Abstract
A conceptual artificial intelligence (AI)-enabled framework is presented in this study involving triangulation of various diagnostic methods for management of coronavirus disease 2019 (COVID-19) and its associated comorbidities in resource-limited settings (RLS). The proposed AI-enabled framework will afford capabilities to harness low-cost polymerase chain reaction (PCR)-based molecular diagnostics, radiological image-based assessments, and end-user provided information for the detection of COVID-19 cases and management of symptomatic patients. It will support self-data capture, clinical risk stratification, explanation-based intelligent recommendations for patient triage, disease diagnosis, patient treatment, contact tracing, and case management. This will enable communication with end-users in local languages through cheap and accessible means, such as WhatsApp/Telegram, social media, and SMS, with careful consideration of the need for personal data protection. The objective of the AI-enabled framework is to leverage multimodal diagnostics of COVID-19 and associated comorbidities in RLS for the diagnosis and management of COVID-19 cases and general support for pandemic recovery. We intend to test the feasibility of implementing the proposed framework through community engagement in sub-Saharan African (SSA) countries where many people are living with pre-existing comorbidities. A multimodal approach to disease diagnostics enabling access to point-of-care testing is required to reduce fragmentation of essential services across the continuum of COVID-19 care.
1. Introduction
Countries in Sub-Saharan Africa (SSA) have been affected by the novel coronavirus 2019 (COVID-19) infection with severe impacts on the people, health systems, and their economies. Many cases of COVID-19 in SSA countries have associated comorbidities such as tuberculosis (TB), HIV/AIDS, diabetes, hypertension, and malaria. Cases of COVID-19 and associated comorbidities are more difficult to manage and have resulted in more deaths [1].
South Africa, Ethiopia, Kenya, and Nigeria are the countries with the highest number of reported COVID-19 cases in SSA [2]. These countries also have a large population of persons with comorbidities such as HIV, TB, and diabetes. In terms of absolute numbers, South Africa has 7.7 million people living with HIV, the highest number in the world. HIV remains prevalent in the population with approximately 20% of people infected with HIV. There is a large number of people with HIV/TB co-infection in South Africa, where TB is recognized as a high burden disease. In 2018, an estimated 63,000 people were reported to have died from active TB, out of which 42,000 were HIV positive. Yearly, TB remains the largest contributor to death in South Africa [3]. Nigeria, with 1.9 million people living with HIV, has the fourth-largest HIV epidemic in the world in term of absolute numbers. Nigeria also has the fourth-largest TB epidemic in the world, with cases of HIV/TB co-infection on the rise. Apart from HIV/AIDS, cases of drug-resistant TB are prevalent in Nigeria. An estimated 407,000 die of TB each year in Nigeria, out of which an estimated 39,000 are HIV positive people [4].
Thus, the existence of a large population of people with comorbidities in SSA countries makes it more difficult to manage cases of COVID-19 worsened by co-infection. For these types of patients, it is more difficult to determine the pattern of progression of their pre-existing conditions in the presence of COVID-19 and the impact these comorbidities have on the best form of personalized care that is required to treat them for their COVID-19 infection [5,6]. Currently, difficulties exist in the area of active case finding due to weak disease surveillance infrastructures, low testing capacity particularly in rural areas, and many RLS in these countries. Additionally, there are delays in the treatment of detected active cases because the traditional healthcare system infrastructures in these countries are overstretched. Experiences from the COVID-19 pandemic in South Africa reveal that cases with comorbidities such as HIV, TB, diabetes, and hypertension were more complicated and difficult to manage [1]. There is limited access to point-of-care (POC) testing and a lack of effective disease surveillance in RLS, which has accounted for a relatively low number of reported COVID-19 cases [7,8]. For example, in South Africa, a significantly greater number of COVID-19 cases have been reported in the more developed provinces such as Western Cape, and Gauteng, compared to more rural provinces such as Limpopo and Eastern Cape with low testing capacity, partly due to inadequate resources. This pattern is repeated in many other SSA countries. The problem of limited access to POC testing opportunities and lack of an effective surveillance system remain major public health challenges that limit timely interventions to contain threats of infectious diseases in SSA.
This scenario makes the need for a multimodal approach to disease diagnostics to increase accessibility to POC testing by different categories of people compelling. It is also essential that the results of diagnostics from diverse sources are effectively harnessed through digital technology to enable effective disease surveillance and clinical management of identified cases. So far, digital health in Africa has been implemented at various levels of maturity and sophistication across regions and countries in SSA, which makes the deployment of digital solutions in many settings difficult. Problems exist in the area of lack of quality data and data curation mechanisms, lack of integration platforms to harness data from relevant sources, poor technology infrastructure, lack of coordination, and generally low digital technology diffusion [9,10].
