Enhancing Clinical Decision Support for Precision Medicine: A Data-Driven Approach
Abstract
1. Introduction
2. Materials and Methods
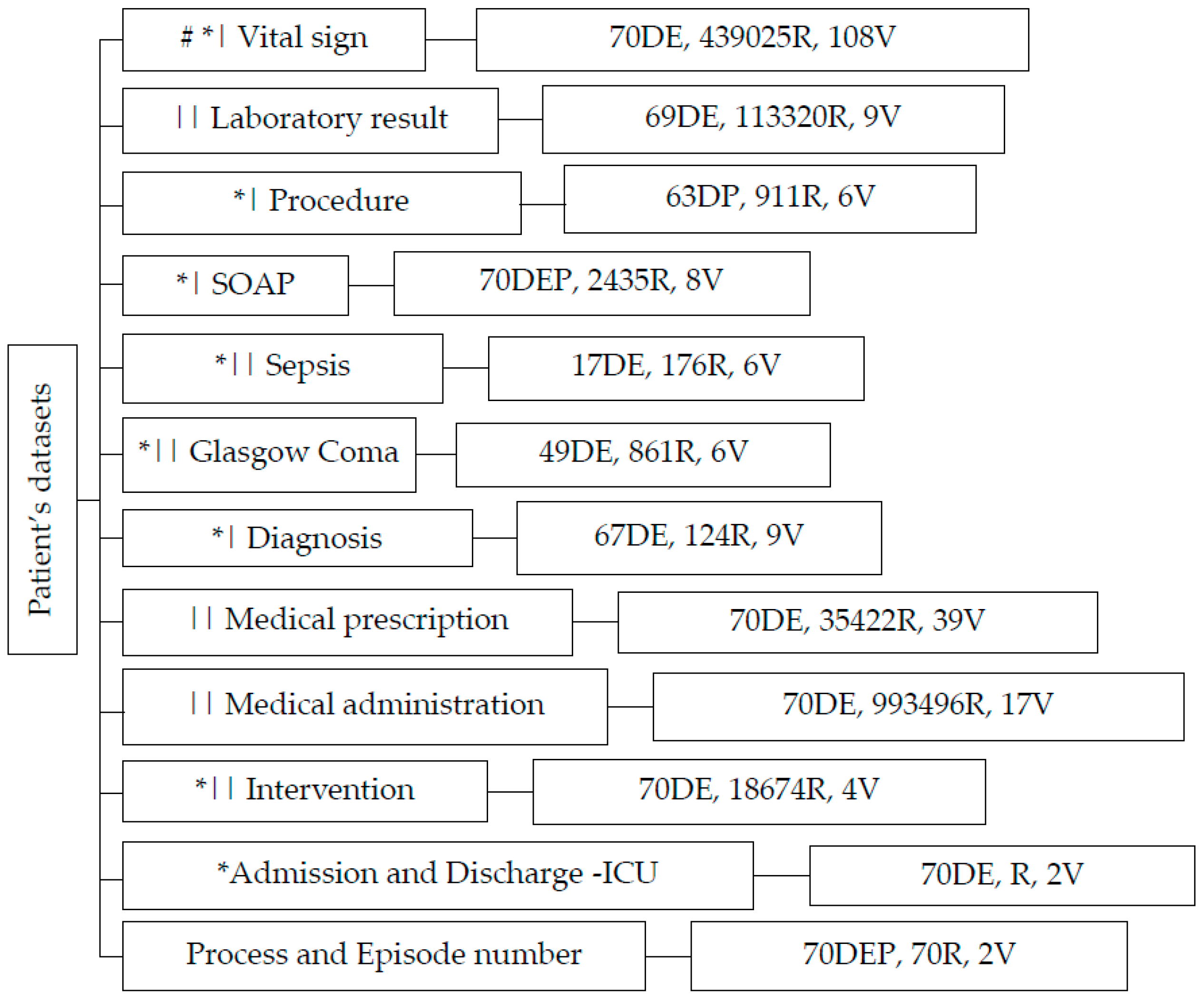
2.1. Data Collection and Exploration
- “Vital Signs”: This dataset contains 439,025 records and 108 biological variables, focusing on vital signs that play a crucial role in assessing a patient’s overall condition.
- “Laboratory Results”: Comprising 113,320 records and 9 variables, this dataset provides information about various laboratory exams conducted, aiding in diagnosing and monitoring patients’ health.
- “Procedures”: With 911 records and 6 variables, the “Procedure” dataset sheds light on the medical actions recommended and prescribed by healthcare professionals.
- “Sepsis (Gravity Score)”: Capturing data from 176 records and 6 variables, this category gauges the severity of patients’ conditions, particularly in cases of sepsis.
- “Glasgow Coma Scale”: Containing 861 records and 6 variables, this dataset evaluates patients’ consciousness levels, a vital indicator of neurological well-being.
- “Diagnosis”: Encompassing 124 records and 9 variables, the “Diagnosis” section focuses on recording signs, symptoms, and potential medical conditions.
- “Medication Prescriptions”: This category, with 35,422 records and 39 variables, provides data on medications prescribed by clinicians, helping track patient treatment plans.
- Medical administration: with 993,496 records and 17 variables associated with drug administration.
- SOAP: encompassing 2435 records and 8 variables, contains critical data related to the Subject, Object, Assessment, and Plan components following the SOAP framework
- “Intervention Actions”: Capturing information about various interventions, this category showcases actions taken to manage patients’ health.
- ICU Admission and Discharge: “Admin-Discharge” houses data about patients’ admissions and discharges from the ICU, facilitating comprehensive patient care management. This dataset consists of the date and time of admission and discharge.
- Reference Dataset: Serving as a point of reference, this dataset includes episode and process numbers, wherein the episode number represents clinical events, while the process number signifies patient identity.
- Feature Selection: Employing a meticulous approach to feature selection, we leveraged domain knowledge, statistical analyses, and data quality assessments. In instances such as the vital sign dataset, variables with over 90% missing data were systematically excluded.
- Feature Engineering: In several datasets, we introduced novel variables to enhance analytical insight and harness valuable information. A prime illustration involves deriving the length of hospital-ICU stay by utilizing admission and discharge dates, thus transforming raw data into meaningful metrics.
- Extraction and Processing: The effectiveness of analytical performance was significantly amplified through rigorous data extraction. An illustrative example is the extraction of demographic details such as age and gender from diverse datasets, culminating in the creation of a consolidated dataset that streamlined subsequent data processing.
- Conditional Columns: Our transformative approach extended to the creation of conditional columns, converting raw data into actionable information. An example is our utilization of laboratory references for each exam, enabling a systematic comparison with exam results to ascertain their normal or abnormal status.
- Grouping and aggregating time-series data for vital indicators, thus addressing sporadic data registration issues encountered with biological sensors in the ICU. By adopting an hourly aggregation approach, we effectively mitigated the challenge of infrequent data updates.
- Cleaning-Missing cells: In addressing gaps within the vital sign dataset, a meticulous strategy was employed: we judiciously populated missing cells by computing the average value from neighboring cells preceding and following the voids. Similarly, within another dataset, a pragmatic approach was taken by eliminating missing cells.
| Key Data Processing | Description|Example |
|---|---|
| Exploratory data analysis | Identify mean, max, min, missing cells, and data quality |
| Feature Selection | Using Knowledge of domain and missing cells |
| Feature engineering | Construct new variables such as length of stay |
| Data extraction | Demography table; Extract age and gender |
| Conditional feature | Create Laboratory result status |
| The correct type of data | Considering categorical, numerical, and also time series data |
| Group and aggregation | For handling infrequent data generation, particularly from biological sensors |
| Missing cells | Imputation method and elimination. |
2.2. Techniques and Metrics
- Clustering: Clustering is a technique used in unsupervised learning to group similar data points together based on certain features or characteristics. It helps identify inherent structures within data without the need for predefined labels.
- K-means Nearest: The K-means algorithm is a popular method for clustering data. It iteratively assigns data points to K clusters based on their proximity to the cluster centroids. The “nearest” aspect refers to how each data point is assigned to the nearest centroid during each iteration.
- Classification: Classification is a supervised learning task where the goal is to assign predefined labels or categories to input data based on their features. It involves training a model on labeled data to make predictions on new, unseen data.
- Random Forest: Random forest is an ensemble learning algorithm that builds multiple decision trees during training. Each tree in the forest independently classifies input data, and the final prediction is determined by a majority vote or averaging.
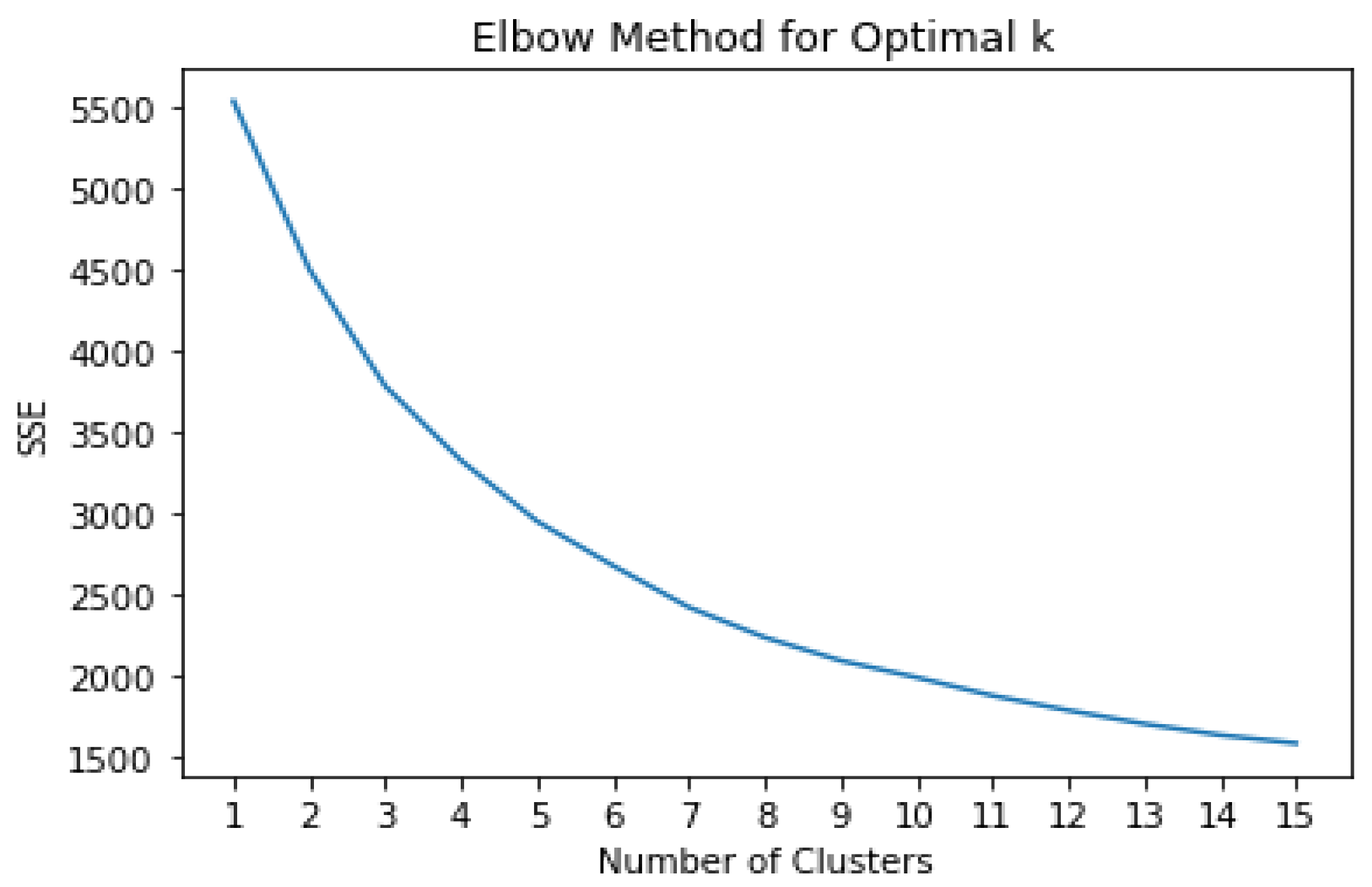
- Elbow Method: The elbow method is a technique used to determine the optimal number of clusters in a dataset for K-means clustering. It involves plotting the within-cluster sum of squares (WCSS) against the number of clusters and identifying the “elbow” point, where the rate of decrease in WCSS slows down.
