Abstract
(1) Background: Tumor-specific standardized data are essential for AI-based progress in research, e.g., for predicting adverse events in patients with melanoma. Although there are oncological Fast Healthcare Interoperability Resources (FHIR) profiles, it is unclear how well these can represent malignant melanoma. (2) Methods: We created a methodology pipeline to assess to what extent an oncological FHIR profile, in combination with a standard FHIR specification, can represent a real-world data set. We extracted Electronic Health Record (EHR) data from a data platform, and identified and validated relevant features. We created a melanoma data model and mapped its features to the oncological HL7 FHIR Basisprofil Onkologie [Basic Profile Oncology] and the standard FHIR specification R4. (3) Results: We identified 216 features. Mapping showed that 45 out of 216 (20.83%) features could be mapped completely or with adjustments using the Basisprofil Onkologie [Basic Profile Oncology], and 129 (60.85%) features could be mapped using the standard FHIR specification. A total of 39 (18.06%) new, non-mappable features could be identified. (4) Conclusions: Our tumor-specific real-world melanoma data could be partially mapped using a combination of an oncological FHIR profile and a standard FHIR specification. However, important data features were lost or had to be mapped with self-defined extensions, resulting in limited interoperability.
1. Introduction
1.1. Background and Significance
Health Level 7 (HL7) Fast Healthcare Interoperability Resources (FHIR) (https://hl7.org/fhir/ (accessed on 6 May 2024)) is one of the most common international standards for interoperability and thus data exchange between healthcare software systems. FHIR describes clinical concepts as patterns using resources and combines them with rules and constraints into profiles [1] for specific use cases. FHIR intentionally implements the most frequently used clinical concept information [2] using the 80/20 rule, meeting 80 percent of the interoperability needs for resources with 20 percent of the requirements [3]. However, FHIR is also intended to be adapted with profiles for more specific use cases [3]. For these adaptations, either standardized or specialized FHIR profiles can be used or (further) developed, or existing ones can be extended in a non-standardized, self-defined manner by using existing generic elements or creating extensions. In Germany, various standardization projects use FHIR, e.g., Medizinische Informationsobjekte [Medical Information Objects] (MIOs) for the elektronische Patientenakte [Personal Health Record] (ePA) [4] and Digitale Gesundheitsanwendungen [Digital Health Applications] (DiGAs) [5], Informationstechnische Systeme in Krankenhäusern [Information Technology in Hospitals] (ISiK) [6], and the Medizininformatik Initiative [Medical Informatics Initiative] (MII) [7] Core Data Set (CDS) [8]. To apply appropriate technical standards, FHIR should be used for clinical data models.
Research in oncology is dynamic, with new methods constantly revealing new biomarkers [9]. Thus, treatment decisions need to be based on actual tumor-specific data. For instance, tumor thickness is the most important prognostic factor for primary melanoma [10] but cannot be applied, e.g., to leukemia. Although there are already recent studies focused on FHIR in oncology [1,11,12,13], and several German studies/projects [14,15] and an international project [16] adapting the standard FHIR for oncology, it is uncertain how well these can represent specific tumors like melanoma. Even more specific FHIR profiles might be required to present individual oncological diseases.
As FHIR has evolved since its introduction in 2011 [1], so has real-world data management within healthcare institutions. For instance, the local FHIR-based healthcare data platform, the Smart Hospital Information Platform (SHIP), of the University Hospital Essen, Germany was initiated over 10 years ago. These data platforms need to be regularly updated to ensure the highest possible level of interoperability by checking new FHIR specifications/profiles for their suitability, implementations, and required mapping efforts. In other medical fields, related work has already demonstrated mappings of German Electronic Health Record (EHR) data to the Observational Medical Outcomes Partnership (OMOP) Common Data Model (CDM) [17] and FHIR [18] standards and extended FHIR profiles [14,19,20,21] for context-specific use. The oncological HL7 FHIR Basisprofil Onkologie [Basic Profile Oncology] [22] cooperates with the German OncoLogical Data Standard (GOLD) [23], combining various German and international oncology data sets into one FHIR-based maximum data set [15]. Unlike GOLD and the MII [8] Modul Onkologie [Module Oncology] that is currently being drafted [24], the Basisprofil Onkologie [Basic Profile Oncology] is already in a released and approved status and therefore ready to use.
