Visual Analytics for Predicting Disease Outcomes Using Laboratory Test Results
Abstract
1. Introduction
2. Background
2.1. Visual Analytics
2.2. Machine Learning Techniques
2.2.1. Frequent Itemset Mining (Eclat)
2.2.2. Extreme Gradient Boosting
3. Materials and Methods
3.1. Design Process and Participants
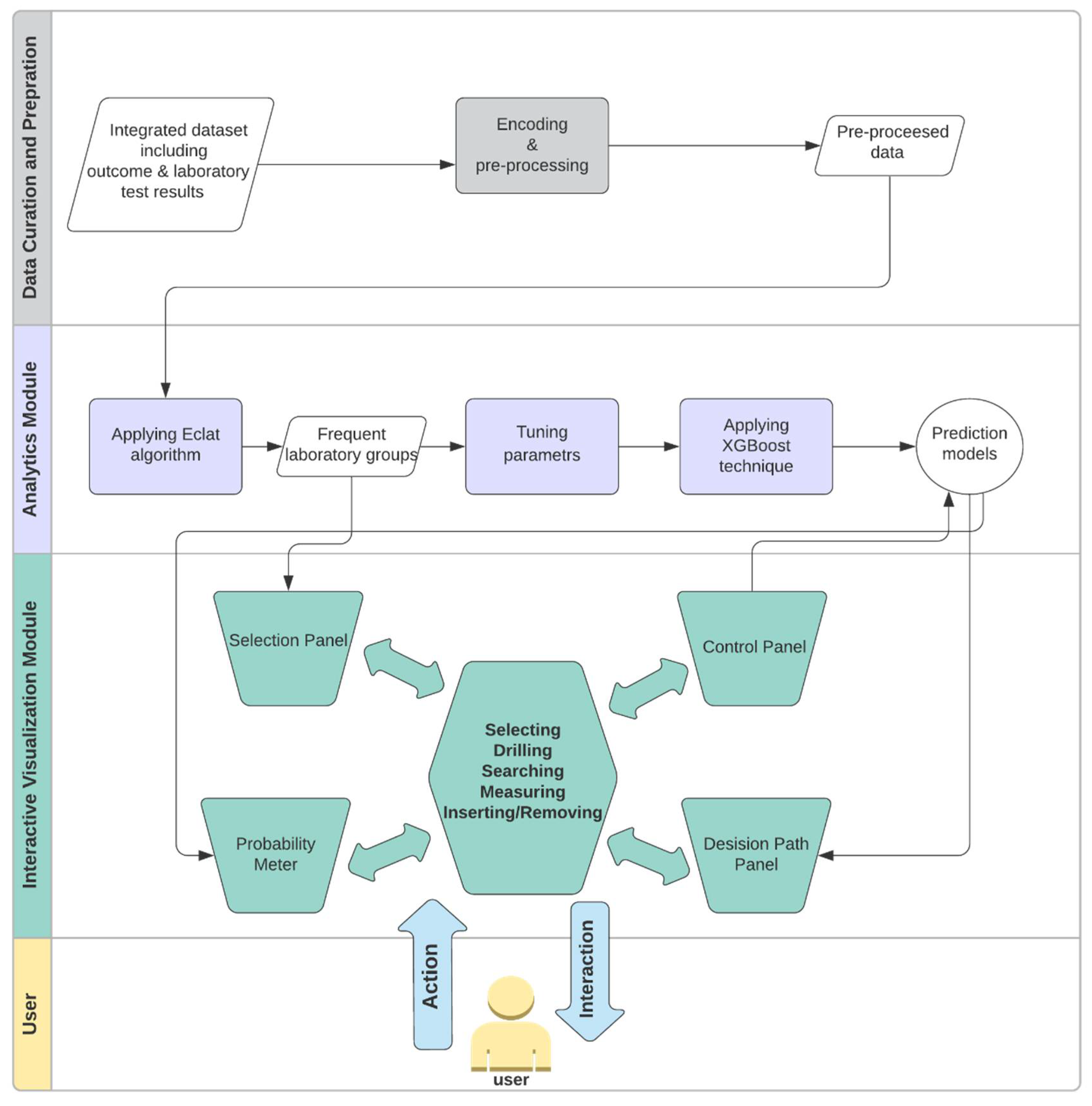
3.2. Workflow
3.3. Analytics Module
3.4. Interactive Visualization Module
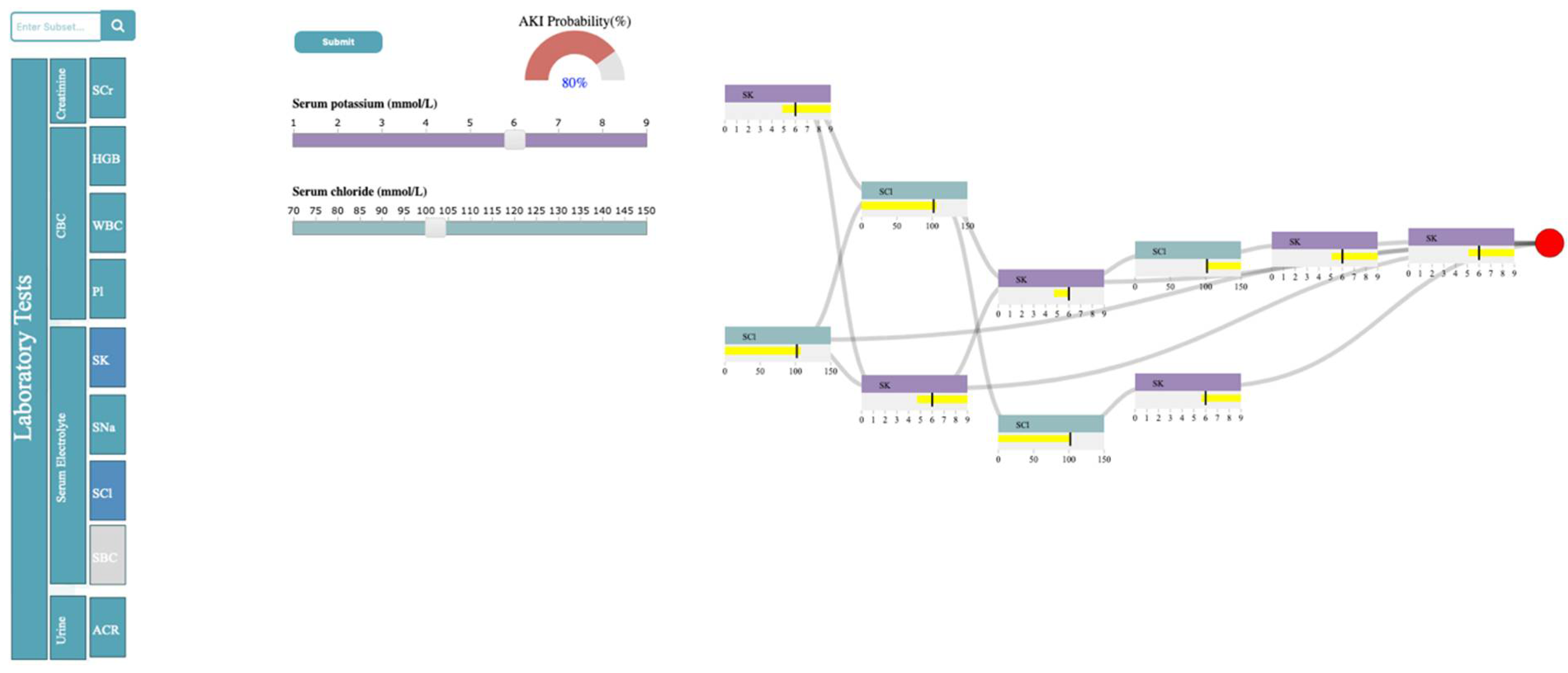
3.4.1. Selection Panel
3.4.2. Control Panel
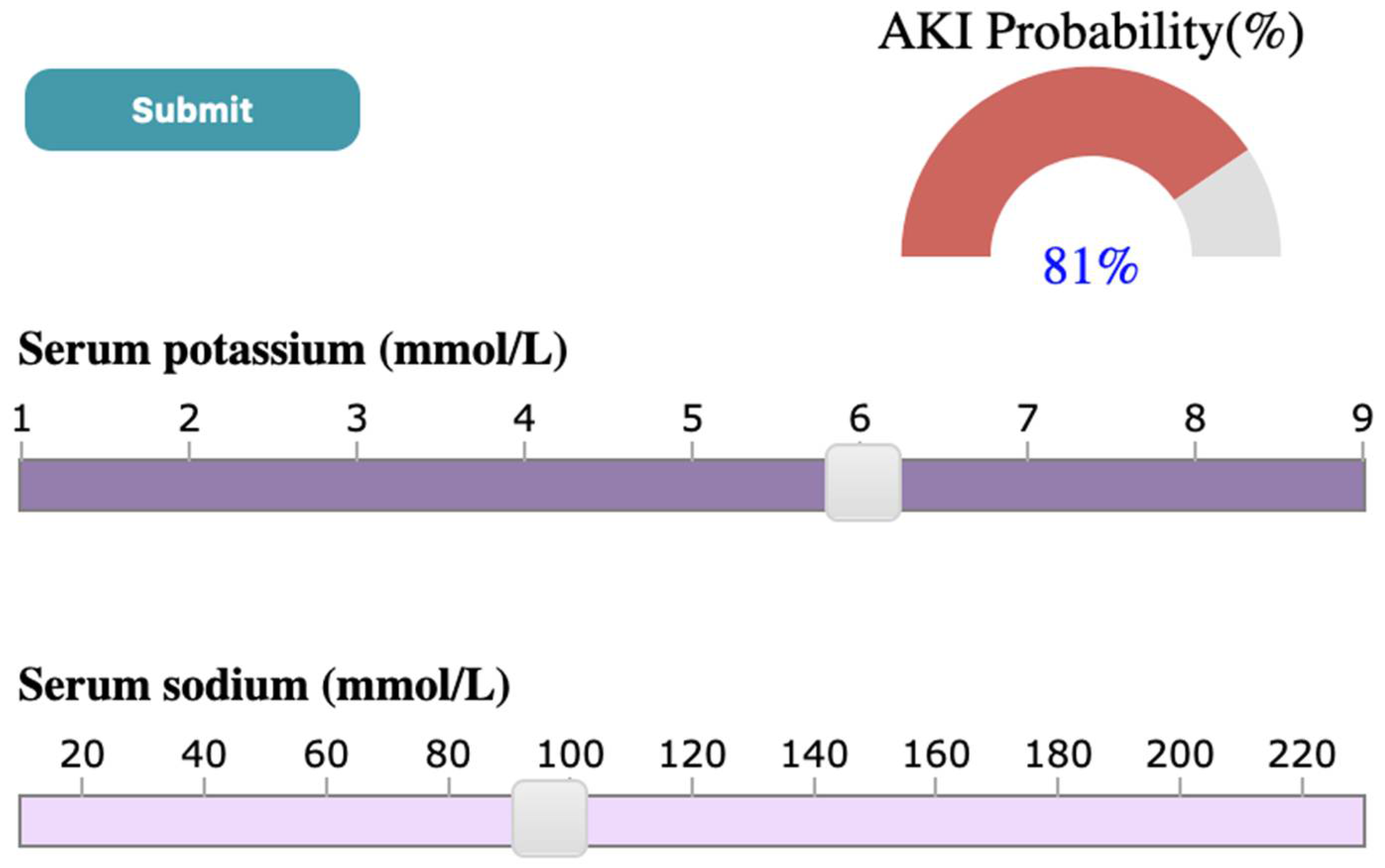
3.4.3. Probability Meter
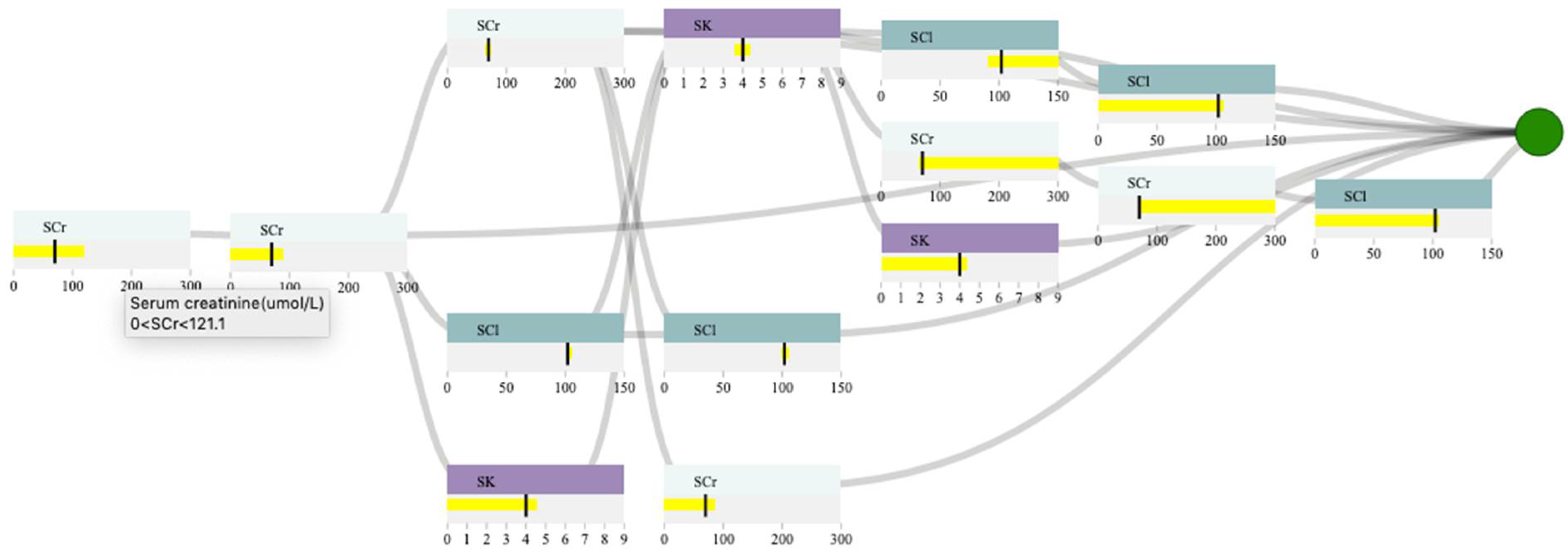
3.4.4. Decision Path Panel
4. Usage Scenario
4.1. Data Description
4.2. Outcome
4.3. Case Study
5. Discussion and Limitations
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A
| Data Source | Description | Study Purpose |
|---|---|---|
| Canadian Institute for Health Information Discharge Abstract Database and National Ambulatory Care Reporting System | The Canadian Institute for Health Information Discharge Abstract Database and the National Ambulatory Care Reporting System collect diagnostic and procedural variables for inpatient stays and ED visits, respectively. Diagnostic and inpatient procedural coding uses the 10th version of the Canadian Modified International Classification of Disease system 10th Revision (after 2002). | Cohort creation, description, exposure, and outcome estimation |
| Dynacare (formerly known as Gamma-Dynacare Medical Laboratories) | Database that contains all outpatient laboratory test results from all Dynacare laboratory locations across Ontario since 2002. Dynacare is one of the three largest laboratory providers in Ontario and contains records on over 59 million tests each year. | Outpatient laboratory tests |
| Group | Laboratory Tests in the Group | AUROC | Max_Depth | Eta | Subsample | Min_Child_Weigth | Gamma |
|---|---|---|---|---|---|---|---|
| 1 | SBC,SCr,SK,SNa | 0.78 | 9 | 0.08 | 0.84 | 0 | 3 |
| 2 | SBC,SCr,SNa | 0.77 | 8 | 0.03 | 0.96 | 10 | 2 |
| 3 | SBC,SK,SNa | 0.66 | 3 | 0.18 | 0.72 | 7 | 4 |
| 4 | SBC,SCr,SK | 0.76 | 7 | 0.07 | 0.97 | 10 | 2 |
| 5 | SBC,SCr | 0.76 | 9 | 0.05 | 0.91 | 5 | 5 |
| 6 | SBC,SK | 0.61 | 3 | 0.29 | 0.96 | 0 | 5 |
| 7 | SBC,SNa | 0.66 | 6 | 0.18 | 0.77 | 0 | 3 |
| 8 | ACr,HGB,Pl,SCl,SCr,SK,SNa,WBC | 0.81 | 7 | 0.16 | 0.99 | 5 | 3 |
| 9 | ACr,Pl,SCl,SCr,SK,SNa,WBC | 0.82 | 8 | 0.12 | 0.75 | 6 | 0 |
| 10 | ACr,HGB,Pl,SCl,SK,SNa,WBC | 0.76 | 3 | 0.27 | 0.72 | 4 | 3 |
| 11 | ACr,HGB,Pl,SCl,SCr,SK,SNa | 0.81 | 10 | 0.29 | 0.82 | 4 | 0 |
| 12 | ACr,Pl,SCl,SCr,SK,SNa | 0.81 | 8 | 0.20 | 0.79 | 0 | 2 |
| 13 | ACr,HGB,Pl,SCl,SK,SNa | 0.75 | 3 | 0.25 | 0.95 | 8 | 2 |
| 14 | ACr,Pl,SCl,SK,SNa,WBC | 0.75 | 4 | 0.30 | 0.74 | 1 | 5 |
| 15 | ACr,HGB,SCl,SCr,SK,SNa,WBC | 0.81 | 7 | 0.27 | 0.76 | 4 | 0 |
| 16 | ACr,SCl,SCr,SK,SNa,WBC | 0.81 | 6 | 0.21 | 0.82 | 9 | 0 |
| 17 | ACr,HGB,SCl,SK,SNa,WBC | 0.77 | 3 | 0.28 | 0.83 | 3 | 2 |
| 18 | ACr,HGB,SCl,SCr,SK,SNa | 0.81 | 9 | 0.28 | 0.95 | 0 | 2 |
| 19 | ACr,SCl,SCr,SK,SNa | 0.80 | 7 | 0.21 | 0.86 | 7 | 0 |
| 20 | ACr,HGB,SCl,SK,SNa | 0.75 | 3 | 0.29 | 0.96 | 0 | 5 |
| 21 | ACr,SCl,SK,SNa,WBC | 0.76 | 10 | 0.22 | 0.94 | 5 | 3 |
| 22 | ACr,Pl,SCl,SK,SNa | 0.74 | 7 | 0.23 | 0.77 | 2 | 5 |
| 23 | ACr,HGB,Pl,SCl,SCr,SNa,WBC | 0.81 | 8 | 0.27 | 0.86 | 3 | 4 |
| 24 | ACr,Pl,SCl,SCr,SNa,WBC | 0.81 | 10 | 0.27 | 0.72 | 0 | 1 |
| 25 | ACr,HGB,Pl,SCl,SNa,WBC | 0.76 | 9 | 0.19 | 0.93 | 7 | 4 |
| 26 | ACr,HGB,Pl,SCl,SCr,SNa | 0.81 | 7 | 0.27 | 0.95 | 1 | 2 |
| 27 | ACr,Pl,SCl,SCr,SNa | 0.80 | 7 | 0.28 | 0.73 | 5 | 4 |
| 28 | ACr,HGB,Pl,SCl,SNa | 0.74 | 3 | 0.28 | 0.71 | 7 | 3 |
| 29 | ACr,Pl,SCl,SNa,WBC | 0.74 | 10 | 0.08 | 0.92 | 5 | 3 |
| 30 | ACr,HGB,SCl,SCr,SNa,WBC | 0.81 | 10 | 0.27 | 0.91 | 6 | 5 |
| 31 | ACr,SCl,SCr,SNa,WBC | 0.81 | 6 | 0.17 | 0.75 | 9 | 2 |
| 32 | ACr,HGB,SCl,SNa,WBC | 0.77 | 4 | 0.26 | 0.77 | 6 | 3 |
| 33 | ACr,HGB,SCl,SCr,SNa | 0.80 | 9 | 0.27 | 0.98 | 5 | 2 |
| 34 | ACr,SCl,SCr,SNa | 0.80 | 9 | 0.03 | 0.71 | 2 | 0 |
| 35 | ACr,HGB,SCl,SNa | 0.75 | 4 | 0.30 | 0.74 | 1 | 5 |
| 36 | ACr,SCl,SNa,WBC | 0.74 | 6 | 0.22 | 0.81 | 3 | 4 |
| 37 | ACr,Pl,SCl,SNa | 0.72 | 3 | 0.26 | 0.70 | 0 | 2 |
| 38 | ACr,SCl,SK,SNa | 0.73 | 5 | 0.28 | 0.82 | 9 | 5 |
| 39 | ACr,HGB,Pl,SCl,SCr,SK,WBC | 0.81 | 9 | 0.11 | 0.98 | 3 | 5 |
| 40 | ACr,Pl,SCl,SCr,SK,WBC | 0.81 | 6 | 0.23 | 0.70 | 9 | 4 |
| 41 | ACr,HGB,Pl,SCl,SK,WBC | 0.78 | 6 | 0.29 | 0.76 | 3 | 4 |
| 42 | ACr,HGB,Pl,SCl,SCr,SK | 0.81 | 10 | 0.19 | 0.73 | 0 | 3 |
| 43 | ACr,Pl,SCl,SCr,SK | 0.8 | 5 | 0.29 | 0.75 | 3 | 0 |
| 44 | ACr,HGB,Pl,SCl,SK | 0.75 | 3 | 0.28 | 0.71 | 7 | 3 |
| 45 | ACr,Pl,SCl,SK,WBC | 0.75 | 3 | 0.27 | 0.92 | 0 | 1 |
| 46 | ACr,HGB,SCl,SCr,SK,WBC | 0.80 | 10 | 0.28 | 0.93 | 7 | 4 |