- Silhouette Score: The silhouette score is a metric used to evaluate the quality of clustering. It measures how similar a data point is to its cluster compared with other clusters. A higher silhouette score indicates better-defined clusters.
- K-Fold Cross Validation: K-Fold cross-validation is a technique used to assess the performance of a machine learning model by splitting the data into K subsets (folds) of equal size. The model is trained K times, each time using K-1 folds for training and the remaining fold for validation. This process is repeated K times, with each fold serving as the validation set exactly once. The performance metrics are then averaged across all K iterations to provide a more reliable estimate of the model’s performance on unseen data and reduce the variance of the evaluation. K-fold validation helps to ensure that the model’s performance is not heavily influenced by the particular random split of data into training and test sets.
- Accuracy: Accuracy is a measure of the proportion of correctly classified instances among all instances. It is calculated as the number of correct predictions divided by the total number of predictions.
- Precision: Precision is a measure of the proportion of true positive instances among all predicted positive instances. It indicates the accuracy of positive predictions.
- Recall: Recall is a measure of the proportion of true positive instances among all actual positive instances. It measures the ability of a classifier to identify all relevant instances.
- Kappa: Kappa is a statistic that measures inter-rater agreement for categorical items. It compares the observed agreement between raters to the agreement expected by chance.
- F1 Score: The F1 Score is the harmonic mean of precision and recall. It balances both precision and recall and is useful when the classes are imbalanced.
- AUC (Area Under Curve): AUC is the area under the receiver operating characteristic (ROC) curve. It measures the performance of a binary classification model across different threshold settings. A higher AUC indicates better model performance.
| Name | Descriptions |
