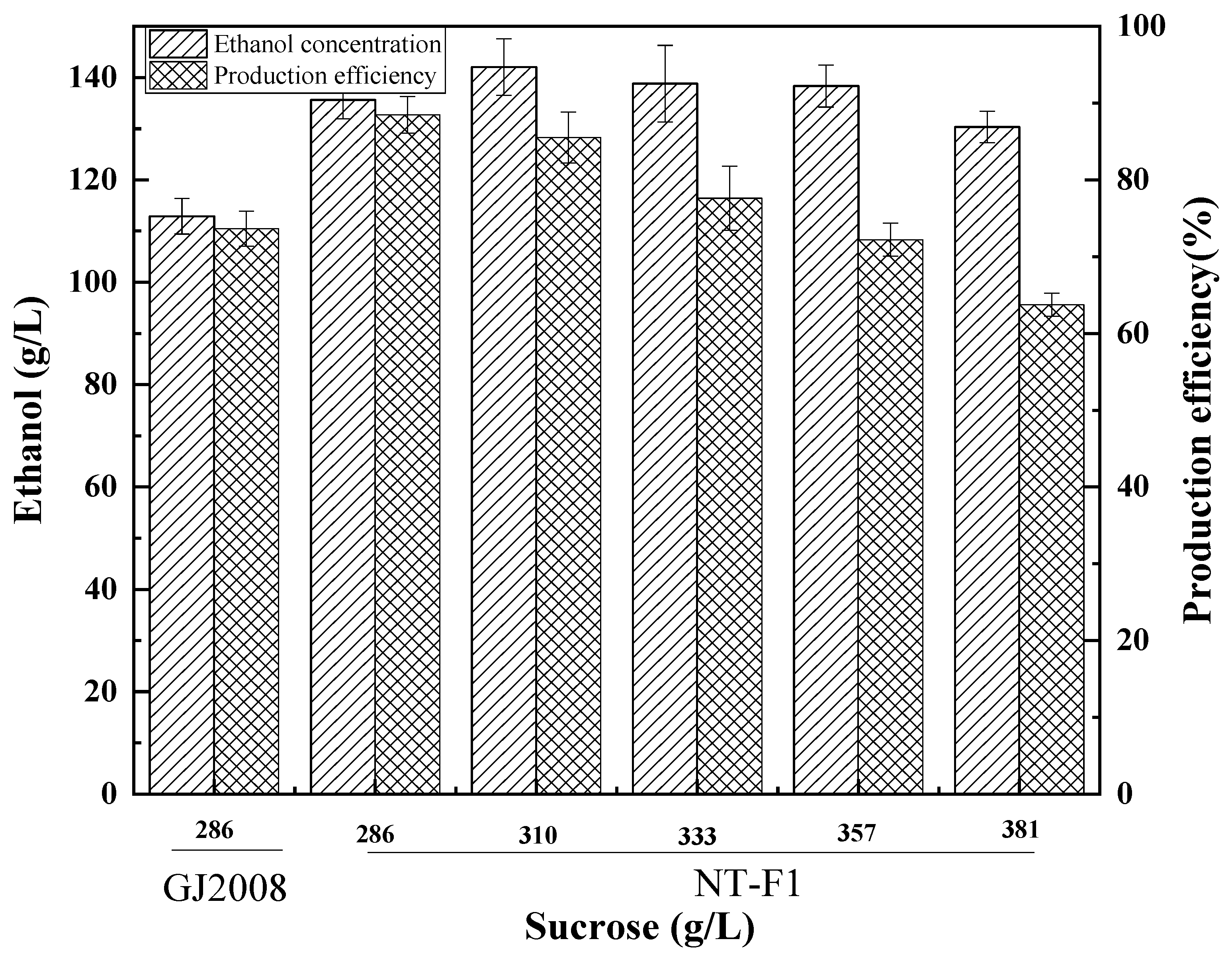
3.2. Tolerance Engineering Improves Tolerance to High Sucrose Levels
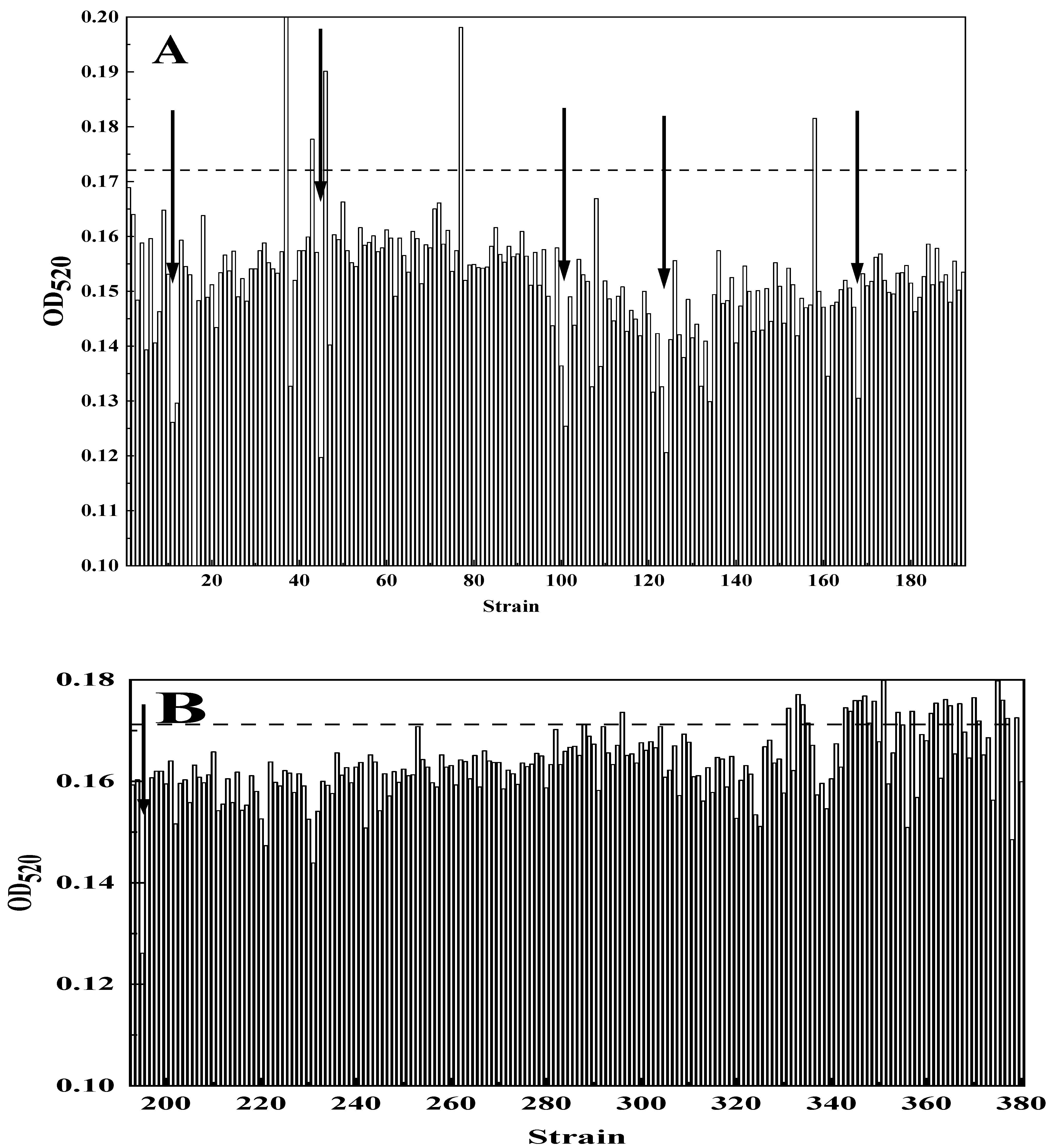
3.3. Isolation of Mutant Strains by TTC and Py-Fe3+
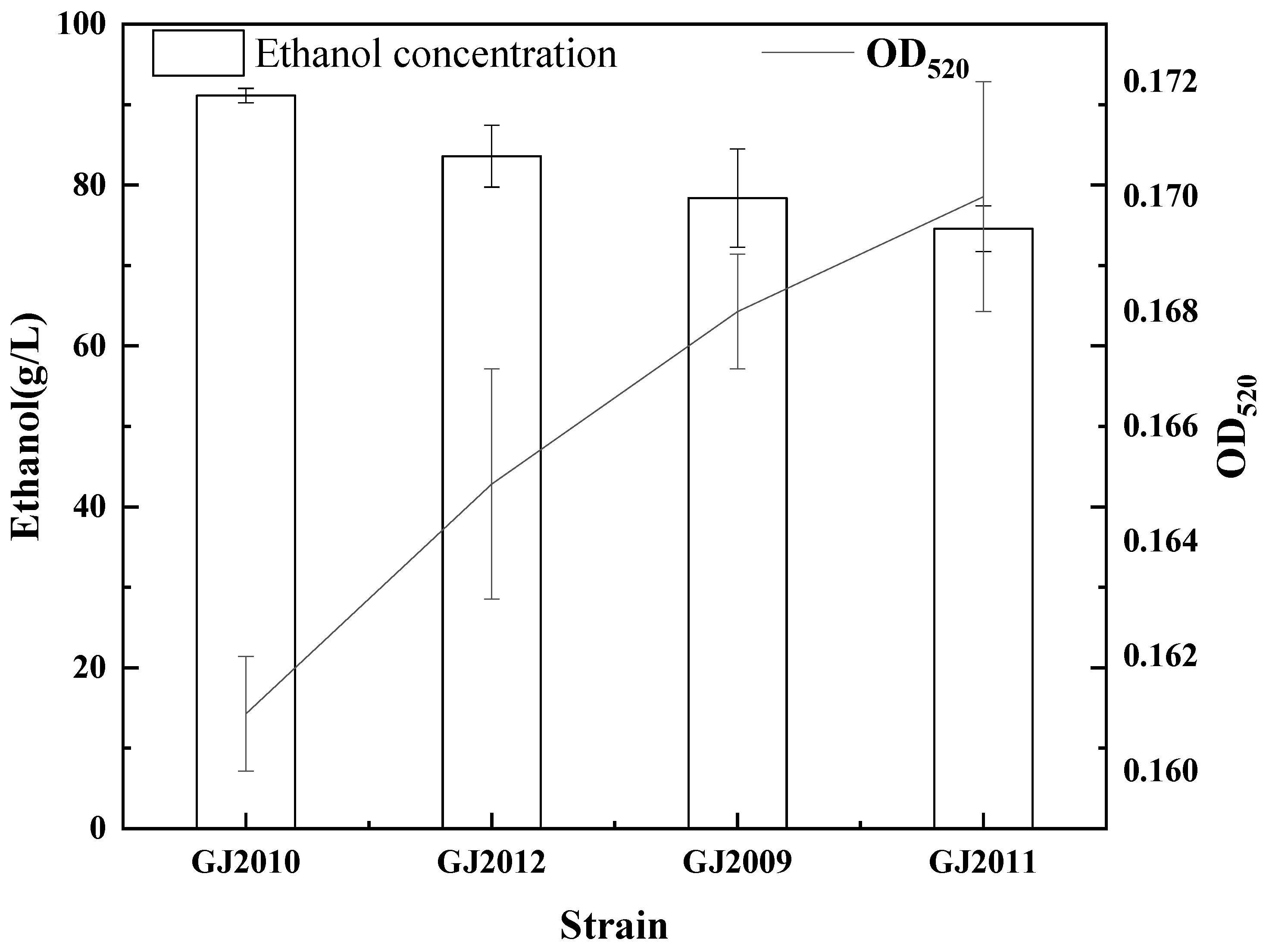
3.4. Comparison of Ethanol Fermentation Capacity and Growth Capacity of Mutant and Non-Mutant Strains
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Competing Interests
Consent for Publication
Availability of Data and Materials
Ethics Approval and Consent to Participate
References
- Bautista, K.; Unpaprom, Y.; Junluthin, P.; Ramaraj, R. Ethanol production from corn stalk juice by Saccharomyces cerevisiae immobilized yeast using a green method. Biomass Convers. Biorefin. 2022, 1–8. [Google Scholar] [CrossRef]
- Almeida, I.C.; Pacheco, T.F.; Machado, F.; Gonalves, S.B. Evaluation of different strains of Saccharomyces cerevisiaefor ethanol production from high-amylopectin BRS AG rice (Oryza sativa L.). Sci. Rep. 2022, 12, 2122. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Li, B.; Su, R.R.; Wang, S.P.; Xia, Z.Y.; Xie, C.Y.; Tang, Y.Q. Screening novel genes by a comprehensive strategy to construct multiple stress-tolerant industrial Saccharomyces cerevisiae with prominent bioethanol production. Biotechnol. Biofuels Bioprod. 2022, 15, 11. [Google Scholar] [CrossRef] [PubMed]
- Qian, L.; Zhao, X.Q.; Chang, A.K.; Zhang, Q.M.; Bai, F.W. Ethanol-induced yeast flocculation directed by the promoter of TPS1 encoding trehalose-6-phosphate synthase 1 for efficient ethanol production. Metab. Eng. 2012, 14, 1–8. [Google Scholar] [CrossRef]