| 47 | ACr,SCl,SCr,SK,WBC | 0.80 | 7 | 0.29 | 0.83 | 3 | 0 |
| 48 | ACr,HGB,SCl,SK,WBC | 0.76 | 3 | 0.29 | 1.00 | 6 | 0 |
| 49 | ACr,HGB,SCl,SCr,SK | 0.81 | 9 | 0.27 | 0.94 | 9 | 1 |
| 50 | ACr,SCl,SCr,SK | 0.79 | 5 | 0.29 | 0.75 | 3 | 0 |
| 51 | ACr,HGB,SCl,SK | 0.75 | 3 | 0.27 | 0.86 | 7 | 1 |
| 52 | ACr,SCl,SK,WBC | 0.75 | 8 | 0.23 | 0.70 | 8 | 1 |
| 53 | ACr,Pl,SCl,SK | 0.74 | 4 | 0.30 | 0.74 | 1 | 5 |
| 54 | ACr,HGB,Pl,SCl,SCr,WBC | 0.81 | 10 | 0.25 | 0.99 | 9 | 0 |
| 55 | ACr,Pl,SCl,SCr,WBC | 0.8 | 8 | 0.23 | 0.83 | 1 | 4 |
| 56 | ACr,HGB,Pl,SCl,WBC | 0.77 | 4 | 0.29 | 0.93 | 4 | 3 |
| 57 | ACr,HGB,Pl,SCl,SCr | 0.80 | 9 | 0.21 | 0.93 | 3 | 1 |
| 58 | ACr,Pl,SCl,SCr | 0.79 | 9 | 0.28 | 0.72 | 1 | 2 |
| 59 | ACr,HGB,Pl,SCl | 0.75 | 3 | 0.28 | 0.76 | 8 | 1 |
| 60 | ACr,Pl,SCl,WBC | 0.74 | 5 | 0.28 | 0.96 | 5 | 2 |
| 61 | ACr,HGB,SCl,SCr,WBC | 0.80 | 7 | 0.27 | 0.77 | 4 | 3 |
| 62 | ACr,SCl,SCr,WBC | 0.80 | 5 | 0.26 | 0.81 | 2 | 4 |
| 63 | ACr,HGB,SCl,WBC | 0.76 | 3 | 0.27 | 0.90 | 6 | 1 |
| 64 | ACr,HGB,SCl,SCr | 0.80 | 10 | 0.19 | 0.73 | 0 | 3 |
| 65 | ACr,SCl,SCr | 0.80 | 8 | 0.30 | 0.99 | 4 | 3 |
| 66 | ACr,HGB,SCl | 0.74 | 3 | 0.22 | 0.83 | 8 | 2 |
| 67 | ACr,SCl,WBC | 0.73 | 5 | 0.29 | 0.73 | 4 | 2 |
| 68 | ACr,Pl,SCl | 0.73 | 6 | 0.22 | 0.89 | 8 | 5 |
| 69 | ACr,SCl,SK | 0.73 | 3 | 0.28 | 0.76 | 8 | 1 |
| 70 | ACr,SCl,SNa | 0.72 | 7 | 0.29 | 0.72 | 2 | 5 |
| 71 | ACr,HGB,Pl,SCr,SK,SNa,WBC | 0.83 | 6 | 0.29 | 0.91 | 2 | 0 |
| 72 | ACr,Pl,SCr,SK,SNa,WBC | 0.82 | 5 | 0.29 | 0.95 | 1 | 4 |
| 73 | ACr,HGB,Pl,SK,SNa,WBC | 0.77 | 4 | 0.29 | 0.77 | 6 | 0 |
| 74 | ACr,HGB,Pl,SCr,SK,SNa | 0.82 | 10 | 0.19 | 0.95 | 1 | 5 |
| 75 | ACr,Pl,SCr,SK,SNa | 0.82 | 6 | 0.25 | 0.76 | 5 | 5 |
| 76 | ACr,HGB,Pl,SK,SNa | 0.74 | 5 | 0.15 | 0.84 | 3 | 1 |
| 77 | ACr,Pl,SK,SNa,WBC | 0.75 | 5 | 0.24 | 0.92 | 6 | 4 |
| 78 | ACr,HGB,SCr,SK,SNa,WBC | 0.83 | 9 | 0.17 | 0.72 | 5 | 2 |
| 79 | ACr,SCr,SK,SNa,WBC | 0.83 | 10 | 0.18 | 0.76 | 5 | 5 |
| 80 | ACr,HGB,SK,SNa,WBC | 0.76 | 8 | 0.08 | 0.73 | 5 | 5 |
| 81 | ACr,HGB,SCr,SK,SNa | 0.81 | 4 | 0.28 | 0.92 | 2 | 2 |
| 82 | ACr,SCr,SK,SNa | 0.8 | 4 | 0.27 | 0.73 | 1 | 3 |
| 83 | ACr,HGB,SK,SNa | 0.75 | 4 | 0.23 | 0.95 | 7 | 0 |
| 84 | ACr,SK,SNa,WBC | 0.75 | 5 | 0.17 | 0.96 | 9 | 0 |
| 85 | ACr,Pl,SK,SNa | 0.72 | 4 | 0.20 | 0.78 | 0 | 4 |
| 86 | ACr,HGB,Pl,SCr,SNa,WBC | 0.82 | 7 | 0.28 | 0.93 | 2 | 4 |
| 87 | ACr,Pl,SCr,SNa,WBC | 0.83 | 7 | 0.27 | 0.77 | 4 | 3 |
| 88 | ACr,HGB,Pl,SNa,WBC | 0.76 | 3 | 0.28 | 0.99 | 0 | 1 |
| 89 | ACr,HGB,Pl,SCr,SNa | 0.82 | 10 | 0.19 | 0.89 | 4 | 5 |
| 90 | ACr,Pl,SCr,SNa | 0.81 | 3 | 0.22 | 0.82 | 8 | 0 |
| 91 | ACr,HGB,Pl,SNa | 0.74 | 3 | 0.10 | 0.86 | 3 | 4 |
| 92 | ACr,Pl,SNa,WBC | 0.74 | 6 | 0.29 | 0.79 | 9 | 5 |
| 93 | ACr,HGB,SCr,SNa,WBC | 0.82 | 9 | 0.28 | 0.95 | 10 | 4 |
| 94 | ACr,SCr,SNa,WBC | 0.83 | 8 | 0.14 | 0.71 | 10 | 1 |
| 95 | ACr,HGB,SNa,WBC | 0.76 | 4 | 0.17 | 0.77 | 6 | 4 |
| 96 | ACr,HGB,SCr,SNa | 0.82 | 7 | 0.28 | 0.93 | 2 | 4 |
| 97 | ACr,SCr,SNa | 0.81 | 3 | 0.30 | 0.99 | 9 | 5 |
| 98 | ACr,HGB,SNa | 0.74 | 4 | 0.04 | 0.73 | 1 | 4 |
| 99 | ACr,SNa,WBC | 0.74 | 5 | 0.22 | 0.80 | 1 | 3 |
| 100 | ACr,Pl,SNa | 0.71 | 3 | 0.27 | 0.90 | 6 | 1 |
| 101 | ACr,SK,SNa | 0.71 | 5 | 0.29 | 0.89 | 10 | 5 |
| 102 | ACr,HGB,Pl,SCr,SK,WBC | 0.83 | 10 | 0.21 | 0.83 | 9 | 2 |
| 103 | ACr,Pl,SCr,SK,WBC | 0.83 | 9 | 0.19 | 0.89 | 3 | 5 |
| 104 | ACr,HGB,Pl,SK,WBC | 0.77 | 7 | 0.23 | 0.77 | 2 | 5 |
| 105 | ACr,HGB,Pl,SCr,SK | 0.82 | 5 | 0.18 | 1.00 | 2 | 4 |
| 106 | ACr,Pl,SCr,SK | 0.82 | 5 | 0.21 | 0.72 | 6 | 3 |
| 107 | ACr,HGB,Pl,SK | 0.75 | 5 | 0.09 | 0.93 | 5 | 4 |
| 108 | ACr,Pl,SK,WBC | 0.75 | 3 | 0.28 | 0.99 | 0 | 1 |
| 109 | ACr,HGB,SCr,SK,WBC | 0.83 | 6 | 0.21 | 0.80 | 1 | 4 |
| 110 | ACr,SCr,SK,WBC | 0.83 | 5 | 0.24 | 0.76 | 1 | 4 |
| 111 | ACr,HGB,SK,WBC | 0.77 | 3 | 0.28 | 0.76 | 8 | 1 |
| 112 | ACr,HGB,SCr,SK | 0.82 | 10 | 0.19 | 0.95 | 1 | 5 |
| 113 | ACr,SCr,SK | 0.81 | 3 | 0.28 | 0.93 | 8 | 1 |
| 114 | ACr,HGB,SK | 0.74 | 4 | 0.03 | 0.83 | 9 | 5 |
| 115 | ACr,SK,WBC | 0.75 | 9 | 0.18 | 0.83 | 10 | 4 |
| 116 | ACr,Pl,SK | 0.72 | 5 | 0.24 | 0.92 | 6 | 4 |
| 117 | ACr,HGB,Pl,SCr,WBC | 0.82 | 8 | 0.25 | 0.81 | 6 | 3 |
| 118 | ACr,Pl,SCr,WBC | 0.82 | 10 | 0.28 | 0.94 | 6 | 4 |
| 119 | ACr,HGB,Pl,WBC | 0.76 | 7 | 0.28 | 0.73 | 5 | 4 |
| 120 | ACr,HGB,Pl,SCr | 0.82 | 10 | 0.19 | 0.81 | 4 | 4 |
| 121 | ACr,Pl,SCr | 0.81 | 3 | 0.28 | 0.83 | 3 | 2 |
| 122 | ACr,HGB,Pl | 0.74 | 4 | 0.20 | 0.72 | 10 | 4 |
| 123 | ACr,Pl,WBC | 0.73 | 5 | 0.25 | 0.89 | 0 | 2 |
| 124 | ACr,HGB,SCr,WBC | 0.81 | 4 | 0.28 | 0.92 | 2 | 2 |
| 125 | ACr,SCr,WBC | 0.82 | 7 | 0.29 | 0.72 | 2 | 5 |
| 126 | ACr,HGB,WBC | 0.75 | 4 | 0.15 | 0.75 | 9 | 5 |
| 127 | ACr,HGB,SCr | 0.82 | 5 | 0.28 | 0.72 | 10 | 1 |
| 128 | ACr,SCr | 0.81 | 4 | 0.26 | 0.77 | 6 | 3 |