|---|---|
| Clustering | Grouping data points based on similarity or proximity. |
| K means nearest | A clustering algorithm that partitions data into K clusters based on centroids. |
| Classification | Assigning labels or categories to data points |
| Random Forest | A machine learning algorithm that builds multiple decision trees to classify data. |
| Elbow Method | A technique to determine the optimal number of clusters in clustering analysis. |
| Silhouette Score | A metric to evaluate the quality of clustering |
| Fold Cross Validation | K-FoldCV repeatedly splits data into K subsets, training on K-1 and validating on one to assess model performance. |
| Accuracy | The proportion of correctly classified instances among all instances. |
| Precision | The proportion of true positive instances among all predicted positive instances. |
| Recall | The proportion of true positive instances among all actual positive instances. |
| Kappa | A statistic that measures inter-rater agreement for categorical items. |
| F1 score | A statistic that measures inter-rater agreement for categorical items |
| AUC | (Area Under Curve): The area under the Receiver Operating Characteristic (ROC) curve indicates model performance for binary classification. |
3. Results
3.1. Crafting CEid and Interrelating Clinical Events
3.2. Analytical Insight through CEid
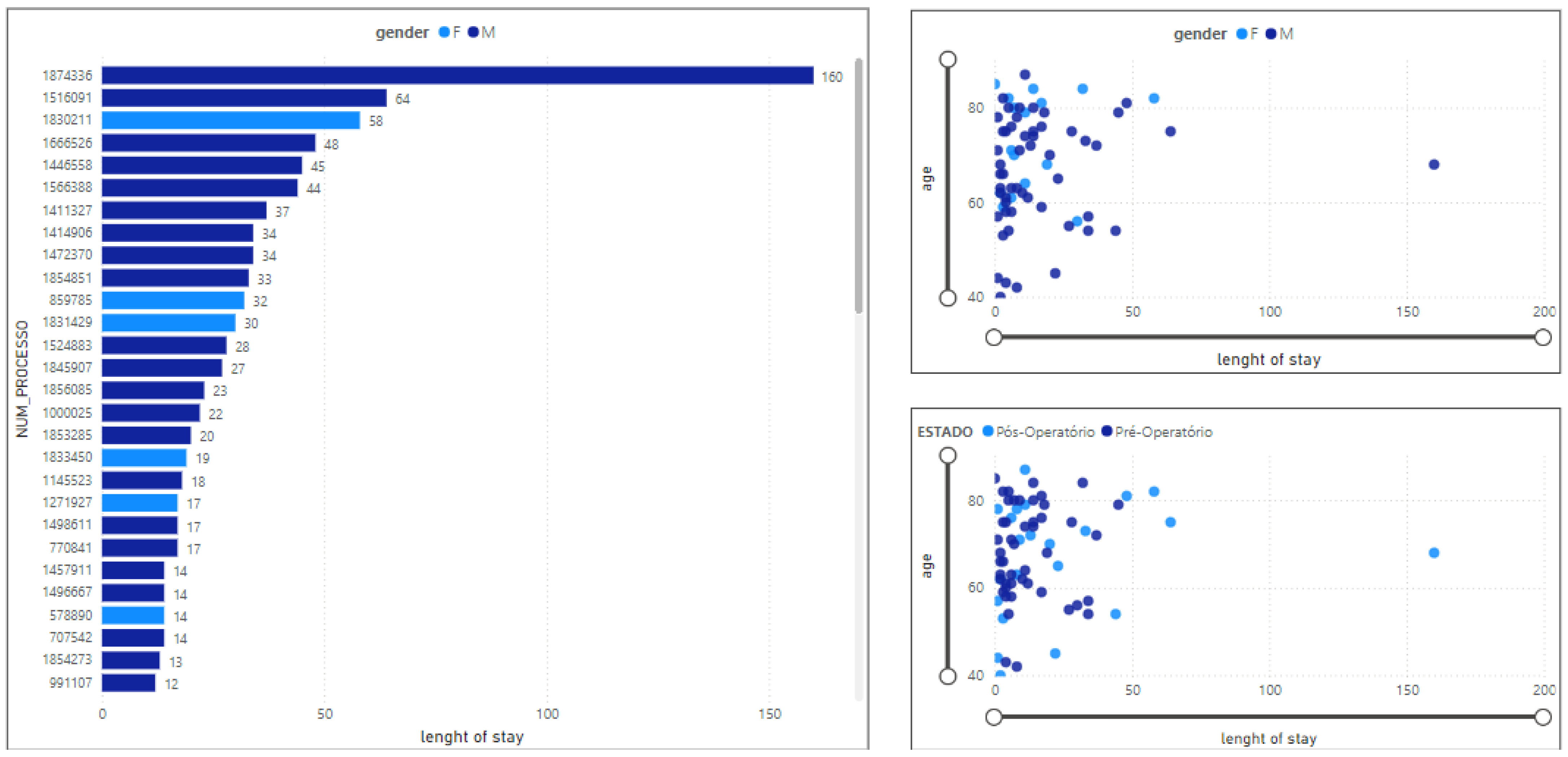
3.2.1. Length of Stay in ICU
3.2.2. Total Number of Clinical Events
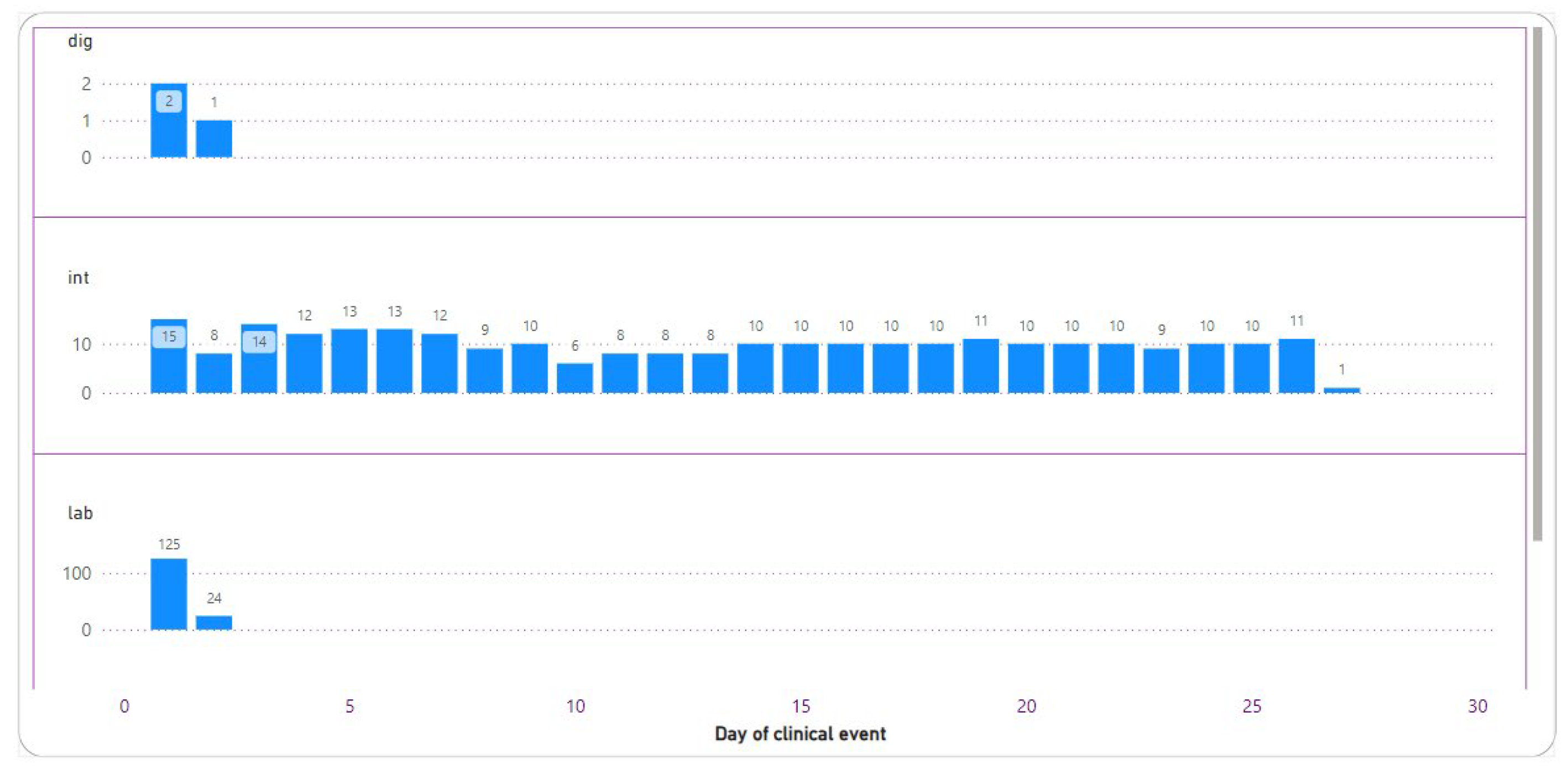
3.2.3. Diagnostics
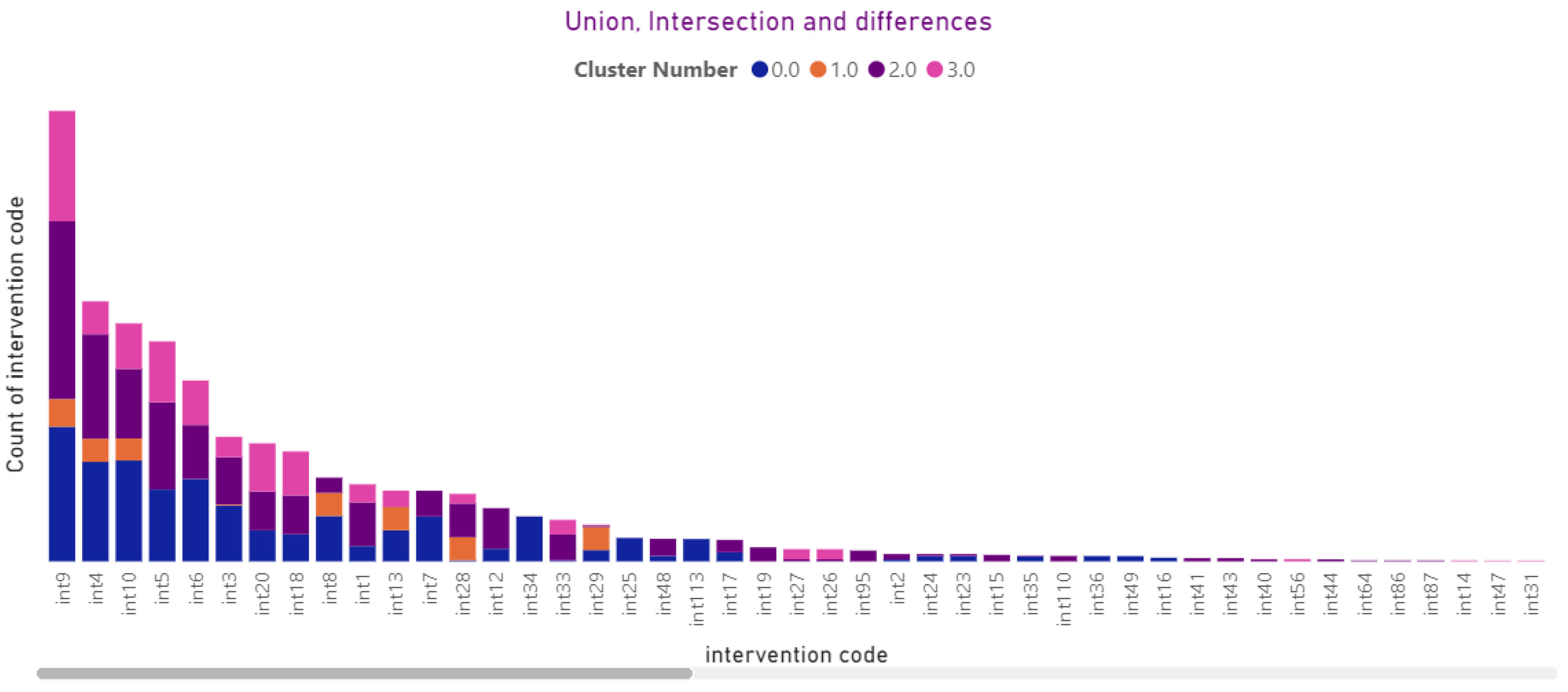
3.2.4. Intervention
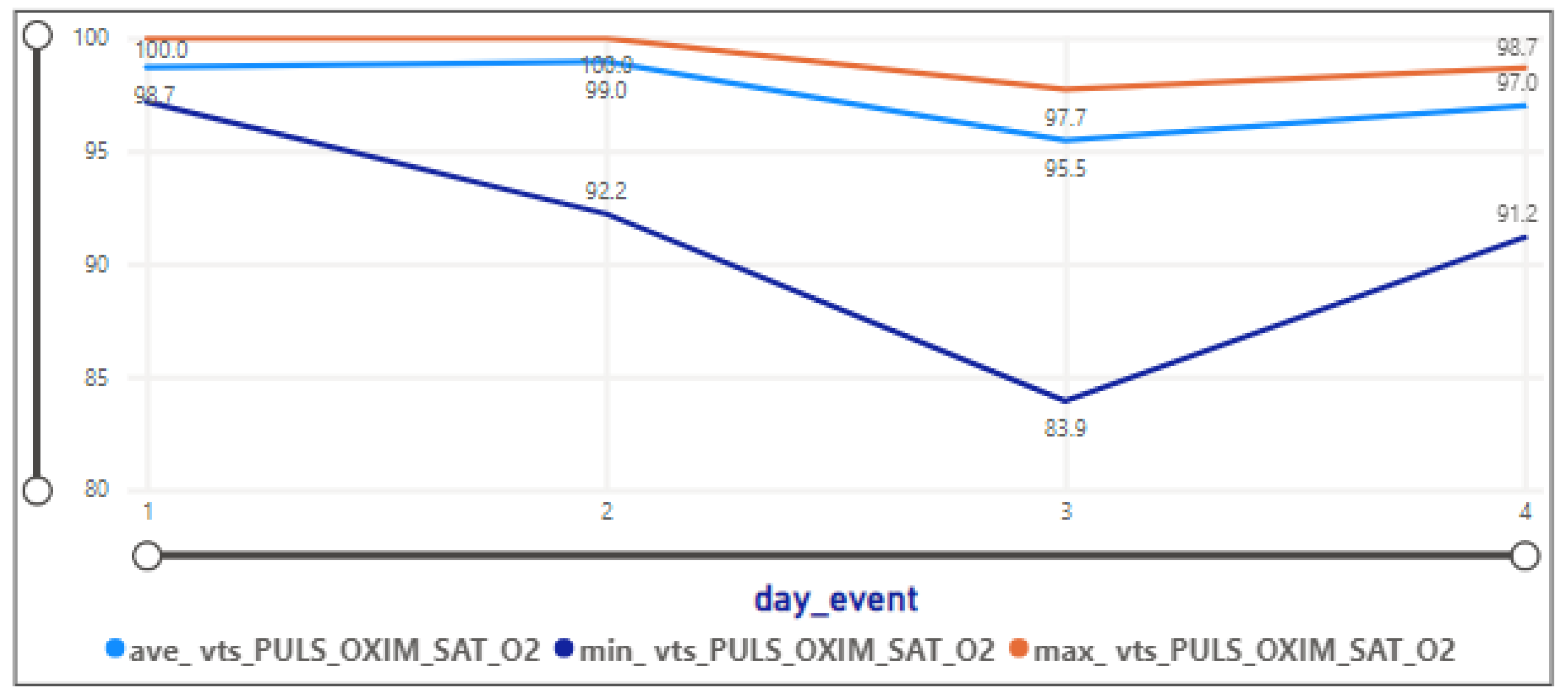
3.2.5. Vital Signs
3.2.6. Laboratory Result
3.2.7. Sepsis
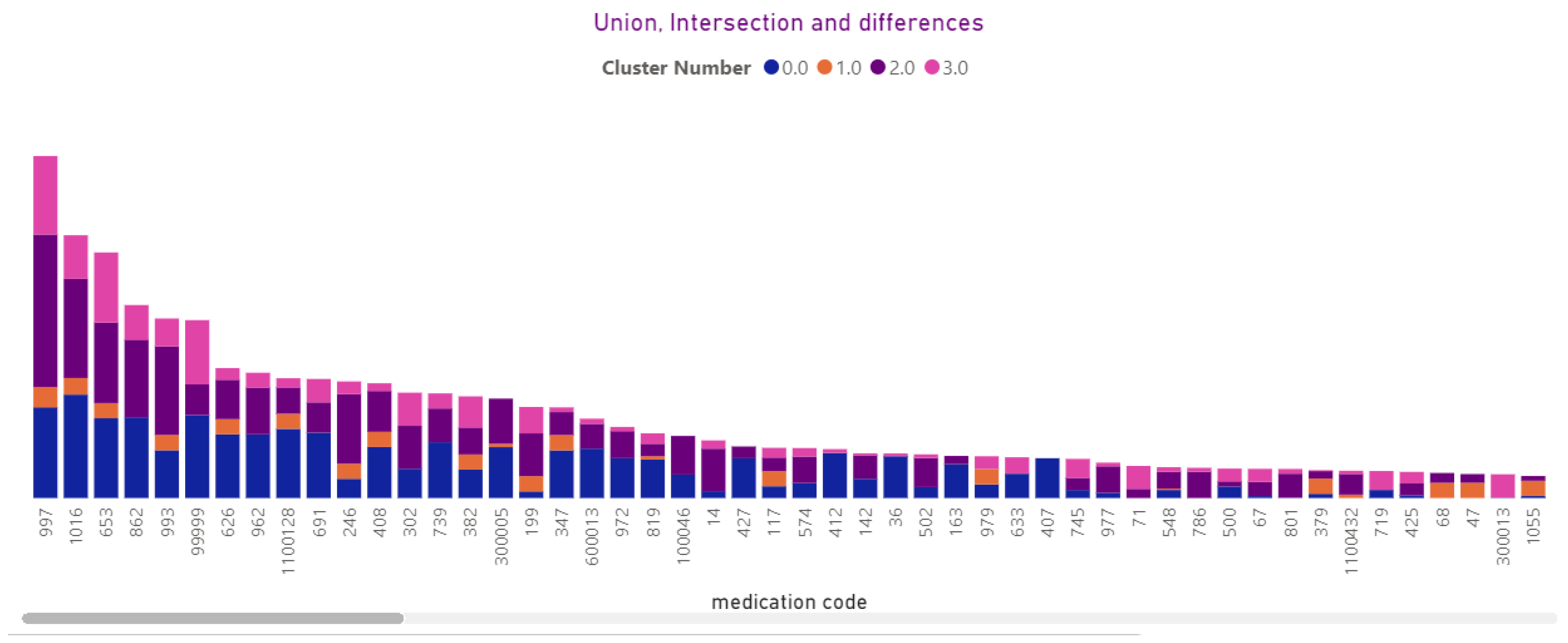
3.2.8. Medication Prescription
3.3. Unveiling Patterns throughout Temporal Clustering Analysis
3.3.1. Identifying Patterns and Cluster References
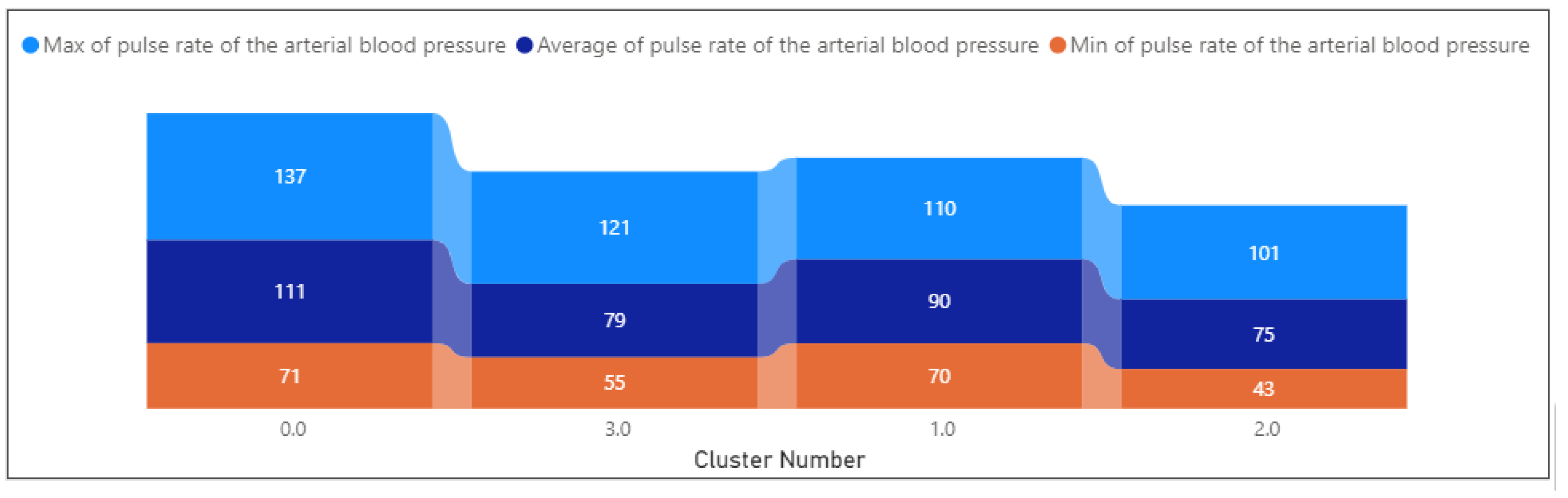
- The key observation of Cluster 0 with the silhouette score equal to 0.23 and data distribution equal to 271 rows shows:
- −
- The average pulse rate of the arterial blood pressure is 100.66, with a standard deviation of 12.20.