- Qin, L.; Zhao, X.; Li, W.C.; Zhu, J.Q.; Liu, L.; Li, B.Z.; Yuan, Y.J. Process analysis and optimization of simultaneous saccharification and co-fermentation of ethylenediamine-pretreated corn stover for ethanol production. Biotechnol. Biofuels 2018, 11, 118. [Google Scholar] [CrossRef]
- Qin, L.; Dong, S.; Yu, J.; Ning, X.; Xu, K.; Zhang, S.-J.; Xu, L.; Li, B.-Z.; Li, J.; Yuan, Y.-J.; et al. Stress-driven dynamic regulation of multiple tolerance genes improves robustness and productive capacity of Saccharomyces cerevisiae in industrial lignocellulose fermentation. Metab. Eng. 2020, 61, 160–170. [Google Scholar] [CrossRef]
- Ye, P.-L.; Wang, X.-Q.; Yuan, B.; Liu, C.-G.; Zhao, X.-Q. Manipulating cell flocculation-associated protein kinases in Saccharomyces cerevisiae enables improved stress tolerance and efficient cellulosic ethanol production. Bioresour. Technol. 2022, 348, 126758. [Google Scholar] [CrossRef]
- Abbott, D.A.; Ingledew, W.M. Buffering capacity of whole corn mash alters concentrations of organic acids required to inhibit growth of Saccharomyces cerevisiae and ethanol production. Biotechnol. Lett. 2004, 26, 1313–1316. [Google Scholar] [CrossRef]
- Elhussieny, N.I.; Bakri, M.M.; Ganash, M.; Ghany, T. Chemical mutagenesis of Saccharomyces cerevisiae for enhancing bioethanol production with ffermentation at very High sugar concentration. Bioresources 2020, 15, 1354–1369. [Google Scholar] [CrossRef]