There is a need to facilitate the integration of patient data from different sources and disparate health platforms by ensuring their interoperability, which will create the basis for data merging, and thereupon the application of artificial intelligence (AI) methods for gaining improved tools for better healthcare in SSA. The augmentation of existing traditional diagnostic and treatment approaches with AI-derived insights will alleviate some of the existing challenges particularly in the resource-limited settings of SSA. This includes, for example, the novel pathology-supported genetic testing (PSGT) approach [11] implemented in South Africa as a case study using the framework recently described by 38 researchers across the African continent (https://www.aasciences.africa/publications/policy-paper-framework-implementation-genomic-medicine-public-health-africa (accessed on 6 September 2021). The capacity of AI technology to augment sound decision-making using PSGT in patients stratified by body mass index [12] and other forms of data (text, image, audio)—be they real-time, recent or older—makes it a viable tool to supplement the capabilities of healthcare systems in SSA in the fight against COVID-19, other highly prevalent infectious diseases, and future pandemics.
The rest of this paper is structured as follows. Section 2 describes the technological challenge and the objectives of the framework. In Section 3, we present the methodology adopted for the design of the framework, while Section 4 discusses the merits of the framework. The paper is concluded in Section 5 with a brief outlook and future work envisaged.
2. The Technological Challenges and Objectives of the Framework
This section briefly presents the technological challenges from the perspective of recent trends identified in the literature and the objectives of the proposed framework using PSGT as a case study of genetic knowledge integration [11].
2.1. The Technological Challenge
Despite the promise of AI and digital technology in advancing infectious disease treatment [8], there is limited evidence on the successful application of AI and digitally connected point-of-care (POC) diagnostics for infectious diseases and associated comorbidities, particularly in rural settings in SSA with a high burden of disease. RLS are characterised by poor and inadequate healthcare infrastructures, shortages of qualified medical personnel, poverty, lack of access to technology, and high cost of healthcare in general [9]. Lately, there has been more interest in smartphone-based POC testing and related diagnostic support that could be suitable for RLS, but few of these applications have been successfully applied and sustained in SSA [13]. The high number of cases of COVID-19 in SSA and the low POC testing capacity in RLS make multiple POC diagnostic approaches that are sufficiently robust, accurate, and readily accessible to different categories of people living in RLS necessary. Increased access to POC testing will engender early detection as well as prompt treatment of cases, and thereby more efficient outbreak control. The potential to generate diverse types of data from various POC diagnostics and other patient data sources for larger populations creates an opportunity to explore the potential of AI-based novel approaches to facilitate and support better healthcare in RLS. Various types of data that may be available per time in a resource-limited setting including pathology data, genomics data, radiological image data, end-user/patient provided data, and electronic medical records will be collected. The types of available data will inform the AI-enabled integration of patient-centred diagnostic methods that are used with a focus on the most common COVID-19 comorbidities underpinned by genomic pathways coincident with the SARS-CoV-2 viral disease [14]. These include the APOE-cholesterol, MTHFR-homocysteine, FII/FV-blood clotting, and HFE-iron metabolism pathways [15] recently combined into a novel POC test kit (https://gtr.ukri.org/projects?ref=103993, accessed on 6 September 2021) for non-communicable diseases (NCDs) ranging from Alzheimer’s disease to cancer and cardiovascular disease (CVD). Thus, this study proposes an AI-enabled framework for multimodal diagnostics of COVID-19 and associated comorbidities at POC to improve access to improved clinical management in resource-limited settings in SSA.
2.2. Objectives of the Proposed Framework
We seek to create an AI-based technical framework that will enable the integration of data from various POC diagnostic approaches for the improved treatment of COVID-19 patients and their associated comorbidities. The better management of COVID-19 cases will entail the flagging of relevant information for prescribing more personalized treatments, predicting the probable pattern of disease progression, and handling post-treatment complications reported by patients. Furthermore, a component of this framework will provide smart and cost-effective disease surveillance tools for RLS within the given regional area of the healthcare providers involved. The framework will be a low-cost, easy-to-implement solution. It will be generic, thereby usable—after localization and adaptation—in many RLS in SSA.