Malignant melanoma is a suitable example of a specific tumor due to its increased observed relevance and its further predicted incidence increase [25] as well as its specific attributes, e.g., of systemic immunotherapies. Immunotherapies, e.g., with the agents nivolumab and pembrolizumab, enhance immune responses [26] and can induce specific immune-related adverse events (irAEs) [27,28] which may result in unpredictable patient-specific life-threatening conditions [29]. Standardized high-qualitative EHR data are needed to improve results.
1.2. Objectives
We aim to assess to what extent an oncological FHIR profile can represent malignant melanoma or whether further, more specialized FHIR profiles are needed.
Our approach provides a methodology pipeline to examine a reference FHIR profile and a standard FHIR specification. The focus is on the coverage of (clinical) data features and the compatibility of the terminologies used. These are relevant indicators for the further development of the standards and potential automation of mapping and updating real-world data to new FHIR specifications and profiles. Therefore, we used real-world data of patients with melanoma from the Department of Dermatology of the University Hospital Essen, Germany and the HL7 oncological FHIR Basisprofil Onkologie [Basic Profile Oncology] in a single-center study. We considered the data features of malignant melanoma required for the prediction of irAEs in first-line immunotherapy.
2. Materials and Methods
Below, the materials and methods used for mapping clinical real-world data of patients with melanoma to oncological FHIR profiles are elaborated. As shown in Figure 1, these include the description of the EHR data used (see Section 2.1); data extraction, pre-processing, and validation (see Section 2.2); clinical concept modeling (see Section 2.3); reference FHIR profile selection (see Section 2.4); and data model mapping with its statistical metrics (see Section 2.5).
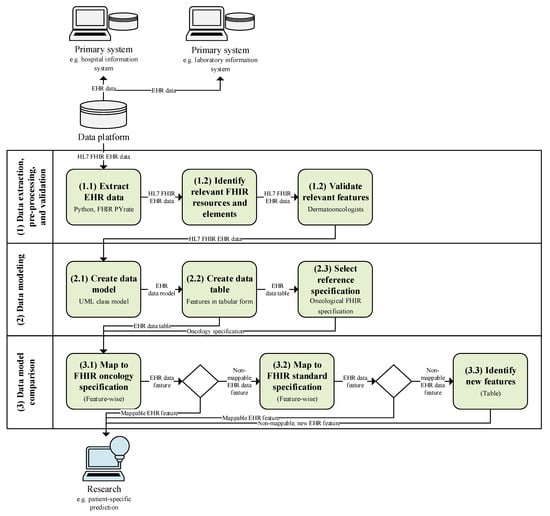
Figure 1.
Study pipeline with initial situation (gray), processing steps of our study (green), and future usage (blue).
2.1. Real-World Electronic Health Record (EHR) Data
In our single-center study, we used German EHR data from patients diagnosed with malignant melanoma from the Department of Dermatology University Hospital Essen, Germany as an example. We used retrospective data documented in the information system Cerner Medico (https://www.cgm.com/deu_de/produkte/krankenhaus/cgm-medico.html (accessed on 6 May 2024)) and Nexus Swisslab (https://www.nexus-ag.de/lab (accessed on 6 May 2024)). As the digital healthcare data platform SHIP of the University Hospital Essen gathers treatment data from primary systems [30] and is based on the FHIR specification R4 (https://hl7.org/fhir/R4/ (accessed on 6 May 2024)), all data used were extracted in a structured form.