- Shen, N.; Wang, Q.; Qin, Y.; Wang, C.; Liao, S.; Zhu, J.; Huang, R. Breeding of a thermotolerant and high ethanol-producing Saccharomyces cerevisiae strain by uv-ntg composite protoplast mutagenesis. Liquor-Making. Sci. Technol. 2011, 5, 23–26. [Google Scholar] [CrossRef]
- Wang, L.; Li, B.; Wang, S.-P.; Xia, Z.-Y.; Gou, M.; Tang, Y.-Q. Improving multiple stress-tolerance of a flocculating industrial Saccharomyces cerevisiae strain by random mutagenesis and hybridization. Process Biochem. 2021, 102, 275–285. [Google Scholar] [CrossRef]
- Arshad, M.; Hussain, T.; Chaudhry, N.; Sadia, H.; Aslam, B.; Tahir, U.; Abbas, M.; Qureshi, N.; Nazir, A.; Rajoka, M.I. Enhancing profitability of ethanol fermentation through gamma ray mutagenesis of Saccharomyces cerevisiae. Pol. J. Environ. Stud. 2019, 28, 35–41. [Google Scholar] [CrossRef]
- Shi, D.J.; Wang, C.L.; Wang, K.M. Genome shuffling to improve thermotolerance, ethanol tolerance and ethanol productivity of Saccharomyces cerevisiae. J. Ind. Microbiol. Biotechnol. 2009, 36, 139–147. [Google Scholar] [CrossRef] [PubMed]
- Li, W.-C.; Zhu, J.-Q.; Zhao, X.; Qin, L.; Xu, T.; Zhou, X.; Li, X.; Li, B.-Z.; Yuan, Y.-J. Improving co-fermentation of glucose and xylose by adaptive evolution of engineering xylose-fermenting Saccharomyces cerevisiae and different fermentation strategies. Renew. Energy 2019, 139, 1176–1183. [Google Scholar] [CrossRef]