| 129 | ACr,HGB | 0.74 | 8 | 0.13 | 0.72 | 0 | 5 |
| 130 | ACr,WBC | 0.73 | 7 | 0.27 | 0.87 | 8 | 4 |
| 131 | ACr,Pl | 0.71 | 3 | 0.29 | 0.81 | 5 | 2 |
| 132 | ACr,SK | 0.7 | 3 | 0.29 | 0.96 | 0 | 5 |
| 133 | ACr,SNa | 0.7 | 8 | 0.22 | 0.76 | 7 | 3 |
| 134 | ACr,SCl | 0.72 | 7 | 0.15 | 0.82 | 5 | 5 |
| 135 | HGB,Pl,SCl,SCr,SK,SNa,WBC | 0.80 | 6 | 0.29 | 0.76 | 3 | 4 |
| 136 | Pl,SCl,SCr,SK,SNa,WBC | 0.79 | 3 | 0.29 | 0.74 | 8 | 2 |
| 137 | HGB,Pl,SCl,SK,SNa,WBC | 0.74 | 9 | 0.28 | 0.79 | 5 | 5 |
| 138 | HGB,Pl,SCl,SCr,SK,SNa | 0.80 | 5 | 0.28 | 0.82 | 9 | 5 |
| 139 | Pl,SCl,SCr,SK,SNa | 0.79 | 5 | 0.23 | 0.72 | 7 | 5 |
| 140 | HGB,Pl,SCl,SK,SNa | 0.72 | 8 | 0.13 | 0.72 | 0 | 5 |
| 141 | Pl,SCl,SK,SNa,WBC | 0.66 | 8 | 0.27 | 0.95 | 1 | 5 |
| 142 | HGB,SCl,SCr,SK,SNa,WBC | 0.80 | 5 | 0.28 | 0.82 | 9 | 5 |
| 143 | SCl,SCr,SK,SNa,WBC | 0.78 | 5 | 0.17 | 0.73 | 0 | 3 |
| 144 | HGB,SCl,SK,SNa,WBC | 0.72 | 3 | 0.26 | 0.70 | 0 | 2 |
| 145 | HGB,SCl,SCr,SK,SNa | 0.80 | 7 | 0.20 | 0.78 | 7 | 4 |
| 146 | SCl,SCr,SK,SNa | 0.78 | 5 | 0.28 | 0.82 | 9 | 5 |
| 147 | HGB,SCl,SK,SNa | 0.71 | 7 | 0.27 | 0.81 | 3 | 5 |
| 148 | SCl,SK,SNa,WBC | 0.65 | 8 | 0.26 | 0.78 | 1 | 5 |
| 149 | Pl,SCl,SK,SNa | 0.65 | 9 | 0.22 | 0.95 | 8 | 3 |
| 150 | HGB,Pl,SCl,SCr,SNa,WBC | 0.80 | 9 | 0.15 | 0.75 | 10 | 5 |
| 151 | Pl,SCl,SCr,SNa,WBC | 0.78 | 6 | 0.20 | 0.93 | 4 | 3 |
| 152 | HGB,Pl,SCl,SNa,WBC | 0.73 | 5 | 0.27 | 0.88 | 3 | 0 |
| 153 | HGB,Pl,SCl,SCr,SNa | 0.79 | 4 | 0.26 | 0.72 | 0 | 3 |
| 154 | Pl,SCl,SCr,SNa | 0.78 | 4 | 0.25 | 0.81 | 4 | 4 |
| 155 | HGB,Pl,SCl,SNa | 0.71 | 7 | 0.13 | 0.75 | 5 | 2 |
| 156 | Pl,SCl,SNa,WBC | 0.65 | 7 | 0.26 | 0.84 | 0 | 3 |
| 157 | HGB,SCl,SCr,SNa,WBC | 0.8 | 5 | 0.13 | 0.92 | 3 | 1 |
| 158 | SCl,SCr,SNa,WBC | 0.78 | 4 | 0.27 | 0.83 | 10 | 4 |
| 159 | HGB,SCl,SNa,WBC | 0.72 | 5 | 0.29 | 0.89 | 10 | 5 |
| 160 | HGB,SCl,SCr,SNa | 0.79 | 5 | 0.14 | 0.74 | 8 | 4 |
| 161 | SCl,SCr,SNa | 0.78 | 9 | 0.28 | 0.79 | 5 | 5 |
| 162 | HGB,SCl,SNa | 0.70 | 10 | 0.23 | 0.78 | 5 | 5 |
| 163 | SCl,SNa,WBC | 0.65 | 8 | 0.26 | 0.78 | 1 | 5 |
| 164 | Pl,SCl,SNa | 0.63 | 10 | 0.19 | 0.95 | 1 | 5 |
| 165 | SCl,SK,SNa | 0.64 | 6 | 0.27 | 0.87 | 1 | 1 |
| 166 | HGB,Pl,SCl,SCr,SK,WBC | 0.81 | 9 | 0.28 | 0.79 | 5 | 5 |
| 167 | Pl,SCl,SCr,SK,WBC | 0.79 | 6 | 0.29 | 0.79 | 9 | 5 |
| 168 | HGB,Pl,SCl,SK,WBC | 0.73 | 4 | 0.23 | 0.95 | 7 | 0 |
| 169 | HGB,Pl,SCl,SCr,SK | 0.80 | 7 | 0.28 | 0.93 | 2 | 4 |
| 170 | Pl,SCl,SCr,SK | 0.78 | 5 | 0.25 | 0.97 | 6 | 0 |
| 171 | HGB,Pl,SCl,SK | 0.71 | 6 | 0.13 | 0.81 | 7 | 4 |
| 172 | Pl,SCl,SK,WBC | 0.66 | 6 | 0.29 | 0.76 | 3 | 4 |
| 173 | HGB,SCl,SCr,SK,WBC | 0.80 | 5 | 0.19 | 0.79 | 6 | 4 |
| 174 | SCl,SCr,SK,WBC | 0.79 | 4 | 0.25 | 0.77 | 7 | 4 |
| 175 | HGB,SCl,SK,WBC | 0.72 | 3 | 0.24 | 0.71 | 9 | 2 |
| 176 | HGB,SCl,SCr,SK | 0.80 | 5 | 0.18 | 0.76 | 3 | 2 |
| 177 | SCl,SCr,SK | 0.78 | 7 | 0.16 | 0.74 | 6 | 5 |
| 178 | HGB,SCl,SK | 0.71 | 9 | 0.29 | 0.85 | 10 | 3 |
| 179 | SCl,SK,WBC | 0.64 | 7 | 0.26 | 0.73 | 0 | 2 |
| 180 | Pl,SCl,SK | 0.63 | 5 | 0.28 | 0.92 | 4 | 3 |
| 181 | HGB,Pl,SCl,SCr,WBC | 0.80 | 7 | 0.05 | 0.75 | 2 | 5 |
| 182 | Pl,SCl,SCr,WBC | 0.78 | 5 | 0.23 | 0.77 | 8 | 2 |
| 183 | HGB,Pl,SCl,WBC | 0.73 | 3 | 0.27 | 0.85 | 5 | 0 |
| 184 | HGB,Pl,SCl,SCr | 0.79 | 5 | 0.27 | 0.88 | 3 | 0 |
| 185 | Pl,SCl,SCr | 0.78 | 6 | 0.15 | 0.76 | 7 | 0 |
| 186 | HGB,Pl,SCl | 0.70 | 7 | 0.18 | 0.88 | 7 | 5 |
| 187 | Pl,SCl,WBC | 0.64 | 9 | 0.22 | 0.81 | 1 | 4 |
| 188 | HGB,SCl,SCr,WBC | 0.8 | 3 | 0.28 | 0.71 | 7 | 3 |
| 189 | SCl,SCr,WBC | 0.78 | 6 | 0.08 | 0.72 | 7 | 5 |
| 190 | HGB,SCl,WBC | 0.71 | 3 | 0.28 | 0.94 | 6 | 0 |
| 191 | HGB,SCl,SCr | 0.80 | 4 | 0.25 | 0.77 | 7 | 4 |
| 192 | SCl,SCr | 0.78 | 3 | 0.23 | 0.84 | 0 | 0 |
| 193 | HGB,SCl | 0.70 | 5 | 0.29 | 0.75 | 3 | 0 |
| 194 | SCl,WBC | 0.62 | 9 | 0.28 | 0.84 | 9 | 5 |
| 195 | Pl,SCl | 0.6 | 8 | 0.30 | 0.82 | 4 | 0 |
| 196 | SCl,SK | 0.61 | 9 | 0.28 | 0.95 | 10 | 4 |
| 197 | SCl,SNa | 0.64 | 9 | 0.23 | 0.87 | 8 | 1 |
| 198 | HGB,Pl,SCr,SK,SNa,WBC | 0.8 | 7 | 0.22 | 0.97 | 1 | 5 |
| 199 | Pl,SCr,SK,SNa,WBC | 0.8 | 4 | 0.29 | 0.77 | 6 | 0 |
| 200 | HGB,Pl,SK,SNa,WBC | 0.73 | 8 | 0.13 | 0.72 | 0 | 5 |
| 201 | HGB,Pl,SCr,SK,SNa | 0.80 | 5 | 0.27 | 0.89 | 5 | 5 |
| 202 | Pl,SCr,SK,SNa | 0.79 | 8 | 0.06 | 0.71 | 0 | 4 |
| 203 | HGB,Pl,SK,SNa | 0.69 | 7 | 0.29 | 0.72 | 2 | 5 |
| 204 | Pl,SK,SNa,WBC | 0.66 | 5 | 0.27 | 0.85 | 0 | 5 |
| 205 | HGB,SCr,SK,SNa,WBC | 0.80 | 5 | 0.20 | 0.74 | 3 | 4 |
| 206 | SCr,SK,SNa,WBC | 0.79 | 4 | 0.20 | 0.78 | 0 | 4 |
| 207 | HGB,SK,SNa,WBC | 0.71 | 7 | 0.29 | 0.72 | 2 | 5 |
| 208 | HGB,SCr,SK,SNa | 0.80 | 5 | 0.29 | 0.89 | 10 | 5 |
| 209 | SCr,SK,SNa | 0.79 | 6 | 0.22 | 0.81 | 3 | 4 |
| 210 | HGB,SK,SNa | 0.69 | 9 | 0.26 | 0.77 | 7 | 5 |
| 211 | SK,SNa,WBC | 0.64 | 5 | 0.28 | 0.92 | 4 | 3 |