2.2. Data Extraction, Pre-Processing, and Validation
We manually analyzed and identified relevant FHIR of patients with melanoma using the internal resource. Our feature selection was adapted to the outcome prediction of irAEs under first-line immunotherapy as an example and validated by conducting unstructured interviews with two dermato-oncologists. Not only were clinically relevant features (e.g., irAE thyroiditis) determined, but also all administrative features (e.g., final status of irAE thyroiditis), and linking features (e.g., irAE thyroiditis of specific systemic infusion therapy) were required for processing. Extraction from SHIP was performed using the Python package FHIR-PYrate (https://github.com/UMEssen/FHIR-Pyrate (accessed on 6 May 2024)) for querying FHIR resources [31] to create an FHIR-based data subset. We specified search parameters for the queries to obtain the appropriate data for our use case.
2.3. Data Modeling
We created and visualized the clinical concept model of our use case to obtain a basis for a subsequent comparison of its features with those of a selected oncological reference FHIR profile. We used a Unified Modeling Language (UML) class diagram for the visualization of our clinical concept model, as a UML class diagram is one FHIR representation type [32] provided by HL7. We determined (resource) classes and represented their features for the prediction of irAEs as FHIR resources, elements, data types, value sets, and references.
Resource Classes: According to their structure and medical content, considering their resource types and internally used identifier systems, we divided the extracted data into 37 different resource classes, e.g., Comorbidity, Melanoma patient, Primary tumor diagnosis, Skin type, and Systemic infusion therapy. For each resource class, we extracted (if available) ten different resource entities using randomly provided resource IDs and examined these resource classes for the data elements they contain to obtain a consistent model.
Logical Areas: For a better overview and understanding of the content, we grouped the resource classes of our data model into logical areas based on similar medical contexts: (i) Biomarker, (ii) Case, (iii) Melanoma patient, (iv) Other Medication, (v) Risk factors, (vi) Systemic infusion therapy, and (vii) Tumor diagnosis. The information in our model can be divided into non-disease-specific and disease-specific information. While the Melanoma patient area contains non-disease-specific information, all other areas contain knowledge for the specific treatment process, with some of them being highly tumor-specific, like Systemic infusion therapy and Tumor diagnosis. The pathological TNM (pTNM) and the clinical TNM (cTNM) stage, the tumor status, and the tumor thickness are, among others, all pieces of information that describe the tumor and the associated diagnosis and are therefore summarized in the logical area Tumor diagnosis. Systemic infusion therapy describes the systemic therapy of malignant melanoma and any irAEs that may occur as a result of this therapy. Any medication other than systemic therapy, including its prescription and/or intake, was grouped as Other medication. All Risk factors were already defined specifically for dermato-oncology within the hospital in consultation with physicians prior to this study. Comorbidities, laboratory values, and tumor markers are features for outcomes and therefore labeled as Biomarkers.
2.4. FHIR Profile Selection
According to Ulrich et al., data sets and specifications from different country origins can differ, e.g., in terms of the terminology used, which can lead to problems [33]. As Table 1 shows, we considered different national (German) FHIR profiles and compared their suitability as a reference national FHIR profile for oncology.

Table 1.
Comparison of characteristics of national German oncological FHIR profiles [15,22,23,24] (as of 27 March 2024).
GOLD combines, among others, various German oncological data sets into one FHIR-based maximum data set, including the Bundeseinheitlicher Onkologischer Basisdatensatz [German Uniform Basic Oncology Data Set] (oBDS) of the Arbeitsgemeinschaft Deutscher Tumorzentren [Association of German Tumor Centers] (ADT) and the Gesellschaft der epidemiologischen Krebsregister in Deutschland [Association of Population-based Cancer Registries in Germany] (GEKID) and the MII CDS, the lung cancer data set of the Nationales Netzwerk Genomische Medizin Lungenkrebs [German National Network Genomic Medicine Lung Cancer] (nNGM) [15].. However, GOLD currently has a draft status and is still undergoing changes.