- Alper, H.; Moxley, J.; Nevoigt, E.; Fink, G.; Stephanopoulos, G. Engineering yeast transcription machinery for improved ethanol tolerance and production. Science 2006, 314, 1565–1568. [Google Scholar] [CrossRef]
- Liu, K.; Yuan, X.; Liang, L.; Fang, J.; Chen, Y.; He, W.; Xue, T. Using CRISPR/Cas9 for multiplex genome engineering to optimize the ethanol metabolic pathway in Saccharomyces cerevisiae. Biochem. Eng. J. 2019, 145, 120–126. [Google Scholar] [CrossRef]
- HamediRad, M.; Lian, J.; Li, H.; Zhao, H. RNAi assisted genome evolution unveils yeast mutants with improved xylose utilization. Biotechnol. Bioeng. 2018, 115, 1552–1560. [Google Scholar] [CrossRef]
- Ping, X.; Qiu, J.; Chao, G.; Ma, C. Biotechnological routes to pyruvate production. J. Biosci. Bioeng. 2008, 105, 169–175. [Google Scholar] [CrossRef]
- Luo, Z.; Zeng, W.; Du, G.; Song, L.; Jian, C. A high-throughput screening procedure for enhancing pyruvate production in Candida glabrata by random mutagenesis. Bioprocess Biosyst. Eng. 2017, 40, 1–9. [Google Scholar] [CrossRef]
- Hu, X.; Lin, B.; Yang, S. Comparison between several chemical detection methods of pyruvate production in fermentation. Cereals. Oils Process 2010, 12, 165–168. [Google Scholar]
- Niu, F.-X.; He, X.; Wu, Y.-Q.; Liu, J.-Z. Enhancing production of pinene in Escherichia coli by using a combination of tolerance, evolution, and modular co-culture engineering. Front. Microbiol. 2018, 9, 1623. [Google Scholar] [CrossRef]
- Zhang, M.; Zhu, R.; Zhang, M.; Wang, S. Creation of an ethanol-tolerant Saccharomyces cerevisiae strain by 266 nm laser radiation and repetitive cultivation. J. Biosci. Bioeng. 2014, 118, 508–513. [Google Scholar] [CrossRef]