Specifically, the AI framework will enable the following tasks:
- Collection of real-world evidence, particularly on diagnosis and treatment of COVID-19 patients
- Creation of an integrated platform for harnessing data from various POC diagnostic methods, including medical imaging-based diagnostics and healthcare IT systems
- Effective decision-making based on screening algorithms informing differential diagnosis and management of COVID-19-associated comorbidities in community-based health services in RLS
- Mobile phone-based contact tracing for COVID 19 and associated comorbidities
- PSGT combining diagnostic RNA-based SARS-CoV-2 testing with DNA-based chronic disease screening for managing potential biochemical abnormalities caused by gene-environment interaction.
The following self-reporting scenario is presented as an example of the PSGT algorithm being developed for feedback to patients by a healthcare practitioner: A patient experiences a reinfection with SARS-CoV-2 [16] three weeks after COVID-19 vaccination (Pfizer, first injection). It concerns a 63-year-old female first diagnosed with asymptomatic COVID-19 pre-surgery in June 2020, which supports the incorporation of COVID-19 status in the patient report. As shown in Table 1, PSGT typically starts with an online questionnaire-based health check including the assessment of body mass index (BMI) and metabolic syndrome features targeting dyslipidaemia associated with COVID-19 severity and co-morbidities. Involvement of this NCD pathway known to be influenced by lifestyle and other environmental factors, which in turn may trigger genetic risk such as the APOE e4 allele identified in this case, should be addressed to prevent cumulative causal effects [17,18]. Vaccine-related adverse effects are typically linked to old age [19] and pre-existing obesity, diabetes, or significant allergies. These are the high-risk subgroups we hope to identify at entry into the multimodal risk management programme for optimal treatment guided by our ever-increasing knowledge base developed in an ongoing way as new discoveries are reported in the literature [20].

Table 1.
Interpretative commenting on the pathology-supported genetic testing performed (April 2011) in the symptomatic COVID-19 reinfected patient based on self-data reporting (July 2021) for clinical follow-up by the treating physician.
3. Materials and Methods
The conceptual design of the proposed AI-enabled framework for multimodal diagnostics of COVID-19 is informed by the set of requirements that have been identified as critical for the practical realization of the envisioned framework requiring integration of data from different health sources. In this Section, these requirements, the conceptual architecture, and a description of key aspects of the architecture are presented.
3.1. Requirements of the AI-Enabled Framework for Multimodal Diagnostics of COVID-19
The proposed framework is envisioned to provide end-to-end support for the management of COVID-19 and pandemic situations. It will support self-data capture, case data capture, explanation-based intelligent recommendations for patient triage, disease diagnosis, patient treatment, contact tracing, and case management. Communication with end-users in local languages through cheap and accessible means such as WhatsApp/Telegram, social media, and SMS are also supported. The key activities that will be supported by the framework include:
- Data collection from patients on symptoms of COVID-19 that have been observed. This will be performed by using diverse sources such as interaction with health care workers via the phone, SMS messaging, WhatsApp messages/call, Telegram message/call. The information collected will be limited to issues relating to COVID-19 and will be stored on a secure system. The security of electronic health records (EHR) during transmission from one point to another shall be secured by using credible encryption methods. The options to consider include the use of scrambled alpha-numeric randomization combined with RSA, as we did in a previous study [23].
- Case documentation by health care workers (HCW) through a mobile app, webform
- Capture of radiological images from patients
- Access and integration with electronic medical records
- Prediction of disease using machine learning (ML)
- AI-generated recommendations with explanations for HCW
- Delivery of AI-generated suggestions to patients through mobile phone via personal calls, SMS messaging, WhatsApp, Telegram, Mobile App. Messages can be conveyed directly through these means, or via a phone call by the HCW
- Regular monitoring of patients (under self-quarantine)
- Aiding decision-making on patient triage and treatment schedule
- Data analytics and reporting for pandemic management by health management systems (Dashboard reporting)—hotspots, cases per community, neighbourhoods, regions
- Contact tracing application based on Bluetooth technology that can track close contact with infected persons (only phone numbers are stored on patient’s mobile phone). The application will be triggered by user consent, and stored content is also shared by user consent.