The MII CDS consists of basic modules and specialized extension modules [8]. In addition to the basic modules person, case, diagnosis, procedure, laboratory test results, and medication [34] that have an active status, the specialized extension module oncology is currently being drafted [22].
The HL7 Deutschland [Germany] Basisprofil Onkologie [Basic Profile Oncology] was developed from 2020 to 2022 and in cooperation with Vision Zero GOLD [22]. Although the Basisprofil Onkologie [Basic Profile Oncology] was also still under development, it has a released status and will serve as the basis for the MII module oncology [22].
Since the profiles, value sets, codes systems, and extensions of Vision Zero GOLD and the MII CDS module oncology are drafts so far, we used the HL7 Basisprofil Onkologie [Basic Profile Oncology] as a reference FHIR profile for oncology.
2.5. Data Model Comparison and Metrics
To verify which features of our data model can be mapped to the Basisprofil Onkologie [Basic Profile Oncology], we compared both data structures in tabular form, resulting in Table 2 and Table S1. Code or semantic mappings to FHIR specifications/profiles shown in tabular form have also been presented by other studies [11,18].
We listed all features on the left-hand side of the table. In the middle part of the table, we showed which FHIR resource and FHIR element(s) represent a feature in our real-world data set. If a feature can also be mapped with the Basisprofil Onkologie [Basic Profile Oncology], we showed its representation by this profile on the right-hand side of the table, using the FHIR resource(s) and element(s) from the profile(s)/extension(s) needed. In addition, we also provided the FHIR data types, terminologies, and specific values (if used for data querying) on the middle and right-hand side of the table. Finally, for better interpretation of our mapping results, we categorized our features using feature types and compared the feature representations using mapping levels. We also considered that features that cannot be explicitly mapped by the reference FHIR profile may be mapped with the standard FHIR specification, or they may be new features.
Mapping Levels: In contrast to Peng et al., we did not only differentiate between mapped and non-mappable/missing elements in the mapping results [18] but we defined five different mapping levels: complete mapping without adjustments (mapping level Y for mapping to reference oncological FHIR profile; R4 for mapping to standard FHIR specification R4), mapping with minor adjustments (mapping level MI), mapping with major adjustments (mapping level MA), or no mapping possible (mapping level N) (see Table 3 and Table S1). We distinguished possible mappings with minor adjustments (MI) from mappings with major adjustments (MA) to highlight differences regarding the required implementation effort. Mapping with minor adjustments (MI) was used if the element(s) used was/were the same and used directly related terminology (e.g., a complete value set and subset). Mapping with major adjustments (MA) was used if the element(s) used differed and/or used a different terminology. Since, in some cases, several elements may be relevant for the mapping of one feature, the lowest applicable mapping level was selected.
Feature Types: For the further evaluation of the technical implementation of the mapping and the clinical content coverage, we determined three types of features in our data set: (i) administrative (A), (ii) clinical (C), and (iii) reference (R).
Mapping Metrics: We determined both the absolute and relative (percentage) values of the features regarding the mapping levels and feature types within the classifications and globally.
3. Results
We created a clinical data model for melanoma containing different resource classes as well as logical areas of the real-world data. The model differentiates between types of features according to the kind of information they represent and therefore their relevance to prediction models. Afterwards, we compared its features with counterparts of the Basisprofil Onkologie [Basic Profile Oncology] and the standard FHIR specification R4. The mappability of our features for the specific tumor towards the oncological FHIR profile was evaluated by determining mappability metrics. Finally, missing mandatory elements required to fully implement the referent specification were addressed.
Below, the results of the data modeling (see Section 3.1) and the data model comparison are reported in detail, including the applicability of the Basisprofil Onkologie [Basic Profile Oncology] to our data model (see Section 3.2), the resulting mappability metrics (see Section 3.3), new data features detected in our data subset (see Section 3.4), and missing mandatory elements for implementing the Basisprofil Onkologie [Basic Profile Oncology] (see Section 3.5).