| 212 | Pl,SK,SNa | 0.64 | 6 | 0.29 | 0.99 | 2 | 3 |
| 213 | HGB,Pl,SCr,SNa,WBC | 0.80 | 3 | 0.29 | 0.81 | 5 | 2 |
| 214 | Pl,SCr,SNa,WBC | 0.79 | 3 | 0.29 | 0.81 | 5 | 2 |
| 215 | HGB,Pl,SNa,WBC | 0.71 | 5 | 0.26 | 0.85 | 6 | 2 |
| 216 | HGB,Pl,SCr,SNa | 0.80 | 4 | 0.26 | 0.72 | 0 | 3 |
| 217 | Pl,SCr,SNa | 0.79 | 5 | 0.19 | 0.79 | 6 | 4 |
| 218 | HGB,Pl,SNa | 0.69 | 7 | 0.21 | 0.73 | 4 | 4 |
| 219 | Pl,SNa,WBC | 0.64 | 4 | 0.29 | 0.77 | 6 | 0 |
| 220 | HGB,SCr,SNa,WBC | 0.80 | 6 | 0.16 | 0.88 | 3 | 5 |
| 221 | SCr,SNa,WBC | 0.79 | 4 | 0.30 | 0.77 | 5 | 2 |
| 222 | HGB,SNa,WBC | 0.70 | 7 | 0.16 | 0.74 | 6 | 5 |
| 223 | HGB,SCr,SNa | 0.79 | 4 | 0.20 | 0.86 | 3 | 5 |
| 224 | SCr,SNa | 0.79 | 3 | 0.17 | 0.73 | 9 | 3 |
| 225 | HGB,SNa | 0.68 | 8 | 0.26 | 0.78 | 1 | 5 |
| 226 | SNa,WBC | 0.61 | 9 | 0.30 | 0.75 | 10 | 3 |
| 227 | Pl,SNa | 0.61 | 9 | 0.19 | 0.95 | 7 | 4 |
| 228 | SK,SNa | 0.63 | 5 | 0.28 | 0.92 | 4 | 3 |
| 229 | HGB,Pl,SCr,SK,WBC | 0.80 | 9 | 0.10 | 0.82 | 4 | 5 |
| 230 | Pl,SCr,SK,WBC | 0.80 | 4 | 0.20 | 0.72 | 10 | 4 |
| 231 | HGB,Pl,SK,WBC | 0.72 | 7 | 0.26 | 0.84 | 0 | 3 |
| 232 | HGB,Pl,SCr,SK | 0.80 | 5 | 0.25 | 0.97 | 6 | 0 |
| 233 | Pl,SCr,SK | 0.79 | 8 | 0.21 | 0.96 | 6 | 5 |
| 234 | HGB,Pl,SK | 0.69 | 9 | 0.16 | 0.95 | 10 | 5 |
| 235 | Pl,SK,WBC | 0.66 | 5 | 0.17 | 0.78 | 3 | 4 |
| 236 | HGB,SCr,SK,WBC | 0.80 | 7 | 0.11 | 0.78 | 0 | 2 |
| 237 | SCr,SK,WBC | 0.79 | 5 | 0.25 | 0.89 | 0 | 2 |
| 238 | HGB,SK,WBC | 0.70 | 5 | 0.29 | 0.95 | 1 | 4 |
| 239 | HGB,SCr,SK | 0.80 | 6 | 0.29 | 0.76 | 3 | 4 |
| 240 | SCr,SK | 0.79 | 6 | 0.08 | 0.79 | 8 | 3 |
| 241 | HGB,SK | 0.68 | 8 | 0.28 | 0.92 | 4 | 0 |
| 242 | SK,WBC | 0.64 | 9 | 0.11 | 0.74 | 10 | 5 |
| 243 | Pl,SK | 0.63 | 9 | 0.20 | 0.89 | 2 | 5 |
| 244 | HGB,Pl,SCr,WBC | 0.8 | 5 | 0.23 | 0.77 | 8 | 2 |
| 245 | Pl,SCr,WBC | 0.79 | 4 | 0.18 | 0.71 | 8 | 5 |
| 246 | HGB,Pl,WBC | 0.71 | 4 | 0.20 | 0.86 | 3 | 5 |
| 247 | HGB,Pl,SCr | 0.79 | 3 | 0.02 | 0.70 | 2 | 2 |
| 248 | Pl,SCr | 0.79 | 5 | 0.18 | 0.99 | 4 | 5 |
| 249 | HGB,Pl | 0.68 | 7 | 0.19 | 0.72 | 8 | 2 |
| 250 | Pl,WBC | 0.63 | 3 | 0.20 | 0.75 | 1 | 0 |
| 251 | HGB,SCr,WBC | 0.8 | 4 | 0.23 | 0.95 | 7 | 0 |
| 252 | SCr,WBC | 0.79 | 4 | 0.26 | 0.77 | 6 | 3 |
| 253 | HGB,WBC | 0.70 | 5 | 0.05 | 0.89 | 10 | 1 |
| 254 | HGB,SCr | 0.79 | 7 | 0.03 | 0.99 | 8 | 5 |
| 255 | SCr | 0.79 | 10 | 0.21 | 0.97 | 8 | 4 |
| 256 | HGB | 0.67 | 8 | 0.27 | 0.77 | 0 | 0 |
| 257 | WBC | 0.60 | 8 | 0.23 | 0.70 | 8 | 1 |
| 258 | Pl | 0.56 | 8 | 0.29 | 0.97 | 3 | 2 |
| 259 | SK | 0.62 | 9 | 0.26 | 0.70 | 1 | 0 |
| 260 | SNa | 0.59 | 6 | 0.13 | 0.86 | 0 | 0 |
| 261 | SCl | 0.60 | 10 | 0.05 | 0.80 | 10 | 4 |
| 262 | ACr | 0.70 | 5 | 0.16 | 0.87 | 3 | 3 |
| 263 | SBC | 0.62 | 3 | 0.14 | 0.91 | 10 | 4 |
References
- Gunčar, G.; Kukar, M.; Notar, M.; Brvar, M.; Černelč, P.; Notar, M.; Notar, M. An Application of Machine Learning to Haematological Diagnosis. Sci. Rep. 2018, 8, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Badrick, T. Evidence-Based Laboratory Medicine. Clin. Biochem. Rev. 2013, 34, 43. [Google Scholar] [PubMed]
- Cabitza, F.; Banfi, G. Machine Learning in Laboratory Medicine: Waiting for the Flood? Clin. Chem. Lab. Med. (CCLM) 2018, 56, 516–524. [Google Scholar] [CrossRef]
- Louis, D.N.; Gerber, G.K.; Baron, J.M.; Bry, L.; Dighe, A.S.; Getz, G.; Higgins, J.M.; Kuo, F.C.; Lane, W.J.; Michaelson, J.S.; et al. Computational Pathology: An Emerging Definition. Arch. Pathol. Lab. Med. 2014, 138, 1133–1138. [Google Scholar] [CrossRef]
- Demirci, F.; Akan, P.; Kume, T.; Sisman, A.R.; Erbayraktar, Z.; Sevinc, S. Artificial Neural Network Approach in Laboratory Test Reporting: Learning Algorithms. Am. J. Clin. Pathol. 2016, 146, 227–237. [Google Scholar] [CrossRef] [PubMed]
- Diri, B.; Albayrak, S. Visualization and Analysis of Classifiers Performance in Multi-Class Medical Data. Expert Syst. Appl. 2008, 34, 628–634. [Google Scholar] [CrossRef]
- Lin, C.; Karlson, E.W.; Canhao, H.; Miller, T.A.; Dligach, D.; Chen, P.J.; Perez, R.N.G.; Shen, Y.; Weinblatt, M.E.; Shadick, N.A. Automatic Prediction of Rheumatoid Arthritis Disease Activity from the Electronic Medical Records. PLoS ONE 2013, 8, e69932. [Google Scholar] [CrossRef]
- Liu, K.E.; Lo, C.-L.; Hu, Y.-H. Improvement of Adequate Use of Warfarin for the Elderly Using Decision Tree-Based Approaches. Methods Inf. Med. 2014, 53, 47–53. [Google Scholar] [CrossRef]
- Razavian, N.; Blecker, S.; Schmidt, A.M.; Smith-McLallen, A.; Nigam, S.; Sontag, D. Population-Level Prediction of Type 2 Diabetes from Claims Data and Analysis of Risk Factors. Big Data 2015, 3, 277–287. [Google Scholar] [CrossRef]