3.2. Architecture of the Proposed Framework for Multimodal Diagnostics of COVID-19
The conceptual architecture of the proposed framework (shown in Figure 1) is influenced by the objectives of the framework. It consists of an integration of existing health systems and components that will enable data from multiple sources and data inputs from various stakeholders to be harnessed for AI-supported decision-making. The self-data capture (1) component allows all data inputs from the patient/user to be collected using the most accessible means for the user. This includes messages sent from a webform, WhatsApp or Telegram, or a voice call in a local language or English. The case data capture (2) component will enable health care workers (HCW) to input all forms of data that pertain to a patient that has been captured through interrogation, clinical examination, or point-of-care testing (PCR-based test, genetic-based testing (PSGT) data, X-ray imaging). All the data from the different sources will be retrieved from the cloud data server by the AI engine component to generate a diagnostic report (3), which will be reviewed by the HCW. Based on the diagnostic report, appropriate information will be sent to the user/patient as the feedback report (4). The feedback could be sent automatically as authorized by the HCW or conveyed personally to the user. An information dashboard (5) that highlights the state of the pandemic, such as case statistics, infection hotspots, surveillance reports, etc., will also be updated regularly based on information that is generated from cases that have been handled via the framework. The AI-enabled framework will rely on the integration of available public health records, accessible private health records, and archival data sources that are available and accessible nationally.
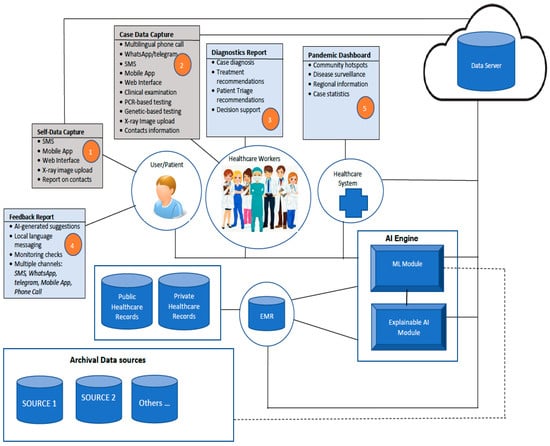
Figure 1.
The architecture of the proposed framework for multimodal diagnostics and management of COVID-19 and comorbidities.
3.3. Description of the Workflow and Configuration of the AI Engine
Let us consider a hypothetical case example of Mrs. AA, whose COVID-19 related data include the following:
- a.
- Socio-demographic and lifestyle characteristics (e.g., age, gender, ethnicity, residential area, occupation, workplace, smoking status, alcohol use, drug use)
- b.
- Exposure history and symptoms (record of the previous contact with infected persons or places; symptoms types, level of severity of symptoms)
- c.
- Comorbidity profile (such as hypertension, COPD, diabetes, heart diseases, TB, and HIV, as well as severity level of the comorbidity)
- d.
- Medical imaging data such as CXR, chest CT of Mrs. AA
- e.
- Laboratory test data such as FBC, ferritin, PCT, MicroRNAs, NT-proBNP, and inflammatory cytokines)
- f.
- Data obtained for PSGT to customise feedback to Mrs. AA based on personal utility and clinical relevance (see Table 1).
- g.
- Vaccine and other Medication data (e.g., steroids, antibiotics, antiviral, antifungal, medication for glycaemic control, antihypertensive)
We now describe how the AI engine can process the data so that a result can be generated by the proposed framework. The AI engine consists of six aspects that determine its workflow (see Figure 2). These are data collection and pre-processing (1), feature extraction and data modelling (2), dataset repository (3), model training (4), explainable recommendations (5), and model evaluation (6). These aspects are described below:
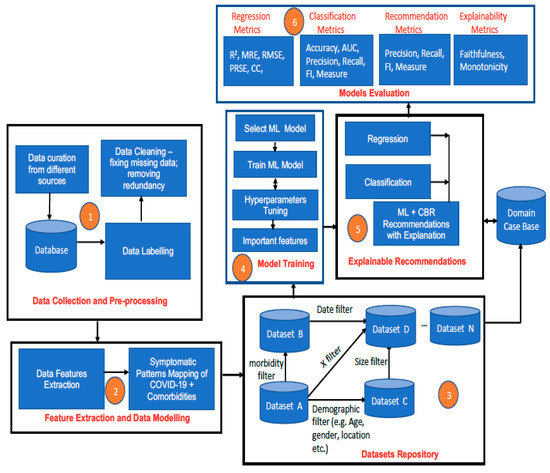
Figure 2.
The architecture of the AI engine of the proposed framework for multimodal diagnostics and management of COVID-19 and comorbidities.
3.3.1. Data Collection and Pre-Processing
- i.
- Archived but anonymized data of patients (like Mrs. AA) with COVID-19 cases are collected from diverse sources—hospital records, medical records databases in South Africa. This similarly applies to any other country in SSA.
- ii.