3.1. Real-World Data Model
We created a data model for the prediction of irAEs of first-line immunotherapy for patients with melanoma using UML, as depicted in Figure 2. Each resource class of our model is displayed as one UML class. An exception is within the logical area of Risk factors, where several classes were combined for better readability. The FHIR resource type, the class descriptor, and the specialized profiles implemented are displayed within the UML class name. Within the classes, relevant features are listed as FHIR elements using UML attributes with bold descriptors, FHIR data types, and value sets. Instead of the initial attribute values usually specified in UML class diagrams, logical filter values that were used as search parameters for querying the data are displayed. FHIR references are visualized as direct associations between FHIR resource classes or logical areas. Multiplicities describe the relationship of the connected classes or areas in the direction of the arrows. To reduce the large number of references to the central areas of Melanoma patient and Case, only logical areas were linked to these instead of the individual resource classes.
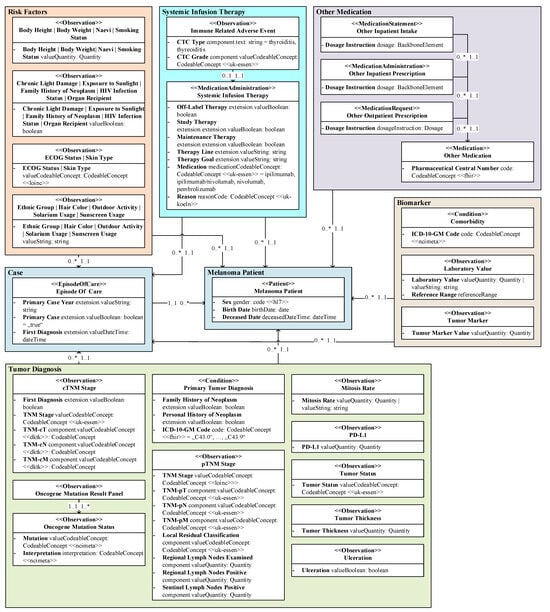
Figure 2.
Simplified HL7 FHIR data model of real-world clinical EHR melanoma patient data as UML class diagram, whereby elements represent prediction-relevant elements and value expressions represent logical data filtering.
Features: Our real-world data model consists of 37 different resource classes with a total of 216 different features. A total of 84 (38.89%) of these 216 features are administrative data (A), 62 (28.70%) are clinical data (C), and 70 (32.41%) are references (R) that are required for a logical linking between the FHIR resources (see Table 3 and Table S1). Administrative features are verification and general status information, date and time information, as well as identifier codes (for example, 21924-6: Tumor marker Cancer) and value codes (for example, C0022917: Lactate Dehydrogenase) of the clinical findings. The clinical features describe the actual clinical findings (for example, 247 U/l) and can be used for medical outcome predictions, as an example.
FHIR Specifications and Profiles: In addition to the standard FHIR specification R4, we identified different specialized FHIR profiles implemented by our real-world data set (see Table S1). The FHIR profiles from the MII core modules were used for the laboratory values (Observation resource), the other inpatient intakes and outpatient prescriptions (MedicationStatement and MedicationAdministration resources), the other medication (Medication resource), and the melanoma patients (Patient resource). In addition, the Patient resource implements the ISiK profile from the Nationale Agentur für Digitale Medizin [National Agency for Digital Medicine] (gematik). Profiles from the nNGM were used for the body height (Observation resource), the body weight (Observation resource), and the medical case (EpisodeOfCare resource).