- −
- Diastolic arterial blood pressure is around 64.18, with a standard deviation of 9.6.
- −
- Systolic arterial blood pressure is notably higher at 118.81, with a standard deviation of 15.46.
- −
- Mean arterial blood pressure is 82.65, with a standard deviation of 11.14.
- −
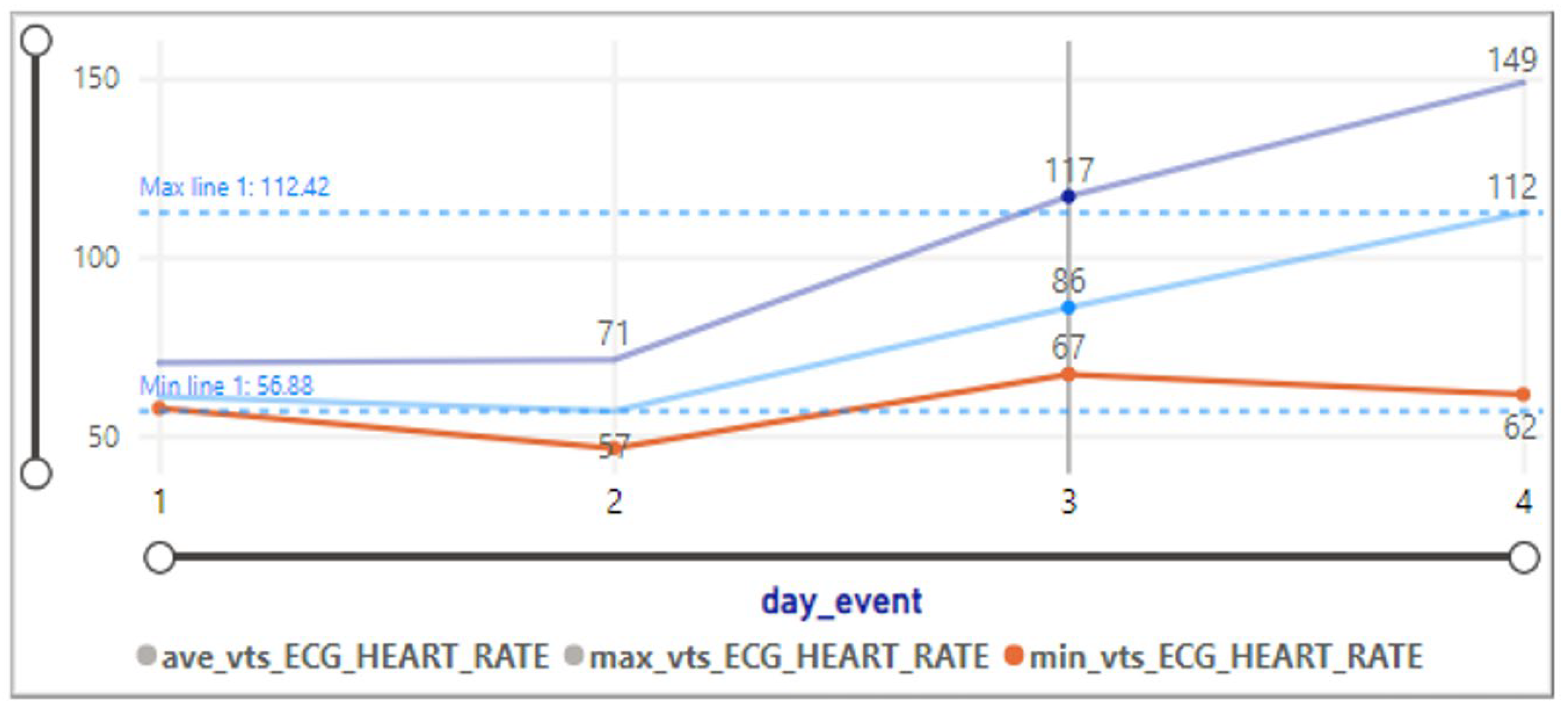
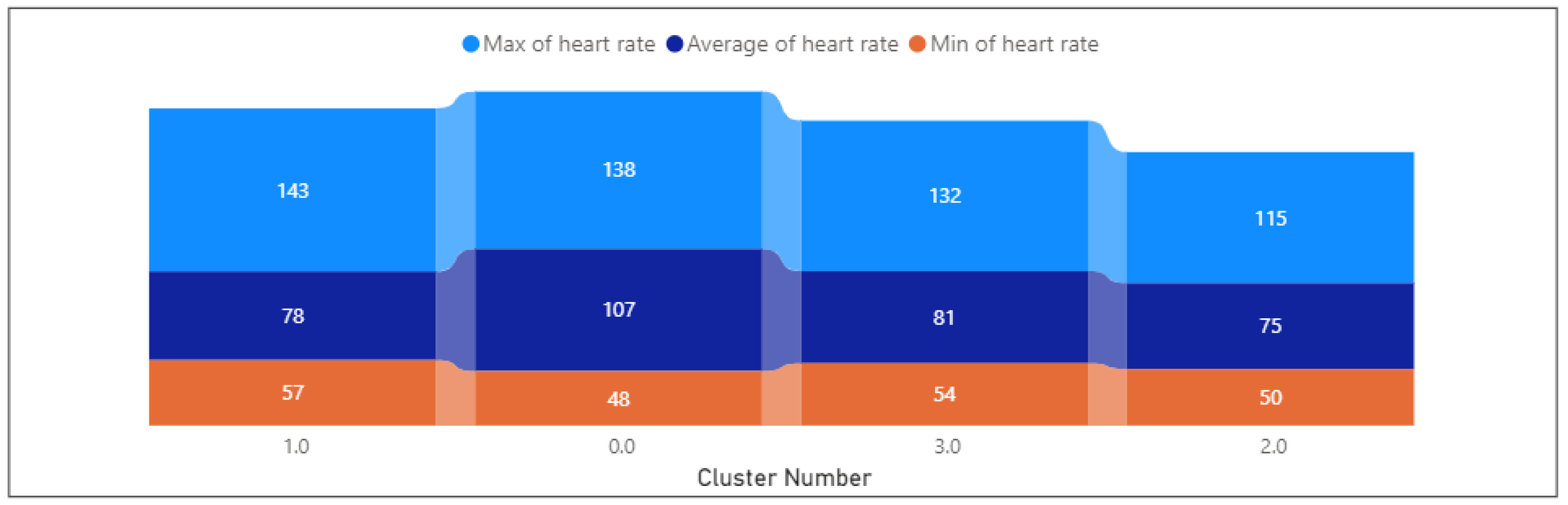
- Heart rate is 99.18, with a standard deviation of 17.49.
- −
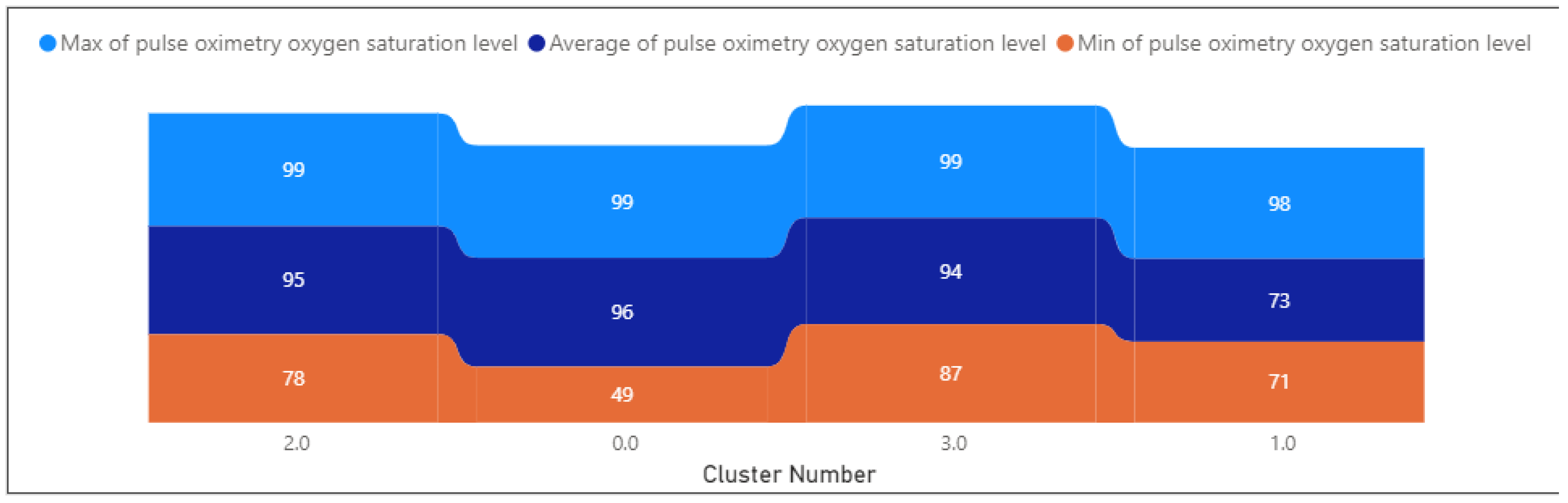
- The pulse oximetry oxygen saturation level is relatively high at 94.21, with a small standard deviation of 4.44.
- −
- Body temperature is around 36.17, with a standard deviation of 1.27.