- The collected data are labelled by using descriptive attribute names that depict the information that they represent. For example, if the objective is to predict the possibility of COVID-19 infection, COVID-19 death, or COVID-19 recovery, then the dependent attributes such as age profile, gender type, symptom severity profile, comorbidity profile, the severity of comorbidity disease, weight profile, blood profile, genetic data, etc. that can influence specific patient outcomes such as infection, death, recovery, survival must be identified and appropriately labelled. The process will be a combination of manual and automated data labelling depending on the inherent quality of collected initial data.
- iii.
- Data pre-processing will be performed to clean up the data, fix missing values, and balance the dataset (if necessary) to reduce bias. Instances of missing numeric values can be replaced by the median or the mean value (for data with a normal distribution), while missing categorical values can be replaced by the mode. When there is an imbalanced dataset due to the under-representation of a specific class, class balancing techniques such as the synthetic minority oversampling technique (SMOTE) [24] will be used to generate additional synthetic data to ensure a balanced dataset.
3.3.2. Feature Extraction and Data Modelling
- iv.
- Data points of interest (features) are extracted from the main database. This will include important biodata such as age, gender, address, demographic data, and key clinical information that pertain to the patient category concerned.
- v.
- Mapping of observed symptoms in patients with COVID-19 and specific morbidities will be performed. This will entail documenting symptoms that are associated with particular instances of COVID-19 and comorbidity. This will seek to answer the question of how the symptomatic patterns of COVID-19 plus HIV are different/similar to those of COVID-19 plus diabetes, for example. This process will allow different patients’ symptomatic profiles to be generated, e.g.:
- COVID-19 + HIV; COVID-19 + TB symptoms
- COVID-19 + HIV +TB symptoms; COVID-19 + diabetes (DB) symptoms
- COVID-19 + hypertension symptoms; COVID-19 + hypertension + DB
- COVID-19 + malaria; COVID-19 + malaria + HIV; COVID-19 + etc.
3.3.3. Dataset Repository
- vi.
- Based on the feature extraction in Step 4 and Step 5, different datasets will be created. It will be possible to filter the created datasets based on some parameters such as date, type of morbidity, age, demography, symptomatic pattern, and other dimensions that are of interest in order to create sub-datasets and variant datasets by using querying techniques. Data can be extracted using SQL statements to create sub-datasets. Examples of different SQL queries that can be used to extract different sub-datasets are shown in Table 2.
 Table 2. Query statements for data extraction and data filtering from the database.
Table 2. Query statements for data extraction and data filtering from the database.
3.3.4. Model Training
- vii.
- Supervised machine learning algorithms (ANN, SVM, deep neural networks, ensemble models—random forest, extreme gradient boosted trees (XGBoost), bagging-gradient boosted trees (B-GBT), etc.) are trained based on selected datasets from the dataset repository and the type of prediction that is required to be made per time. Appropriate hyperparameter tuning methods such as cross-validation and regularisation shall be applied to prevent overfitting during training and better generalization of trained ML models. Methods such as k-fold validation, dropout regularization, ridge regression regularization (L2-norm), lasso regression regularization (L1-norm), entropy regularization, and noise injection are good options that shall be explored to achieve this [25]. Experiments on batch training using gradient descent algorithms and minibatch training with stochastic gradient descent using different activation functions shall also be performed to ensure that the most efficient methods are used for training so that the best-trained models are eventually used for predictions [26].
- viii.
- Trained predictive models are stored in the cloud-based machine learning server.
- ix.
- The most important features of trained models (highest weighted) are also stored. For tree-based ML models such as decision tree, random forest, extreme gradient boosted trees (XGBoost), and bagging-gradient boosted trees (B-GBT), this can be obtained from the feature importance property of the tree model [27]. For regression models, this could be obtained by using univariate selection, while for other ML models, dimensionality reduction methods such as principal component analysis (PCA) can be used to determine feature selection and feature importance [28]. Knowing the most important features of an ML model is particularly useful for the explainability of the ML model, as it reveals the features that have the strongest relationships with the output variable the most [29].
3.3.5. Real-Time Explainable ML-CBR Recommendations by the AI Engine
- x.
- New case data is received that requires prediction.
- xi.
- Required AI operation is selected by the user (HCW). This could be a regression task to estimate something, classify/identify something, or obtain a recommendation.
- xii.
- ML prediction is generated by the AI engine.
- xiii.
- The result of the ML algorithm is sent to a case-based reasoning module (CBR).
- CBR uses the most important features as a basis to retrieve similar cases to the current case from the case base. The case base is linked to the original dataset used to train the ML model.