Terminologies: We identified terminology bindings used by our data set (see Table S1). In addition to system-internal and in-house terminologies of Cerner Medico and Nexus Swisslab, the terminologies Logical Observation Identifiers Names and Codes (LOINC, http://loinc.org (accessed on 6 May 2024)), SNOMED Clinical Terms (SNOMED CT, https://www.snomed.org/value-of-snomedct (accessed on 6 May 2024)), National Cancer Institute meta thesaurus (NCIm, http://ncimeta.nci.nih.gov (accessed on 6 May 2024)), as well as value sets from HL7 and terminologies of the German University Hospital Cologne, such as nNGM, are used.
3.2. Applicability of Standardized Data Model
Figure 3 demonstrates the simplified example mapping concept of the primary tumor diagnosis of a fictitious patient to the oncological FHIR profile.
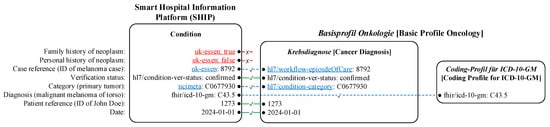
Figure 3.
A sketched mapping concept of the clinical data features (left) of the primary tumor diagnosis from SHIP (middle) to the Basisprofil Onkologie [Basic Profile Oncology] [22] (right) of a fictitious patient. The differences are underlined. The lines and font colors indicate the mapping types: no mapping (red font color, solid red line with a cross symbol), complete mapping (solid green line with a check mark symbol), and mapping with adjustments (blue font color, dashed blue line with a check mark symbol).
Based on our data model, we mapped all features in tabular form. Figure 4 shows the (highly detailed) data model section of the resource class Primary tumor diagnosis as an example, which resulted in the features on the left-hand side in Table 2 and their FHIR representation in the middle of Table 2. The feature mapping of the Basisprofil Onkologie [Basic Profile Oncology] is shown on the right-hand side in Table 2. The mapping type information and, if applicable, a marker indicating that it is a new feature (*), resulted from the comparison of both FHIR representations.
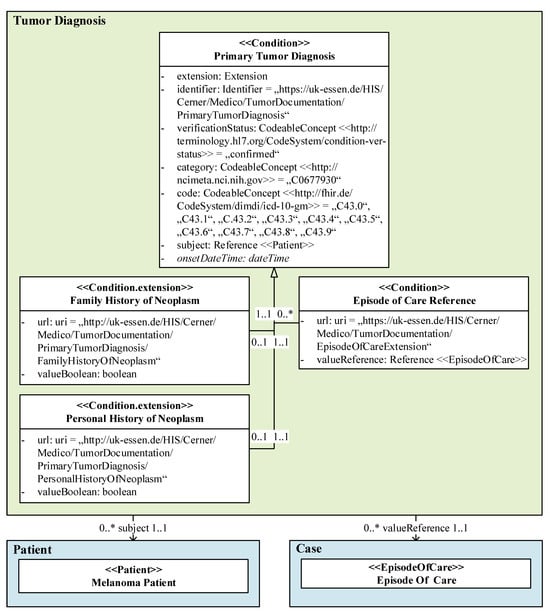
Figure 4.
A detailed data model excerpt of the primary tumor diagnosis as a UML class diagram, whereby the elements are prediction-relevant, and the value expressions represent query parameters.
The clinical features (feature type C) Family history of neoplasm and Personal history of neoplasm of our resource class Primary tumor diagnosis cannot be mapped by profiles or extensions of the Basisprofil Onkologie [Basic Profile Oncology] nor the standard FHIR specification; therefore, these features are classified as mapping level N (not possible). Since the representations of the clinical feature ICD-10-GM code differ regarding the required FHIR elements, mapping is only possible with major adjustments (MA), although the terminologies used are the same.
While the administrative features Verification and Date can be completely mapped with the reference FHIR profile (Y), different terminologies are used for the feature Category primary neoplasm, which requires major adjustments (MA).
The representation of the Patient reference of our data model and the reference FHIR profile match completely (Y). Finally, major adjustments (MA) are required to map the Episode of care reference feature due to different Uniform Resource Locators (URLs).