- −
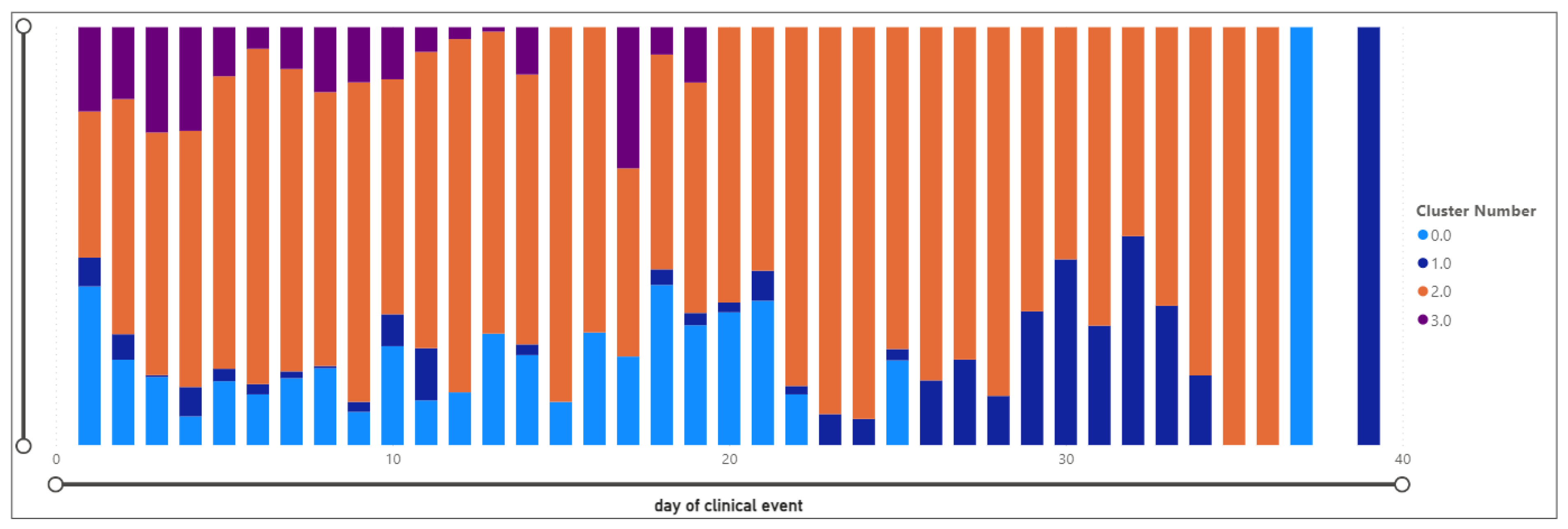
- The day of the clinical event is, on average, 10.60, with a relatively high standard deviation of 8.00.
- Cluster 1, with a silhouette score equal to 0.12 and data distribution of 44, has a lower silhouette score, indicating less cohesion and separation.
- −
- Variables exhibit higher standard deviations, suggesting greater variability within this cluster.
- −
- Body temperature has a relatively high standard deviation of 2.72, indicating variability in this aspect.
- Cluster 2 silhouette has a higher silhouette score, suggesting better-defined clusters (score: 0.26), and the data distribution is equal to 285 rows. In this cluster
- −
- Variables such as systolic arterial blood pressure and mean arterial blood pressure show significant variability with standard deviations of 15.13 and 12.20, respectively.
- −
- The pulse oximetry oxygen saturation level has a low standard deviation, indicating more consistency.
- Cluster 3 has the highest silhouette score, indicating well-defined clusters (silhouette score: 0.36), and data distribution is equal to 92 rows. The key observations show:
- −
- Systolic arterial blood pressure is notably high at 129.29, with a standard deviation of 15.13.
- Cluster 3 stands out with the highest silhouette score, indicating the most distinct cluster.
- −
- Systolic arterial blood pressure is a key differentiator among clusters, with Cluster 3 having the highest values.
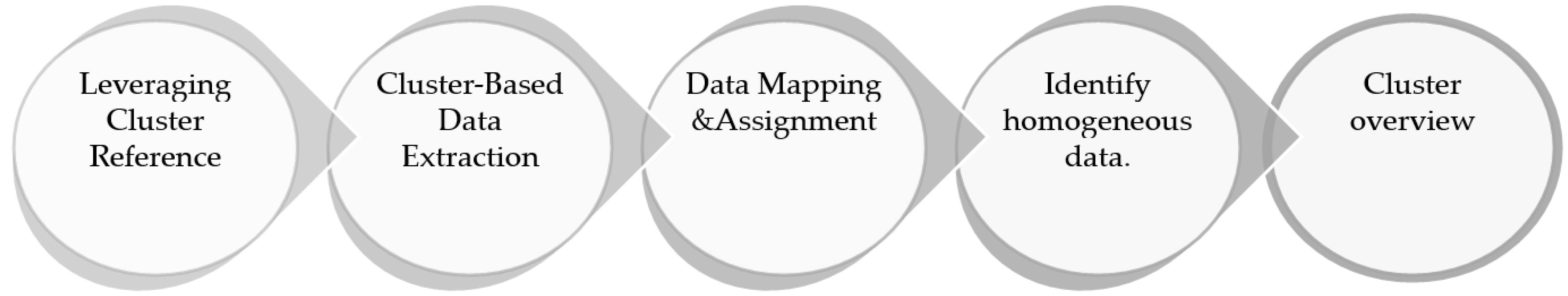
3.3.2. Cluster-Based Mapping and Assignment
3.3.3. Cluster Overview
- Cluster 0: 598,266 rows
- Cluster 1: 110,042 rows
- Cluster 2: 593,192 rows
- Cluster 3: 325,289 rows
3.3.4. Exploring Health Profiles
- Cluster 0:
- −
- The day of the clinical event ranges from 1 to 37, with a mean of 1.06. events are spread across the observed period.
- −
- Glasgow Coma Scale: Varies between 3 and 15, with a mean of 12.27. Indicates a range of consciousness levels, potentially reflecting diverse patient conditions.
- −
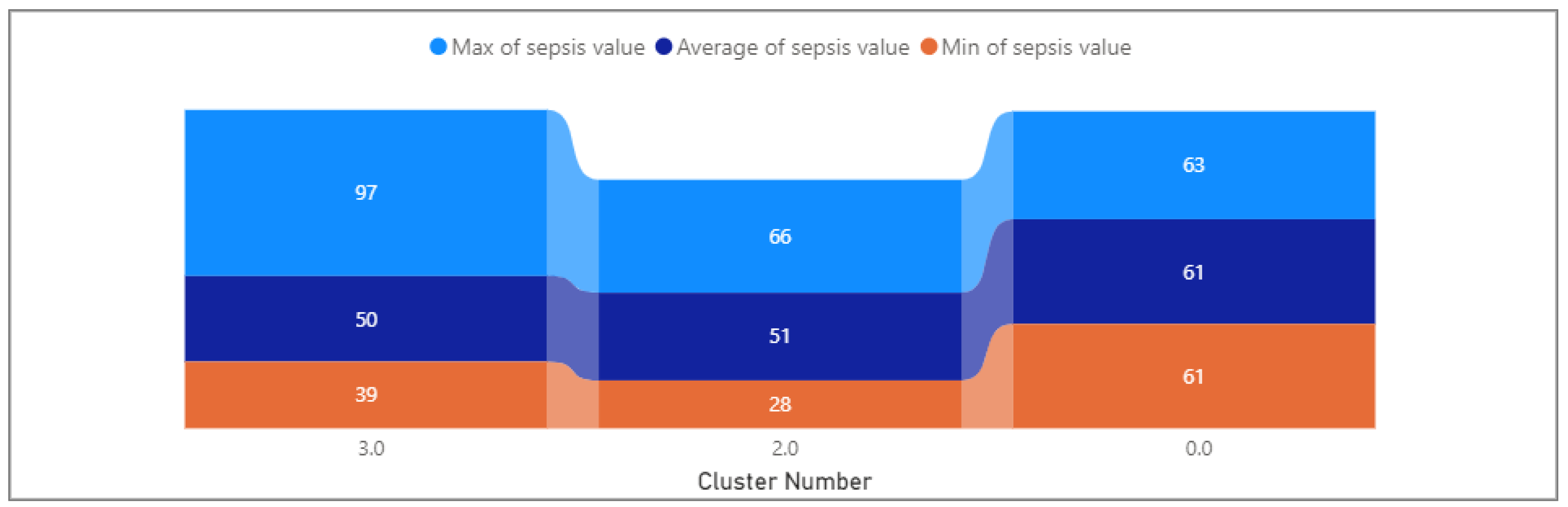
- Sepsis: Ranged from 61 to 63, with a mean of 61.13. This variable shows minimal variability within the cluster.
- −
- Arterial blood pressure ranges from 70.54 to 136.56, with a mean of 111.04. Suggests a spectrum of blood pressure levels, potentially indicating varying severity of cardiovascular conditions.
- −
- Systolic arterial blood pressure varies between 77.92 and 114.91, with a mean of 108.65. Indicates a range of systolic blood pressure levels.
- −
- Mean arterial blood pressure ranges from 48.81 to 104.75, with a mean of 96.04. Indicates variations in overall blood pressure within the cluster.
- −
- Diastolic arterial blood pressure varies from 31.27 to 38.01, with a mean of 64.57. Shows a range of diastolic blood pressure levels.
- −
- Temperature: Consistent at a mean of 36.52, reflecting stability in body temperature within the cluster.
- −
- Oxygen saturation levels range from 49.18 to 99.13, with a mean of 96.04. Indicates variability in oxygen saturation.
- −
- Heart rate varies between 47.72 and 137.94, with a mean of 107.05. Shows a range of heart rates within the cluster.
- −
- The laboratory results display a wide range, spanning from −0.6 to 88,033, with an average value of 109.55. This considerable variability suggests a spectrum of diverse clinical conditions.
- Cluster 1:
- −
- Event Day: Similar to Cluster 0, with a mean of 1.09, indicating events spread across the observed period.
- −
- Glasgow Coma Scale: Consistently low at 5.33, indicating a uniform level of consciousness within the cluster.
- −
- Sepsis: Not provided, limiting insights into this variable.
- −
- arterial blood pressure ranges from 45.15 to 88.72, with a mean of 90.43. Indicates a moderate range of blood pressure levels.
- −
- Systolic arterial blood pressure varies between 31.65 and 60.13, with a mean of 55.58. Indicates a range of systolic blood pressure levels.
- −
- Mean arterial blood pressure ranges from 10.83 to 53.51, with a mean of 27.02. Shows variability in overall blood pressure within the cluster.
- −
- Diastolic arterial blood pressure varies from 23.54 to 38.64, with a mean of 35.65. Shows a range of diastolic blood pressure levels.
- −
- Temperature: Consistent at a mean of 27.02, reflecting stability in body temperature within the cluster.
- −
- Oxygen saturation levels range from 57.39 to 142.82, with a mean of 73.15. Indicates variability in oxygen saturation.
- −
- Heart rate varies widely from 0 to 59,065.7, with a mean of 77.68. The wide range may be due to potential outliers.
- Cluster 2:
- −
- Day of clinical event: Similar to Clusters 0 and 1, with a mean of 1.09, indicating events spread across the observed period.
- −
- Glasgow Coma Scale: Varies between 7 and 15, with a mean of 14.52. Indicates a higher level of consciousness within the cluster.