- The feature values of cases that are similar to the current case are used to construct an explanation to justify the prediction generated by the ML model, thereby utilizing a post hoc approach to ensure the explainability of the ML result.
- xiv.
- Every predicted case is stored in the case base for future reuse.
3.3.6. Model Evaluation
The performance of the selected ML models in terms of accuracy of prediction will be evaluated by using standard metrics for regression, classification, and recommendation algorithms. The quality of explanation can also be evaluated. The evaluation metrics are mainly quantitative metrics that will be used to assess the performance of AI results by using human expert judgement as a gold standard. The usability, reliability, and trustworthiness assessment of the AI will be conducted through field trials that capture the perspective of individuals that have used the system to support their operations.
3.4. Utilization of Generated Results
The generated result and explanation will be sent to the HCW for approval. If, after review, the generated result and explanation are approved by the HCW, then they will be sent to the patient directly by the system or relayed to the patient by the HCW through the most accessible means to reach the patient. This could be via WhatsApp, Telegram, SMS, or a phone call.
4. The Merits of the Proposed Framework
This framework was conceived by a consortium of experts in AI, medical sciences, laboratory sciences, and business that cuts across academia and industry. There is a plan to conduct a full-scale study on the applicability of the framework using study sites in South Africa and Nigeria. The implementation of the AI-enabled framework is expected to yield the following merits:
- Harness the different types of clinically relevant data for decision-making on diagnosis and treatment of COVID-19 and comorbidities;
- Enable differentiated and personalised treatment for patients with COVID-19 and comorbidities in terms of patient triage, treatment, and post-infection management;
- Use multimodal diagnostic tools to guide the management of patients with COVID-19 and comorbidities (HIV/TB/malaria) during treatment, determining the pattern of disease progression and handling of posttreatment complications;
- Use clinical diagnosis from different disciplines to guide management of PRDs in the presence of COVID-19;
- Determine the possibility of patients with comorbidity developing COVID-19 if there is exposure due to their risk factors;
- Generate intelligent and explanation-rich recommendations on detected cases for use by healthcare workers (HCW) to aid decision-making, thus enabling trustworthy AI for healthcare;
- Support the process of disease surveillance in RLS through multi-sensory contact tracing, thus helping with outbreak control;
- Generate timely analytical reports on key factors that influence spread, infection recovery rates, and other factors of interest; and
- Aid the formulation of policies and strategies for containing and combating COVID-19.
5. Conclusions
In this paper, we have presented the conceptual overview and the merits of an AI-based framework for multimodal diagnostics of COVID-19 and its associated comorbidities, which could be applied in RLS in sub-Sharan Africa. The notion of the framework is novel as it is the first that is based on the triangulation of multiple diagnostic methods (in-vivo and molecular-based testing) such that data from several methods can be harnessed through AI to facilitate COVID-19 detection, treatment, and management in RLS. Our immediate next objectives are to source funds for the implementation of this framework and to draw vital lessons of relevance to RLS in Africa and other, similar, contexts in developing countries.
Author Contributions
Conceptualization, O.D., P.N. and T.M.-T.; methodology, O.D., P.N., T.M.-T., T.M., S.B., J.N., M.J.K. and V.C.O.; investigation, O.D., P.N., T.M.-T., T.M., S.B., J.N., M.J.K., L.W. and V.C.O.; writing—original draft preparation, O.D.; writing—review and editing, O.D., M.J.K., L.W., P.N., T.M.-T., V.C.O., A.H. and K.S. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding. The APC was funded by NRF, South Africa and implementation of PSGT by the Technology Innovation Agency of South Africa.
Institutional Review Board Statement
The study was conducted according to the guidelines of the Declaration of Helsinki, and approved by the Faculty Ethics Committee (FEC) of the Faculty of Informatics and Design of the Cape Peninsula University of Technology, SA, USA (30 July 2020). For the application of personalised medicine using an integrated service and research approach, the Health and Research Ethics Review Committee of Stellenbosch University, SA, approved assessment of comorbidities and treatment response in COVID-19 cases during follow-up (protocol code N09/08/224-7545, 2 August 2021).
Informed Consent Statement
Informed consent is obtained on a case-by-case basis for implementation of PSGT by a multi-disciplinary healthcare team.
Data Availability Statement
The knowledge database supporting the PSGT case study (Table 1) is available to registered users at https://www.gknowmix.org.
Acknowledgments
We acknowledge Kelebogile E. Moremi at the Division of Chemical Pathology, Stellenbosch University, for her contribution to the incorporation of COVID-19 risk assessment into the PSGT case study. We acknowledge support from the National Research Foundation (NRF), South Africa; the South African Medical Research Council (SAMRC).