- Putin, E.; Mamoshina, P.; Aliper, A.; Korzinkin, M.; Moskalev, A.; Kolosov, A.; Ostrovskiy, A.; Cantor, C.; Vijg, J.; Zhavoronkov, A. Deep Biomarkers of Human Aging: Application of Deep Neural Networks to Biomarker Development. Aging 2016, 8, 1021–1030. [Google Scholar] [CrossRef]
- Yuan, C.; Ming, C.; Chengjin, H. UrineCART, a Machine Learning Method for Establishment of Review Rules Based on UF-1000i Flow Cytometry and Dipstick or Reflectance Photometer. Clin. Chem. Lab. Med. (CCLM) 2012, 50, 2155–2161. [Google Scholar] [CrossRef] [PubMed]
- Goldstein, B.A.; Navar, A.M.; Carter, R.E. Moving beyond Regression Techniques in Cardiovascular Risk Prediction: Applying Machine Learning to Address Analytic Challenges. Eur. Heart J. 2017, 38, 1805–1814. [Google Scholar] [CrossRef] [PubMed]
- Surinova, S.; Choi, M.; Tao, S.; Schüffler, P.J.; Chang, C.-Y.; Clough, T.; Vysloužil, K.; Khoylou, M.; Srovnal, J.; Liu, Y. Prediction of Colorectal Cancer Diagnosis Based on Circulating Plasma Proteins. EMBO Mol. Med. 2015, 7, 1166–1178. [Google Scholar] [CrossRef] [PubMed]
- Richardson, A.; Signor, B.M.; Lidbury, B.A.; Badrick, T. Clinical Chemistry in Higher Dimensions: Machine-Learning and Enhanced Prediction from Routine Clinical Chemistry Data. Clin. Biochem. 2016, 49, 1213–1220. [Google Scholar] [CrossRef]
- Somnay, Y.R.; Craven, M.; McCoy, K.L.; Carty, S.E.; Wang, T.S.; Greenberg, C.C.; Schneider, D.F. Improving Diagnostic Recognition of Primary Hyperparathyroidism with Machine Learning. Surgery 2017, 161, 1113–1121. [Google Scholar] [CrossRef]
- Nelson, D.W.; Rudehill, A.; MacCallum, R.M.; Holst, A.; Wanecek, M.; Weitzberg, E.; Bellander, B.-M. Multivariate Outcome Prediction in Traumatic Brain Injury with Focus on Laboratory Values. J. Neurotrauma 2012, 29, 2613–2624. [Google Scholar] [CrossRef]
- Kumar, Y.; Sahoo, G. Prediction of Different Types of Liver Diseases Using Rule Based Classification Model. Technol. Health Care 2013, 21, 417–432. [Google Scholar] [CrossRef]
- Lu, W.; Ng, R. Automated Analysis of Public Health Laboratory Test Results. AMIA Jt Summits Transl. Sci. 2020, 2020, 393–402. [Google Scholar]
- Yang, H.S.; Hou, Y.; Vasovic, L.V.; Steel, P.; Chadburn, A.; Racine-Brzostek, S.E.; Velu, P.; Cushing, M.M.; Loda, M.; Kaushal, R.; et al. Routine Laboratory Blood Tests Predict SARS-CoV-2 Infection Using Machine Learning. Clin. Chem. 2020, 66, 1396–1404. [Google Scholar] [CrossRef]
- Han, J.; Kamber, M.; Pei, J. Data Mining Concepts and Techniques Third Edition. In The Morgan Kaufmann Series in Data Management Systems; Elsevier: Amsterdam, The Netherlands, 2011; pp. 83–124. [Google Scholar]
- Krause, J.; Perer, A.; Bertini, E. Using Visual Analytics to Interpret Predictive Machine Learning Models. arXiv 2016, arXiv:1606.05685. [Google Scholar]
- Keim, D.A.; Mansmann, F.; Thomas, J. Visual Analytics: How Much Visualization and How Much Analytics? SIGKDD Explor. Newsl. 2010, 11, 5. [Google Scholar] [CrossRef]
- Kehrer, J.; Hauser, H. Visualization and Visual Analysis of Multifaceted Scientific Data: A Survey. IEEE Trans. Vis. Comput. Graph. 2013, 19, 495–513. [Google Scholar] [CrossRef]
- Ola, O.; Sedig, K. Discourse with Visual Health Data: Design of Human-Data Interaction. Multimodal Technol. Interact. 2018, 2, 10. [Google Scholar] [CrossRef]
- Munzner, T. Visualization Analysis and Design; CRC Press: Boca Raton, FL, USA, 2014; ISBN 978-1-4987-5971-7. [Google Scholar]
- Treisman, A. Preattentive Processing in Vision. Comput. Vis. Graph. Image Processing 1985, 31, 156–177. [Google Scholar] [CrossRef]
- Ware, C. Information Visualization: Perception for Design; Morgan Kaufmann: Burlington, MA, USA, 2019; ISBN 978-0-12-812876-3. [Google Scholar]
- Simpao, A.F.; Ahumada, L.M.; Gálvez, J.A.; Rehman, M.A. A Review of Analytics and Clinical Informatics in Health Care. J. Med. Syst. 2014, 38, 45. [Google Scholar] [CrossRef] [PubMed]
- Saffer, J.D.; Burnett, V.L.; Chen, G.; van der Spek, P. Visual Analytics in the Pharmaceutical Industry. IEEE Comput. Graph. Appl. 2004, 24, 10–15. [Google Scholar] [CrossRef] [PubMed]
- Abdullah, S.S.; Rostamzadeh, N.; Sedig, K.; Garg, A.X.; McArthur, E. Multiple Regression Analysis and Frequent Itemset Mining of Electronic Medical Records: A Visual Analytics Approach Using VISA_M3R3. Data 2020, 5, 33. [Google Scholar] [CrossRef]
- Abdullah, S.S.; Rostamzadeh, N.; Sedig, K.; Garg, A.X.; McArthur, E. Visual Analytics for Dimension Reduction and Cluster Analysis of High Dimensional Electronic Health Records. Informatics 2020, 7, 17. [Google Scholar] [CrossRef]
- Parsons, P.; Sedig, K.; Mercer, R.; Khordad, M.; Knoll, J.; Rogan, P. Visual Analytics for Supporting Evidence-Based Interpretation of Molecular Cytogenomic Findings. In VAHC ’15: Proceedings of the 2015 Workshop on Visual Analytics in Healthcare; Association for Computing Machinery: New York, NY, USA, 2015. [Google Scholar]