- −
- Sepsis: Ranges from 28 to 66, with a mean of 51.18. This variable shows variability within the cluster.
- −
- Arterial blood pressure ranges from 42.69 to 101.35, with a mean of 79.01. Indicates a moderate range of blood pressure levels.
- −
- Systolic arterial blood pressure varies between 67.61 and 149.12, with a mean of 101.39. Indicates a range of systolic blood pressure levels.
- −
- Mean arterial blood pressure ranges from 37.01 to 98.28, with a mean of 93.86. Shows variability in overall blood pressure within the cluster.
- −
- Diastolic arterial blood pressure varies from 31.04 to 38.64, with a mean of 56.27. Shows a range of diastolic blood pressure levels.
- −
- Temperature: Consistent at a mean of 30.01, reflecting stability in body temperature within the cluster.
- −
- Oxygen saturation levels range from 57.85 to 142.82, with a mean of 80.84. Indicates variability in oxygen saturation.
- −
- Heart rate varies between −1 and 44,302.5, with a mean of 75.21. The wide range may be due to potential outliers.
- Cluster 3:
- −
- Day of clinical event: Similar to Clusters 0 and 1, with a mean of 1.22, indicating events spread across the observed period.
- −
- Glasgow Coma Scale: Varies between 3 and 15, with a mean of 14.11. Indicates a higher level of consciousness within the cluster.
- −
- Sepsis: Ranges from 39 to 97, with a mean of 49.97. This variable shows variability within the cluster.
- −
- Arterial blood pressure ranges from 55.28 to 121.17, with a mean of 74.88. Indicates a moderate range of blood pressure levels.
- −
- Systolic arterial blood pressure varies between 58.62 and 103.86, with a mean of 128.07. Indicates a range of systolic blood pressure levels.
- −
- The mean arterial blood pressure ranges from 43.49 to 84.62, with a mean of 95.41. Shows variability in overall blood pressure within the cluster.
- −
- Diastolic arterial blood pressure varies from 16.75 to 32.63, with a mean of 64.98. Shows a range of diastolic blood pressure levels.
- −
- Temperature: Consistent at a mean of 36.53, reflecting stability in body temperature within the cluster.
- −
- Oxygen saturation levels range from 86.72 to 99, with a mean of 95.41. Indicates variability in oxygen saturation.
- −
- Heart rate varies between −0.35 and 49,300, with a mean of 80.84. The wide range may be due to potential outliers.
3.4. Insight into the Data Behavior within Clusters
3.4.1. Glasgow Coma Scale (GSC)
3.4.2. Systolic Arterial Blood Pressure
3.4.3. Oxygen Saturation
3.4.4. Heart Rate
3.4.5. Temperature
3.4.6. Sepsis
3.4.7. Days of Clinical Events
3.4.8. Temporal Distribution of Clusters
3.4.9. Laboratory Exams and Results
3.4.10. Interventions
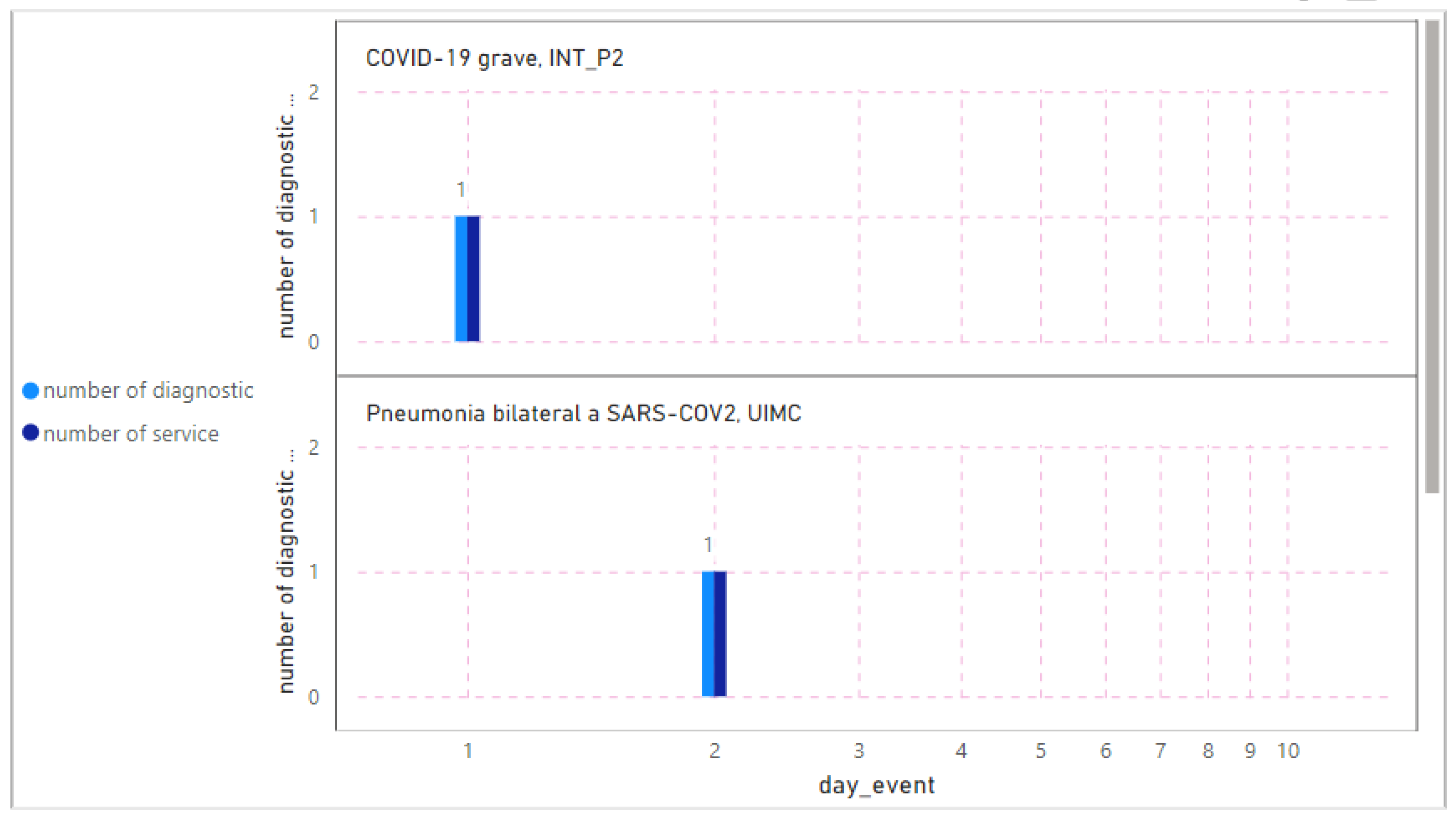
3.4.11. Diagnostic
3.4.12. Medications
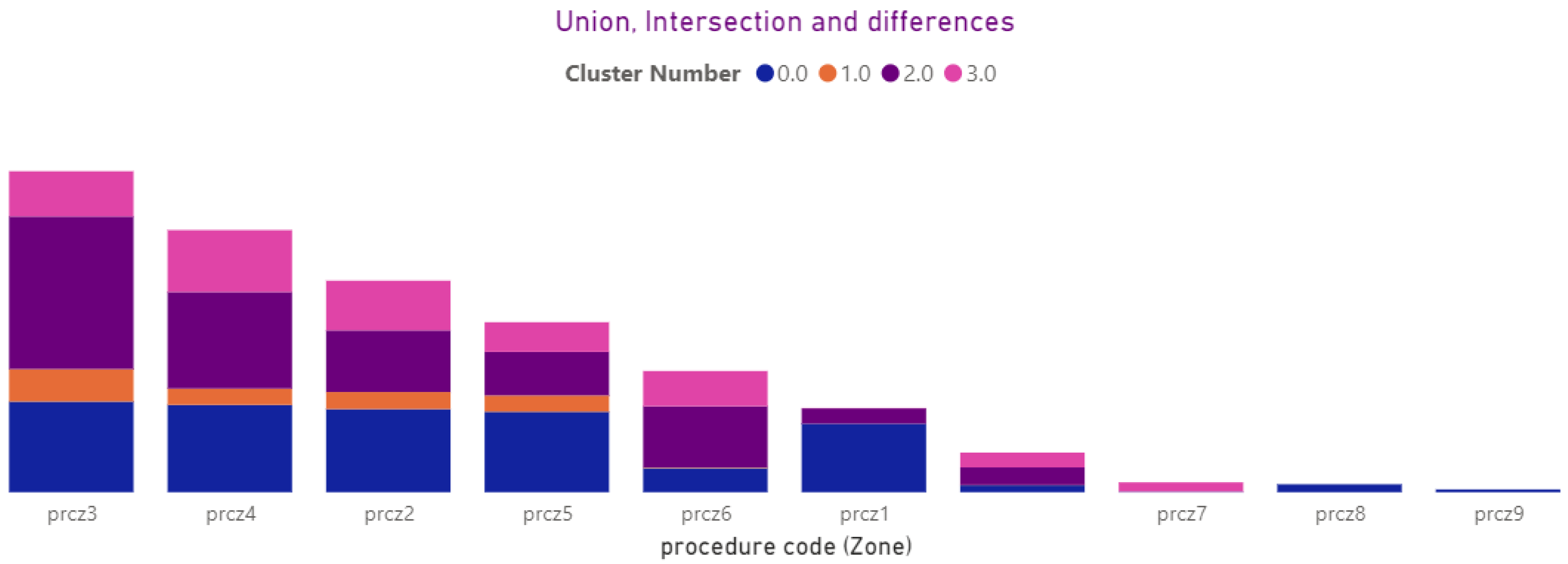
3.4.13. Procedures (Local, Zona)
3.5. Predicting BehaviorBased Cluster
3.5.1. Classification
- Mean Accuracy: 0.946, with a standard deviation of 0.078. This indicates that, on average, the model correctly predicts the target class around 94.6% of the time.
- Mean Precision: 0.961, with a standard deviation of 0.052. Precision measures the accuracy of the positive predictions. This result suggests that, on average, around 96.1% of the predicted positive cases are positive.
- Mean Recall: 0.946, with a standard deviation of 0.078. Recall measures the ability of the model to identify the positive cases correctly. This value indicates that around 94.6% of the actual positive cases are correctly identified by the model.
- Mean F1 Score: 0.946, with a standard deviation of 0.077. The F1 score is the harmonic mean of precision and recall and provides a balance between them. This value suggests that the model achieves a good balance between precision and recall.
- Mean Cohen’s Kappa: 0.912, with a standard deviation of 0.126. Cohen’s Kappa measures the agreement between predicted and actual class labels, considering the possibility of the agreement occurring by chance. This value indicates a substantial level of agreement beyond chance.
- The mean AUC (Area Under the Curve) value of approximately 0.997 indicates that, on average, the classifier has an excellent ability to discriminate between different classes in the dataset. AUC values close to 1 suggest that the classifier can effectively separate positive and negative instances, making it highly reliable in distinguishing between classes.