Conflicts of Interest
M.J.K. and L.W. are shareholders of Gknowmix (Pty) Ltd., M.J.K. is a non-executive director of Gknowmix (Pty) Ltd., which has developed a genomics database and point-of-care assays using PSGT for research translation under the auspices of the SAMRC. J.N. is a shareholder of Envisionit Deep AI Pty LTD, a medical technology company using AI as a clinical decision support tool in imaging diagnosis. The other authors declare no conflict of interest.
References
- Davies, M.-A. HIV and risk of COVID-19 death: A population cohort study from the Western Cape Province, South Africa. medRxiv 2020. [Google Scholar] [CrossRef]
- COVID Live Update: 166,632,552 Cases and 3,460,798 Deaths from the Coronavirus—Worldometer. Available online: https://www.worldometers.info/coronavirus/ (accessed on 22 May 2021).
- TB Facts—Tests, Drugs, Statistics & Lots more about TB Disease. Available online: http://www.tbfacts.org (accessed on 22 May 2021).
- TB in Nigeria—Funding, Children, Diagnosing TB, HIV/TB. Available online: https://tbfacts.org/tb-nigeria/ (accessed on 22 May 2021).
- Chehade, M.J.; Yadav, L.; Jayatilaka, A.; Gill, T.K.; Palmer, E. Personal digital health hubs for multiple conditions. Bull. World Health Organ. 2020, 98, 569–575. [Google Scholar] [CrossRef] [PubMed]
- Moyo, S.; Boffa, J.; Mhlaba, T.; Sulis, G.; Sifumba, Z.; Pai, M.; Daftary, A. COVID-19 and tuberculosis in South Africa: A dangerous combination. S. Afr. Med. J. 2020, 110, 341–342. [Google Scholar]
- Kavanagh, M.M.; Erondu, N.A.; Tomori, O.; Dzau, V.J.; Okiro, E.A.; Maleche, A.; Aniebo, I.C.; Rugege, U.; Holmes, C.B.; Gostin, L.O. Access to lifesaving medical resources for African countries: COVID-19 testing and response, ethics, and politics. Lancet 2020, 395, 1735–1738. [Google Scholar] [CrossRef]
- Mashamba-Thompson, T.P.; Crayton, E.D. Blockchain and artificial intelligence technology for novel Coronavirus disease-19 self-testing. Diagnostics 2020, 10, 198. [Google Scholar] [CrossRef] [Green Version]
- Daramola, O.; Moser, T. Semantic integration of multiple health data for treatment decision-making in low-resource settings. In Proceedings of the Multi Conference on Computer Science and Information Systems. In Proceedings of the MCCSIS 2019—Proceedings of the International Conference on e-Health 2019, Porto, Portugal, 16–19 July 2019. [Google Scholar]
- Stroetmann, K.A. Digital Health Ecosystems for African Countries—A Guide for Public and Private Actors for Establishing Holistic Digital Health Ecosystems in Africa; Federal Ministry for Economic Cooperation and Development and Strategic: Bonn, Germany, 2018. [Google Scholar]
- Kotze, M.J.; Van Velden, D.P.; Botha, K.; Badenhorst, C.H.; Avenant, H.; Van Rensburg, S.J.; Cronjé, F.J. Pathology-supported genetic testing directed at shared disease pathways for optimized health in later life. Per. Med. 2013, 10, 497–507. [Google Scholar] [CrossRef]
- Kwok, S.; Adam, S.; Ho, J.H.; Iqbal, Z.; Turkington, P.; Razvi, S.; Le Roux, C.W.; Soran, H.; Syed, A.A. Obesity: A critical risk factor in the COVID-19 pandemic. Clin. Obes. 2020, 10, e12403. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Geng, Z.; Fan, Z.; Liu, J.; Chen, H. Point-of-care testing based on smartphone: The current state-of-the-art (2017–2018). Biosens. Bioelectron. 2019, 132, 17–37. [Google Scholar] [CrossRef] [PubMed]
- Dolan, M.E.; Hill, D.P.; Mukherjee, G.; McAndrews, M.S.; Chesler, E.J.; Blake, J.A. Investigation of COVID-19 comorbidities reveals genes and pathways coincident with the SARS-CoV-2 viral disease. Sci. Rep. 2020, 10, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Kotze, M. Application of advanced molecular technology in the diagnosis and management of genetic disorders in South Africa. S. Afr. Med. J. 2016, 106, S114–S118. [Google Scholar] [CrossRef] [Green Version]