- Ola, O.; Sedig, K. The Challenge of Big Data in Public Health: An Opportunity for Visual Analytics. Online J. Public Health Inform. 2014, 5, 223. [Google Scholar] [CrossRef][Green Version]
- Baytas, I.M.; Lin, K.; Wang, F.; Jain, A.K.; Zhou, J. PhenoTree: Interactive Visual Analytics for Hierarchical Phenotyping From Large-Scale Electronic Health Records. IEEE Trans. Multimed. 2016, 18, 2257–2270. [Google Scholar] [CrossRef]
- Perer, A.; Sun, J. MatrixFlow: Temporal Network Visual Analytics to Track Symptom Evolution during Disease Progression. AMIA Annu. Symp Proc. 2012, 2012, 716–725. [Google Scholar] [PubMed]
- Ninkov, A.; Sedig, K. VINCENT: A Visual Analytics System for Investigating the Online Vaccine Debate. Online J. Public Health Inform. 2019, 11, e5. [Google Scholar] [CrossRef] [PubMed]
- Perer, A.; Wang, F.; Hu, J. Mining and Exploring Care Pathways from Electronic Medical Records with Visual Analytics. J. Biomed. Inform. 2015, 56, 369–378. [Google Scholar] [CrossRef]
- Klimov, D.; Shknevsky, A.; Shahar, Y. Exploration of Patterns Predicting Renal Damage in Patients with Diabetes Type II Using a Visual Temporal Analysis Laboratory. J. Am. Med. Inform. Assoc. 2015, 22, 275–289. [Google Scholar] [CrossRef] [PubMed]
- Mane, K.K.; Bizon, C.; Schmitt, C.; Owen, P.; Burchett, B.; Pietrobon, R.; Gersing, K. VisualDecisionLinc: A Visual Analytics Approach for Comparative Effectiveness-Based Clinical Decision Support in Psychiatry. J. Biomed. Inform. 2012, 45, 101–106. [Google Scholar] [CrossRef] [PubMed]
- Gotz, D.H.; Sun, J.; Cao, N. Multifaceted Visual Analytics for Healthcare Applications. IBM J. Res. Dev. 2012, 56, 1–6. [Google Scholar] [CrossRef]
- Mittelstädt, S.; Hao, M.C.; Dayal, U.; Hsu, M.C.; Terdiman, J.; Keim, D.A. Advanced Visual Analytics Interfaces for Adverse Drug Event Detection. In Proceedings of the Workshop on Advanced Visual Interfaces AVI; Association for Computing Machinery: New York, NY, USA, 2014; pp. 237–244. [Google Scholar]
- Basole, R.C.; Braunstein, M.L.; Kumar, V.; Park, H.; Kahng, M.; Chau, D.H.; Tamersoy, A.; Hirsh, D.A.; Serban, N.; Bost, J.; et al. Understanding Variations in Pediatric Asthma Care Processes in the Emergency Department Using Visual Analytics. J. Am. Med. Inform. Assoc. 2015, 22, 318–323. [Google Scholar] [CrossRef]
- Rostamzadeh, N.; Abdullah, S.S.; Sedig, K. Visual Analytics for Electronic Health Records: A Review. Informatics 2021, 8, 12. [Google Scholar] [CrossRef]
- Abdullah, S. Visual Analytics of Electronic Health Records with a Focus on Acute Kidney Injury. Ph.D. Thesis, The University of Western Ontario, London, UK, 2020. [Google Scholar]
- Abdullah, S.S.; Rostamzadeh, N.; Sedig, K.; Lizotte, D.J.; Garg, A.X.; McArthur, E. Machine Learning for Identifying Medication-Associated Acute Kidney Injury. Informatics 2020, 7, 18. [Google Scholar] [CrossRef]
- Rostamzadeh, N.; Abdullah, S.S.; Sedig, K.; Garg, A.X.; McArthur, E. VERONICA: Visual Analytics for Identifying Feature Groups in Disease Classification. Information 2021, 12, 344. [Google Scholar] [CrossRef]
- Thomas, J.J.; Cook, K.A. Illuminating the Path: The Research and Development Agenda for Visual Analytics; Pacific Northwest National Lab.(PNNL): Richland, WA, USA, 2005. [Google Scholar]
- Endert, A.; Hossain, M.S.; Ramakrishnan, N.; North, C.; Fiaux, P.; Andrews, C. The Human Is the Loop: New Directions for Visual Analytics. J. Intell. Inf. Syst. 2014, 43, 411–435. [Google Scholar] [CrossRef]
- Sedig, K.; Parsons, P. Design of Visualizations for Human-Information Interaction: A Pattern-Based Framework. Synth. Lect. Vis. 2016, 4, 1–185. [Google Scholar] [CrossRef][Green Version]
- Angulo, D.A.; Schneider, C.; Oliver, J.H.; Charpak, N.; Hernandez, J.T. A Multi-Facetted Visual Analytics Tool for Exploratory Analysis of Human Brain and Function Datasets. Front. Neuroinform. 2016, 10. [Google Scholar] [CrossRef] [PubMed]
- Zhao, K.; Ward, M.; Rundensteiner, E.; Higgins, H. MaVis: Machine Learning Aided Multi-Model Framework for Time Series Visual Analytics. Electron. Imaging 2016, 2016, 1–10. [Google Scholar] [CrossRef]
- Sedig, K.; Parsons, P.; Babanski, A. Towards a Characterization of Interactivity in Visual Analytics. J. Multimed. Processing Technol. 2012, 3, 12–28. [Google Scholar]
- Didandeh, A.; Sedig, K. Externalization of Data Analytics Models. In Proceedings of the Human Interface and the Management of Information: Information, Design and Interaction; Yamamoto, S., Ed.; Springer International Publishing: Cham, Switzerland, 2016; pp. 103–114. [Google Scholar]
- Keim, D.A.; Munzner, T.; Rossi, F.; Verleysen, M. Bridging Information Visualization with Machine Learning (Dagstuhl Seminar 15101). Dagstuhl Rep. 2015, 5, 1–27. [Google Scholar] [CrossRef]