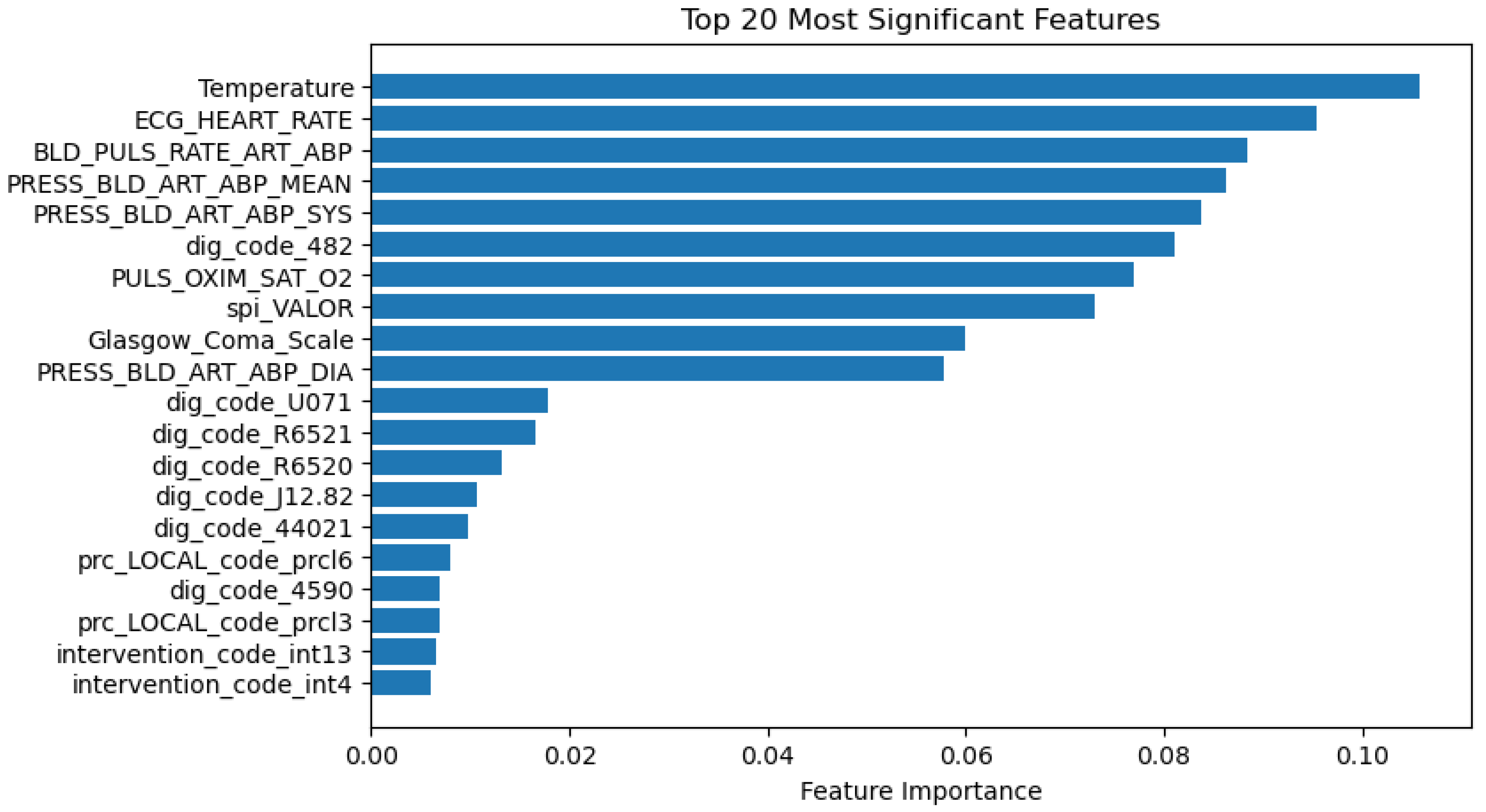
3.5.2. Significance of Predictors
4. Discussion
4.1. CEid Formulation and Its Transformative Impacts
4.2. Analytical Insight
4.3. Cluster Analysis
- Identification of Extreme Values
- Min and Max Values: Examining the minimum and maximum values within each cluster helps identify extreme values or outliers. These outliers may represent unique cases or anomalies that can provide valuable information about exceptional health conditions or atypical patient responses.
- Understanding Range and Variability
- Differential Health Characteristics
- Clinical Relevance
- Feature Importance in Predictive Modelling
- Quality Control and Data Integrity
4.4. Classification
4.5. Application
4.6. Challenges
4.7. Theoretical Reference—Suboptimal Decision-Making
4.8. Application in PM
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kosorok, M.R.; Laber, E.B. Precision Medicine. Annu. Rev. Stat. Its Appl. 2019, 6, 263–286. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Duffy, A.H.B.; Whitfield, R.I.; Boyle, I.M. Integration of decision support systems to improve decision support performance. Knowl. Inf. Syst. 2010, 22, 261–286. [Google Scholar] [CrossRef]
- Banappagoudar, S.B.; Santhosh, S.U.; Thangam, M.M.N.; David, S.; Shamili, G.S. The Advantages of Precision Medicine over Traditional Medicine in Improving Public Health Outcomes. J. ReAttach Ther. Dev. Divers. 2023, 6, 272–277. [Google Scholar]
- Beckmann, J.S.; Lew, D. Reconciling evidence-based medicine and precision medicine in the era of big data: Challenges and opportunities. Genome Med. 2016, 8, 134. [Google Scholar] [CrossRef] [PubMed]
- Pencina, M.J.; Peterson, E.D. Moving from clinical trials to precision medicine: The role for predictive modeling. JAMA—J. Am. Med. Assoc. 2016, 315, 1713–1714. [Google Scholar] [CrossRef] [PubMed]
- Naithani, N.; Sinha, S.; Misra, P.; Vasudevan, B.; Sahu, R. Precision medicine: Concept and tools. Med. J. Armed Forces India 2021, 77, 249–257. [Google Scholar] [CrossRef] [PubMed]
- Gopal, G.; Suter-Crazzolara, C.; Toldo, L.; Eberhardt, W. Digital transformation in healthcare—Architectures of present and future information technologies. Clin. Chem. Lab. Med. 2019, 57, 328–335. [Google Scholar] [CrossRef]
- Alqenae, F.A.; Steinke, D.; Keers, R.N. Prevalence and Nature of Medication Errors and Medication-Related Harm Following Discharge from Hospital to Community Settings: A Systematic Review. Drug Saf. 2020, 43, 517–537. [Google Scholar] [CrossRef]
- Alzheimer’s Association. 2017 Alzheimer’s disease facts and figures. Alzheimer’s Dement. 2017, 13, 325–373. [Google Scholar] [CrossRef]
- Maglaveras, N.; Kilintzis, V.; Koutkias, V.; Chouvarda, I. Integrated Care and Connected Health Approaches Leveraging Personalised Health through Big Data Analytics; Studies in Health Technology and Informatics; IOP Press: Amsterdam, The Netherlands, 2016; Volume 224, pp. 117–122. [Google Scholar] [CrossRef]
- Francis, S.; Collins, M.D.; Varmus, M.D. A commentary on ‘A new initiative on precision medicine’. Front. Psychiatry 2015, 6, 88. [Google Scholar] [CrossRef][Green Version]
- Haque, M.; Islam, T.; Sartelli, M.; Abdullah, A.; Dhingra, S. Prospects and challenges of precision medicine in lower-and middle-income countries: A brief overview. Bangladesh J. Med. Sci. 2020, 19, 32–47. [Google Scholar] [CrossRef]
- Bonkhoff, A.K.; Grefkes, C. Precision medicine in stroke: Towards personalized outcome predictions using artificial intelligence. Brain 2022, 145, 457–475. [Google Scholar] [CrossRef] [PubMed]
- Mosavi, N.S.; Santos, M.F. Unveiling Precision Medicine with Data Mining: Discovering Patient Subgroups and Patterns. In Proceedings of the 2023 IEEE Symposium Series on Computational Intelligence, SSCI 2023, Mexico City, Mexico, 5–8 December 2023. [Google Scholar] [CrossRef]
- Boettger, J.; Deussen, O.; Ziezold, H. Apparatus and Methods for Controlling and Applying Flash Lamp Radiation. US10881872B1, 2014. [Google Scholar]
- Awwalu, J.; Garba, A.G.; Ghazvini, A.; Atuah, R. Artificial Intelligence in Personalized Medicine Application of AI Algorithms in Solving Personalized Medicine Problems. Int. J. Comput. Theory Eng. 2015, 7, 439–443. [Google Scholar] [CrossRef]
- König, I.R.; Fuchs, O.; Hansen, G.; von Mutius, E.; Kopp, M.V. What is precision medicine? Eur. Respir. J. 2017, 50, 1700391. [Google Scholar] [CrossRef]
- Jameson, J.L.; Longo, D.L. Precision medicine—Personalized, problematic, and promising. N. Engl. J. Med. 2015, 372, 2229–2234. [Google Scholar] [CrossRef] [PubMed]
- LSanchez-Pinto, L.N.; Bhavani, S.V.; Atreya, M.R.; Sinha, P. Leveraging Data Science and Novel Technologies to Develop and Implement Precision Medicine Strategies in Critical Care. Crit. Care Clin. 2023, 39, 627–646. [Google Scholar] [CrossRef]
- Pelter, M.N.; Druz, R.S. Precision medicine: Hype or hope? Trends Cardiovasc. Med. 2024, 34, 120–125. [Google Scholar] [CrossRef]
- Szelka, J.; Wrona, Z. Knowledge Discovery in Data KDD Meets Big Data. Arch. Civ. Eng. 2016, 62, 217–228. [Google Scholar] [CrossRef][Green Version]
- MBekbolatova, M.; Mayer, J.; Ong, C.W.; Toma, M. Transformative Potential of AI in Healthcare: Definitions, Applications, and Navigating the Ethical Landscape and Public Perspectives. Healthcare 2024, 12, 125. [Google Scholar] [CrossRef]
- Lee, D.; Yoon, S.N. Application of artificial intelligence-based technologies in the healthcare industry: Opportunities and challenges. Int. J. Environ. Res. Public Health 2021, 18, 271. [Google Scholar] [CrossRef]
- Sutton, R.T.; Pincock, D.; Baumgart, D.C.; Sadowski, D.C.; Fedorak, R.N.; Kroeker, K.I. An overview of clinical decision support systems: Benefits, risks, and strategies for success. NPJ Digit. Med. 2020, 3, 17. [Google Scholar] [CrossRef] [PubMed]
- Mosavi, N.S.; Santos, M.F. Adoption of Precision Medicine; Limitations and Considerations. Comput. Sci. Inf. Technol. 2021, 13–24. [Google Scholar] [CrossRef]
- Mesko, B. The role of artificial intelligence in precision medicine. Expert Rev. Precis. Med. Drug Dev. 2017, 2, 239–241. [Google Scholar] [CrossRef]
- Sriram, R.D.; Subrahmanian, E. Transforming Health Care through Digital Revolutions. J. Indian Inst. Sci. 2020, 100, 753–772. [Google Scholar] [CrossRef] [PubMed]
- Watson, H.J. The Cognitive Decision-Support Generation. Bus. Intell. J. 2017, 22, 3–6. [Google Scholar]
- Watson, H.J. Preparing for the cognitive generation of decision support. MIS Q. Exec. 2017, 16, 3–6. [Google Scholar]
- Johnson, K.B.; Wei, W.; Weeraratne, D.; Frisse, M.E.; Misulis, K.; Rhee, K.; Zhao, J.; Snowdon, J.L. Precision Medicine, AI, and the Future of Personalized Health Care. Clin. Transl. Sci. 2021, 14, 86–93. [Google Scholar] [CrossRef]
- Jabbar, M.A.; Samreen, S.; Aluvalu, R. The future of health care: Machine learning. Int. J. Eng. Technol. 2018, 7, 23–25. [Google Scholar] [CrossRef][Green Version]
- Tarassoli, S.P. Artificial intelligence, regenerative surgery, robotics? What is realistic for the future of surgery? Ann. Med. Surg. 2019, 41, 53–55. [Google Scholar] [CrossRef] [PubMed]
- Leone, D.; Schiavone, F.; Appio, F.P.; Chiao, B. How does artificial intelligence enable and enhance value co-creation in industrial markets? An exploratory case study in the healthcare ecosystem. J. Bus. Res. 2021, 129, 849–859. [Google Scholar] [CrossRef]
- Abbaoui, W.; Retal, S.; El Bhiri, B.; Kharmoum, N.; Ziti, S. Towards revolutionizing precision healthcare: A systematic literature review of artificial intelligence methods in precision medicine. Inform. Med. Unlocked 2024, 46, 101475. [Google Scholar] [CrossRef]
- Ahmed, Z.; Mohamed, K.; Zeeshan, S.; Dong, X. Artificial intelligence with multi-functional machine learning platform development for better healthcare and precision medicine. Database 2020, 2020, baaa010. [Google Scholar] [CrossRef]
- Chang, D.; Chang, D.; Pourhomayoun, M. Risk Prediction of critical vital signs for ICU patients using recurrent neural network. In Proceedings of the 6th Annual Conference on Computational Science and Computational Intelligence, CSCI, Las Vegas, NV, USA, 5–7 December 2019; pp. 1003–1006. [Google Scholar] [CrossRef]
- Fenning, S.J.; Smith, G.; Calderwood, C. Realistic Medicine: Changing culture and practice in the delivery of health and social care. Patient Educ. Couns. 2019, 102, 1751–1755. [Google Scholar] [CrossRef] [PubMed]
- Carra, G.; Salluh, J.I.; Ramos, F.J.d.S.; Meyfroidt, G. Data-driven ICU management: Using Big Data and algorithms to improve outcomes. J. Crit. Care 2020, 60, 300–304. [Google Scholar] [CrossRef]
- Gupta, N.S.; Kumar, P. Perspective of artificial intelligence in healthcare data management: A journey towards precision medicine. Comput. Biol. Med. 2023, 162, 107051. [Google Scholar] [CrossRef] [PubMed]
- Johnson, N.; Parbhoo, S.; Ross, A.S.; Doshi-Velez, F. Learning Predictive and Interpretable Timeseries Summaries from ICU Data. AMIA Annu. Symp. Proc. 2021, 2021, 581–590. Available online: https://arxiv.org/abs/2109.11043v1 (accessed on 10 January 2022).