- Mehrotra, R.-N.; Hasan, T. Detection and spectrophotometric determination of pyruvic acid. Anal. Lett. 1986, 19, 1713–1724. [Google Scholar] [CrossRef]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, W.-Y.; Wu, S.-H.; Xie, Y.-H.; Zhong, M.; Wei, M.-L.; Li, Z.-Y.; Long, X.-F.; Niu, F.-X. A High-Throughput Screening Procedure (Py-Fe3+) for Enhancing Ethanol Production by Saccharomyces cerevisiae Using ARTP Random Mutagenesis. Processes 2022, 10, 2186. https://doi.org/10.3390/pr10112186
Wang W-Y, Wu S-H, Xie Y-H, Zhong M, Wei M-L, Li Z-Y, Long X-F, Niu F-X. A High-Throughput Screening Procedure (Py-Fe3+) for Enhancing Ethanol Production by Saccharomyces cerevisiae Using ARTP Random Mutagenesis. Processes. 2022; 10(11):2186. https://doi.org/10.3390/pr10112186
Chicago/Turabian StyleWang, Wei-Yang, Shi-Hua Wu, Yuan-Han Xie, Miao Zhong, Man-Li Wei, Ze-Yang Li, Xiu-Feng Long, and Fu-Xing Niu. 2022. "A High-Throughput Screening Procedure (Py-Fe3+) for Enhancing Ethanol Production by Saccharomyces cerevisiae Using ARTP Random Mutagenesis" Processes 10, no. 11: 2186. https://doi.org/10.3390/pr10112186
APA StyleWang, W.-Y., Wu, S.-H., Xie, Y.-H., Zhong, M., Wei, M.-L., Li, Z.-Y., Long, X.-F., & Niu, F.-X. (2022). A High-Throughput Screening Procedure (Py-Fe3+) for Enhancing Ethanol Production by Saccharomyces cerevisiae Using ARTP Random Mutagenesis. Processes, 10(11), 2186. https://doi.org/10.3390/pr10112186