- Cavanaugh, A.M.; Spicer, K.B.; Thoroughman, D.; Glick, C.; Winter, K. Reduced risk of reinfection with SARS-CoV-2 after COVID-19 vaccination—Kentucky, May–June 2021. MMWR Morb. Mortal. Wkly. Rep. 2021, 70, 1081–1083. [Google Scholar] [CrossRef] [PubMed]
- Lückhoff, H.K.; Kidd, M.; van Rensburg, S.J.; van Velden, D.P.; Kotze, M.J. Apolipoprotein E genotyping and questionnaire-based assessment of lifestyle risk factors in dyslipidemic patients with a family history of Alzheimer’s disease: Test development for clinical application. Metab. Brain Dis. 2016, 31, 213–224. [Google Scholar] [CrossRef] [PubMed]
- Marais, A.D.; Kotze, M.J.; Raal, F.J.; Khine, A.A.; Talmud, P.J.; Humphries, S.E. Familial hypercholesterolaemia workshop for leveraging point-of-care testing and personalised medicine in association with the Lipid and Atherosclerosis Society of Southern Africa. Cardiovasc. J. Afr. 2019, 30, 297–304. [Google Scholar] [CrossRef] [PubMed]
- Wyller, T.B.; Kittang, B.R.; Ranhoff, A.H.; Harg, P.; Myrstad, M. Nursing home deaths after COVID-19 vaccination. Tidsskr. Nor. Laegeforen. 2021, 141. [Google Scholar] [CrossRef]
- Sorokin, A.V.; Karathanasis, S.K.; Yang, Z.-H.; Freeman, L.; Kotani, K.; Remaley, A.T. COVID-19-Associated dyslipidemia: Implications for mechanism of impaired resolution and novel therapeutic approaches. FASEB J. 2020, 34, 9843–9853. [Google Scholar] [CrossRef]
- Kumar, P.; Kumar, M.; Bedi, O.; Gupta, M.; Kumar, S.; Jaiswal, G.; Rahi, V.; Yedke, N.G.; Bijalwan, A.; Sharma, S.; et al. Role of vitamins and minerals as immunity boosters in COVID-19. Inflammopharmacology 2021, 29, 1001–1016. [Google Scholar] [CrossRef]
- Kuo, C.L.; Pilling, L.C.; Atkins, J.L.; Masoli, J.A.; Delgado, J.; Kuchel, G.A.; Melzer, D. APOE e4 genotype predicts severe COVID-19 in the UK biobank community cohort. J. Gerontol. A Biol. Sci. Med. Sci. 2020, 75, 2231–2232. [Google Scholar] [CrossRef]
- Osamor, V.C.; Edosomwan, I.B. Employing scrambled alpha-numeric randomization and RSA algorithm to ensure enhanced encryption in electronic medical records. Inform. Med. Unlocked. 2021, 25, 100672. [Google Scholar] [CrossRef]
- Gonzalez-Cuautle, D.; Hernandez-Suarez, A.; Sanchez-Perez, G.; Toscano-Medina, L.K.; Portillo-Portillo, J.; Olivares-Mercado, J.; Perez-Meana, H.M.; Sandoval-Orozco, A.L. Synthetic Minority Oversampling Technique for optimizing classification tasks in botnet and intrusion-detection-system datasets. Appl. Sci. 2020, 10, 794. [Google Scholar] [CrossRef] [Green Version]
- Nusrat, I.; Jang, S.-B. A comparison of regularization techniques in deep neural networks. Symmetry 2018, 10, 648. [Google Scholar] [CrossRef] [Green Version]
- Domingos, E.; Ojeme, B.; Daramola, O. Experimental analysis of hyperparameters for deep learning-based churn prediction in the banking sector. Computation 2021, 9, 34. [Google Scholar] [CrossRef]
- Brownlee, J. Data Preparation for Machine Learning: Data Cleaning, Feature Selection, and Data Transforms in Python; Machine Learning Mastery: San Francisco, CA, USA, 2020. [Google Scholar]
- Khalid, S.; Khalil, T.; Nasreen, S. A survey of feature selection and feature extraction techniques in machine learning. In Proceedings of the 2014 Science and Information Conference, London, UK, 27–29 August 2014; IEEE: Manhattan, NY, USA, 2014. [Google Scholar]
- Saarela, M.; Jauhiainen, S. Comparison of feature importance measures as explanations for classification models. SN Appl. Sci. 2021, 3, 1–12. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).