- Yu, Y.; Long, J.; Liu, F.; Cai, Z. Machine Learning Combining with Visualization for Intrusion Detection: A Survey. In Proceedings of the Modeling Decisions for Artificial Intelligence; Torra, V., Narukawa, Y., Navarro-Arribas, G., Yañez, C., Eds.; Springer International Publishing: Cham, Switzerland, 2016; pp. 239–249. [Google Scholar]
- Jeong, D.H.; Ji, S.Y.; Suma, E.A.; Yu, B.; Chang, R. Designing a Collaborative Visual Analytics System to Support Users’ Continuous Analytical Processes. Hum. Cent. Comput. Inf. Sci. 2015, 5, 5. [Google Scholar] [CrossRef]
- Han, J.; Kamber, M. Data Mining: Concepts and Techniques; Elsevier: Amsterdam, The Netherlands, 2011. [Google Scholar]
- Rostamzadeh, N.; Abdullah, S.S.; Sedig, K. Data-Driven Activities Involving Electronic Health Records: An Activity and Task Analysis Framework for Interactive Visualization Tools. Multimodal Technol. Interact. 2020, 4, 7. [Google Scholar] [CrossRef]
- Salomon, G. Distributed Cognitions: Psychological and Educational Considerations; Cambridge University Press: Cambrige, UK, 1997; ISBN 978-0-521-57423-5. [Google Scholar]
- Liu, Z.; Nersessian, N.; Stasko, J. Distributed Cognition as a Theoretical Framework for Information Visualization. IEEE Trans. Vis. Comput. Graph. 2008, 14, 1173–1180. [Google Scholar] [CrossRef]
- Agrawal, R.; Srikant, R. Fast Algorithms for Mining Association Rules in Large Databases. In Proceedings of the 20th International Conference on Very Large Data Bases, San Francisco, CA, USA, 12–15 September 1994; Morgan Kaufmann Publishers Inc.: San Francisco, CA, USA, 1994; pp. 487–499. [Google Scholar]
- Fernando, B.; Fromont, E.; Tuytelaars, T. Effective Use of Frequent Itemset Mining for Image Classification. In European Conference on Computer Vision; Fitzgibbon, A., Lazebnik, S., Perona, P., Sato, Y., Schmid, C., Eds.; Springer: Berlin/Heidelberg, Germany, 2012; pp. 214–227. [Google Scholar]
- Naulaerts, S.; Meysman, P.; Bittremieux, W.; Vu, T.N.; Vanden Berghe, W.; Goethals, B.; Laukens, K. A Primer to Frequent Itemset Mining for Bioinformatics. Brief. Bioinform. 2015, 16, 216–231. [Google Scholar] [CrossRef]
- Brauckhoff, D.; Dimitropoulos, X.; Wagner, A.; Salamatian, K. Anomaly Extraction in Backbone Networks Using Association Rules. IEEE/ACM Trans. Netw. 2012, 20, 1788–1799. [Google Scholar] [CrossRef]
- Glatz, E.; Mavromatidis, S.; Ager, B.; Dimitropoulos, X. Visualizing Big Network Traffic Data Using Frequent Pattern Mining and Hypergraphs. Computing 2014, 96, 27–38. [Google Scholar] [CrossRef]
- Mukherjee, A.; Liu, B.; Glance, N. Spotting Fake Reviewer Groups in Consumer Reviews. In Proceedings of the 21st International Conference on World Wide Web; Association for Computing Machinery: New York, NY, USA, 2012; pp. 191–200. [Google Scholar]
- Liu, Y.; Zhao, Y.; Chen, L.; Pei, J.; Han, J. Mining Frequent Trajectory Patterns for Activity Monitoring Using Radio Frequency Tag Arrays. IEEE Trans. Parallel Distrib. Syst. 2012, 23, 2138–2149. [Google Scholar] [CrossRef]
- Ordonez, C. Association Rule Discovery with the Train and Test Approach for Heart Disease Prediction. IEEE Trans. Inf. Technol. Biomed. 2006, 10, 334–343. [Google Scholar] [CrossRef]
- Ilayaraja, M.; Meyyappan, T. Efficient Data Mining Method to Predict the Risk of Heart Diseases Through Frequent Itemsets. Proc. Comput. Sci. 2015, 70, 586–592. [Google Scholar] [CrossRef]
- Zaki, M.J. Scalable Algorithms for Association Mining. IEEE Trans. Knowl. Data Eng. 2000, 12, 372–390. [Google Scholar] [CrossRef]
- Chen, T.; Guestrin, C. XGBoost: A Scalable Tree Boosting System. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining; ACM: San Francisco, CA, USA, 2016; pp. 785–794. [Google Scholar] [CrossRef]
- Pavlyshenko, B.M. Linear, Machine Learning and Probabilistic Approaches for Time Series Analysis. In Proceedings of the 2016 IEEE First International Conference on Data Stream Mining and Processing (DSMP); Institute of Electrical and Electronics Engineers (IEEE): Piscataway, NJ, USA, 2016; pp. 377–381. [Google Scholar]
- Tamayo, D.; Silburt, A.; Valencia, D.; Menou, K.; Ali-Dib, M.; Petrovich, C.; Huang, C.X.; Rein, H.; van Laerhoven, C.; Paradise, A.; et al. A machine learns to predict the stability of tightly packed planetary systems. Astrophys. J. Lett. 2016, 832, L22. [Google Scholar] [CrossRef]
- Möller, A.; Ruhlmann-Kleider, V.; Leloup, C.; Neveu, J.; Palanque-Delabrouille, N.; Rich, J.; Carlberg, R.; Lidman, C.; Pritchet, C. Photometric Classification of Type Ia Supernovae in the SuperNova Legacy Survey with Supervised Learning. J. Cosmol. Astropart. Phys. 2016, 2016, 008. [Google Scholar] [CrossRef]
- Friedman, J.H. Stochastic Gradient Boosting. Comput. Stat. Data Anal. 2002, 38, 367–378. [Google Scholar] [CrossRef]