- McPadden, J.; Durant, T.J.; Bunch, D.R.; Coppi, A.; Price, N.; Rodgerson, K.; Torre, C.J., Jr.; Byron, W.; Hsiao, A.L.; Krumholz, H.M.; et al. Health care and precision medicine research: Analysis of a scalable data science platform. J. Med. Internet Res. 2019, 21, e13043. [Google Scholar] [CrossRef]
- Gligorijević, V.; Malod-Dognin, N.; Pržulj, N. Integrative methods for analyzing big data in precision medicine. Proteomics 2016, 16, 741–758. [Google Scholar] [CrossRef]
- Hulsen, T.; Jamuar, S.S.; Moody, A.R.; Karnes, J.H.; Varga, O.; Hedensted, S.; Spreafico, R.; Hafler, D.A.; McKinney, E.F. From big data to precision medicine. Front. Med. 2019, 6, 34. [Google Scholar] [CrossRef]
- Delen, D. Prescriptive Analytics: The Final Frontier for Evidence—Based Management and Optimal Decision; Pearson Education, Inc.: London, UK, 2020. [Google Scholar]
- March, J.G. Bounded Rationality, Ambiguity, and the Engineering of Choice. Bell J. Econ. 1978, 9, 587–608. [Google Scholar] [CrossRef]
- Simon, H. Theories of Decision-Making and Behavioral Science. Am. Econ. Rev. 1959, 49, 253–283. [Google Scholar]
- Sharda, R.; Delen, D.; Turban, E.; Aronson, J.E.; Liang, T.-P.; King, D. Business Intelligence and Analytics: Systems for Decision Support; Pearson: London, UK, 2014. [Google Scholar]
- Palanisamy, V.; Thirunavukarasu, R. Implications of big data analytics in developing healthcare frameworks—A review. J. King Saud Univ. -Comput. Inf. Sci. 2019, 31, 415–425. [Google Scholar] [CrossRef]
- Mosavi, N.S.; Santos, M.F. How prescriptive analytics influences decision making in precision medicine. Procedia Comput. Sci. 2020, 177, 528–533. [Google Scholar] [CrossRef]
- Mosavi, N.S.; Santos, M.F. Data Engineering to Support Intelligence for Precision Medicine in Intensive Care. In Proceedings of the 2022 International Conference on Computational Science and Computational Intelligence (CSCI), Las Vegas, NV, USA, 14–16 December 2022. [Google Scholar] [CrossRef]
- Mosavi, N.; Santos, M. Intelligent Decision Support System for Precision Medicine: Time Series Multi-variable Approach for Data Processing. In Proceedings of the International Joint Conference on Knowledge Discovery, Knowledge Engineering and Knowledge Management, IC3K, Valletta, Malta, 24–26 October 2022; Volume 3, pp. 231–238. [Google Scholar] [CrossRef]

















| Technique | Focused Area | Limitations |
|---|---|---|
| attention scores | feature importance | feature selection, time series data, nonlinear features |
| post-hoc explanation techniques | Modeling and features relations | Interpretation-black box |
| summaries of patient time series data | Data extraction-time-series clinical data | vital signs and lab data |
| Filtering outliers | Data cleaning | Time series data-vital signs |
| Electron | Data extraction | Physiologic Signal Monitoring and Analysis |
| Topological Data Analysis (TDAs) | Dimensionality Reduction | |
| anytime algorithms | Velocity | learn from streaming data |
| GNMTF | Variety | Complex when data types increase |
| Indicator—Numerical (692 rows, 8 columns) | CLUSTER 0 | CLUSTER 1 | CLUSTER 2 | CLUSTER 3 | ||||
|---|---|---|---|---|---|---|---|---|
| Silhouette Score: 0.23 | Silhouette Score: 0.12 | Silhouette Score 0.26 | Silhouette Score: 0.36 | |||||
| Data Distribution: 271 | Data Distribution: 44 | Data Distribution: 285 | Data Distribution: 92 | |||||
| Min | Max | Min | Max | Min | Max | Min | Max | |
| the pulse rate of the arterial blood pressure | 70.54 | 144.32 | 70.34 | 123.46 | 42.69 | 101.43 | 55.27 | 124.39 |
| diastolic arterial blood pressure | 48.80 | 138.37 | 10.83 | 53.50 | 37.01 | 101.60 | 40.51 | 88.03 |
| systolic arterial blood pressure | 82.88 | 173.66 | 45.15 | 96.03 | 98.27 | 179.98 | 74.60 | 164.71 |
| mean arterial blood pressure | 51.20 | 143.86 | 31.65 | 61.22 | 67.61 | 165.66 | 58.62 | 108.86 |
| heart rate | 47.71 | 144.65 | 57.38 | 149.02 | 49.63 | 116.32 | 54.46 | 133.74 |
| pulse oximetry oxygen saturation level | 49.18 | 99.46 | 71.31 | 98.75 | 77.85 | 99.54 | 65.00 | 99.03 |
| body temperature | 30.65 | 39.00 | 23.53 | 38.67 | 29.64 | 37.91 | 16.74 | 32.63 |
| day of the clinical event | 1 | 37 | 1 | 39 | 1 | 29 | 1 | 19 |
| Numerical Variables | C0 | C1 | C2 | C3 | |
|---|---|---|---|---|---|
| the pulse rate of the arterial blood pressure | average | 111.03 | 90.42 | 74.87 | 79.01 |
| minimum | 70.54 | 70.34 | 42.69 | 55.27 | |
| maximum | 136.56 | 109.50 | 101.35 | 121.17 | |
| diastolic arterial blood pressure | average | 64.56 | 35.64 | 64.98 | 56.27 |
| minimum | 48.80 | 10.83 | 37.01 | 43.49 | |
| maximum | 104.75 | 53.50 | 98.27 | 84.61 | |
| systolic arterial blood pressure | average | 108.64 | 55.57 | 128.06 | 101.38 |
| minimum | 82.88 | 45.15 | 98.27 | 74.60 | |
| maximum | 172.13 | 88.72 | 178.96 | 158.90 | |
| mean arterial blood pressure | average | 77.92 | 42.80 | 85.31 | 70.51 |
| minimum | 51.20 | 31.65 | 67.61 | 58.62 | |
| maximum | 114.90 | 60.12 | 149.11 | 103.85 | |
| pulse oximetry oxygen saturation level | average | 96.04 | 73.14 | 95.40 | 93.85 |
| minimum | 49.18 | 71.31 | 77.85 | 86.71 | |
| maximum | 99.13 | 97.93 | 99.46 | 99.00 | |
| Heart rate | average | 107.04 | 77.68 | 75.20 | 80.84 |
| minimum | 47.71 | 57.38 | 49.63 | 54.46 | |
| maximum | 137.93 | 142.82 | 114.80 | 131.86 | |
| Body temperature | average | 36.52 | 27.02 | 36.53 | 30.01 |
| minimum | 31.27 | 23.53 | 31.03 | 16.74 | |
| maximum | 38 | 38.63 | 37.90 | 32.62 | |
| Day of clinical event | average | 1.05 | 1.09 | 1.21 | 1.08 |
| minimum | 1 | 1 | 1 | 1 | |
| maximum | 37 | 39 | 36 | 19 | |
| Glasgow Coma Scale | average | 2.6 | 5.33 | 14.51 | 14.10 |
| minimum | 3.00 | 5.33 | 7.00 | 3.00 | |
| maximum | 15.00 | 5.33 | 15.00 | 15.00 |
| Metrics | Value (Mean) |
|---|---|
| Accuracy | 95.36% |
| Precision | 0.961 |
| Recall | 0.946 |
| Kappa | 0.912 |
| F1 score | 0.946 |
| AUC | 0.997 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mosavi, N.S.; Santos, M.F. Enhancing Clinical Decision Support for Precision Medicine: A Data-Driven Approach. Informatics 2024, 11, 68. https://doi.org/10.3390/informatics11030068
Mosavi NS, Santos MF. Enhancing Clinical Decision Support for Precision Medicine: A Data-Driven Approach. Informatics. 2024; 11(3):68. https://doi.org/10.3390/informatics11030068
Chicago/Turabian StyleMosavi, Nasim Sadat, and Manuel Filipe Santos. 2024. "Enhancing Clinical Decision Support for Precision Medicine: A Data-Driven Approach" Informatics 11, no. 3: 68. https://doi.org/10.3390/informatics11030068
APA StyleMosavi, N. S., & Santos, M. F. (2024). Enhancing Clinical Decision Support for Precision Medicine: A Data-Driven Approach. Informatics, 11(3), 68. https://doi.org/10.3390/informatics11030068