- Zhao, X.; Wu, Y.; Lee, D.L.; Cui, W. IForest: Interpreting Random Forests via Visual Analytics. IEEE Trans. Vis. Comput. Graph. 2019, 25, 407–416. [Google Scholar] [CrossRef]
- Hettinger, A.Z.; Roth, E.M.; Bisantz, A.M. Cognitive Engineering and Health Informatics: Applications and Intersections. J. Biomed. Inform. 2017, 67, 21–33. [Google Scholar] [CrossRef] [PubMed]
- Benyon, D. Designing Interactive Systems: A Comprehensive Guide to HCI, UX and Interaction Design, 3rd ed.; Pearson: London, UK, 2013; ISBN 978-1-4479-2011-3. [Google Scholar]
- Sedig, K.; Naimi, A.; Haggerty, N. Aligning information technologies with evidencebased health-care activities: A design and evaluation framework. Hum. Technol. 2017, 13, 180–215. [Google Scholar] [CrossRef][Green Version]
- Leighton, J.P. Defining and Describing Reason. In The Nature of Reasoning; Cambridge University Press: Cambrige, UK, 2004; pp. 3–11. ISBN 0-521-81090-6. [Google Scholar]
- Sedig, K.; Parsons, P. Interaction Design for Complex Cognitive Activities with Visual Representations: A Pattern-Based Approach. AIS Trans. Hum. Comput. Interact. 2013, 5, 84–133. [Google Scholar] [CrossRef]
- Bergstra, J.; Bengio, Y. Random Search for Hyper-Parameter Optimization. J. Mach. Learn. Res. 2012, 13, 281–305. [Google Scholar]
- Ferri, C.; Hernández-Orallo, J.; Modroiu, R. An Experimental Comparison of Performance Measures for Classification. Pattern Recognit. Lett. 2009, 30, 27–38. [Google Scholar] [CrossRef]
- Garcıa, V.; Sánchez, J.S.; Mollineda, R.A. On the Suitability of Numerical Performance Measures for Class Imbalance Problems. In Proceedings of the 1st International Conference on Pattern Recognition Applications and Methods; SciTePress—Science and and Technology Publications: Setubal, Portugalia, 2012; pp. 310–313. [Google Scholar]
- Parikh, R.; Mathai, A.; Parikh, S.; Chandra Sekhar, G.; Thomas, R. Understanding and Using Sensitivity, Specificity and Predictive Values. Indian J. Ophthalmol. 2008, 56, 45–50. [Google Scholar] [CrossRef] [PubMed]
- Waikar, S.S.; Curhan, G.C.; Wald, R.; McCarthy, E.P.; Chertow, G.M. Declining Mortality in Patients with Acute Renal Failure, 1988 to 2002. JASN 2006, 17, 1143–1150. [Google Scholar] [CrossRef]
- Liangos, O.; Wald, R.; O’Bell, J.W.; Price, L.; Pereira, B.J.; Jaber, B.L. Epidemiology and Outcomes of Acute Renal Failure in Hospitalized Patients: A National Survey. CJASN 2006, 1, 43–51. [Google Scholar] [CrossRef]
- Waikar, S.S.; Curhan, G.C.; Ayanian, J.Z.; Chertow, G.M. Race and Mortality after Acute Renal Failure. J. Am. Soc. Nephrol. 2007, 18, 2740–2748. [Google Scholar] [CrossRef]
- Chen, D.-N.; Du, J.; Xie, Y.; Li, M.; Wang, R.-L.; Tian, R. Relationship between Early Serum Sodium and Potassium Levels and AKI Severity and Prognosis in Oliguric AKI Patients. Int. Urol. Nephrol. 2021, 53, 1171–1187. [Google Scholar] [CrossRef]
- Lim, S.Y.; Park, Y.; Chin, H.J.; Na, K.Y.; Chae, D.-W.; Kim, S. Short-Term and Long-Term Effects of Low Serum Bicarbonate Level at Admission in Hospitalised Patients. Sci. Rep. 2019, 9, 2798. [Google Scholar] [CrossRef]
- Oh, H.J.; Kim, S.; Park, J.T.; Kim, S.-J.; Han, S.H.; Yoo, T.-H.; Ryu, D.-R.; Kang, S.-W.; Chung, Y.E. Baseline Chloride Levels Are Associated with the Incidence of Contrast-Associated Acute Kidney Injury. Sci. Rep. 2017, 7, 17431. [Google Scholar] [CrossRef] [PubMed]
- Du Cheyron, D.; Parienti, J.-J.; Fekih-Hassen, M.; Daubin, C.; Charbonneau, P. Impact of Anemia on Outcome in Critically Ill Patients with Severe Acute Renal Failure. Intensive Care Med. 2005, 31, 1529–1536. [Google Scholar] [CrossRef] [PubMed]
- Gameiro, J.; Lopes, J.A. Complete Blood Count in Acute Kidney Injury Prediction: A Narrative Review. Ann. Intensive Care 2019, 9, 87. [Google Scholar] [CrossRef] [PubMed]






Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rostamzadeh, N.; Abdullah, S.S.; Sedig, K.; Garg, A.X.; McArthur, E. Visual Analytics for Predicting Disease Outcomes Using Laboratory Test Results. Informatics 2022, 9, 17. https://doi.org/10.3390/informatics9010017
Rostamzadeh N, Abdullah SS, Sedig K, Garg AX, McArthur E. Visual Analytics for Predicting Disease Outcomes Using Laboratory Test Results. Informatics. 2022; 9(1):17. https://doi.org/10.3390/informatics9010017
Chicago/Turabian StyleRostamzadeh, Neda, Sheikh S. Abdullah, Kamran Sedig, Amit X. Garg, and Eric McArthur. 2022. "Visual Analytics for Predicting Disease Outcomes Using Laboratory Test Results" Informatics 9, no. 1: 17. https://doi.org/10.3390/informatics9010017
APA StyleRostamzadeh, N., Abdullah, S. S., Sedig, K., Garg, A. X., & McArthur, E. (2022). Visual Analytics for Predicting Disease Outcomes Using Laboratory Test Results. Informatics, 9(1), 17. https://doi.org/10.3390/informatics9010017