Table 2.
Mapping of primary tumor diagnosis features (left) of real-world data (middle) to Basisprofil Onkologie [Basic Profile Oncology] [22] (right) with simplified Uniform Resource Locators (URLs).
Table 2.
Mapping of primary tumor diagnosis features (left) of real-world data (middle) to Basisprofil Onkologie [Basic Profile Oncology] [22] (right) with simplified Uniform Resource Locators (URLs).
| Feature | Real-World Data | HL7 Basisprofil Onkologie [Basic Profile Oncology] | |||
|---|---|---|---|---|---|
| (Type, Mapping Level) | Resource.Element | Type = Value | Profile or Extension | Resource.Element | Type = Value |
| Family history of neoplasm (C, N) * | Condition | ||||
| .extension | Extension | ||||
| .extension.url | uri = uk-essen | ||||
| .extension. valueBoolean | boolean | ||||
| Personal history of neoplasm (C, N) * | Condition | ||||
| .extension | Extension | ||||
| .extension.url | uri = uk-essen | ||||
| .extension. valueBoolean | boolean | ||||
| Episode of care reference (R, MA) | Condition | Condition | |||
| .extension | Extension | Krebsdiagnose [Cancer Diagnosis] | .extension:Fall | Extension | |
| Extension | |||||
| .extension.url | uri = uk-essen | Krebsdiagnose [Cancer Diagnosis] | .url | uri = hl7/workflow- episodeOfCare | |
| .extension. valueReference | Reference (EpisodeOfCare) | Episode of Care | .valueReference | Reference (EpisodeOfCare) | |
| Verification (A, Y) | Condition | Condition | |||
| .verification Status | CodeableConcept (hl7/conditionver- status) = “confirmed” | Krebsdiagnose [Cancer Diagnosis] | .verification Status | CodeableConcept (hl7/conditionver- status) = “confirmed” | |
| Category of primary neoplasm (A, MA) | Condition | Condition | |||
| .category | CodeableConcept (ncimeta) = “C0677930” | Krebsdiagnose [Cancer Diagnosis] | .category | CodeableConcept(hl7/condition-category) | |
| ICD-10- GM code (C, MA) | Condition | Condition | |||
| .code | CodeableConcept (fhir/icd-10-gm) = C43.0–C43.9 | Krebsdiagnose [Cancer Diagnosis] | .code.coding | fhir/CodingICD10 GM | |
| Data Type | |||||
| Coding-Profil für ICD-10-GM [Coding Profile for ICD-10-GM] | .system | uri = fhir/icd-10-gm | |||
| Coding-Profil für ICD-10-GM [Coding Profile for ICD-10-GM] | .code | code | |||
| Patient reference (R, Y) | Condition | Condition | |||
| .subject | Reference(Patient) | Krebsdiagnose [Cancer Diagnosis] | .subject | Reference(Patient) | |
| Date (A, Y) | Condition | Condition | |||
| .onsetDateTime | dateTime | Krebsdiagnose [Cancer Diagnosis] | .onsetDateTime | dateTime | |
Feature categories: A = administrative, C = clinical, R = reference; mapping levels: Y = completely possible to reference FHIR profile, MA = with major adjustments to reference FHIR profile, N = not possible to reference FHIR profile/standard FHIR specification R4; * new feature.
3.3. Mappability Metrics
Table 3 and Figure 5a show the statistical evaluation of our mapping tables (see Table S1). Out of 37 resource classes, 7 resource classes (18.92%) could be mapped (partially) to the Basisprofil Onkologie [Basic Profile Oncology], and 30 (81.08%) could not be mapped to the Basisprofil Onkologie [Basic Profile Oncology] but they could be mapped to the standard FHIR specification R4. A closer look at the clinical features shows that the reference oncological FHIR profile is able to map content in the logical areas Risk factors (1/16, 6.25%), Systemic infusion therapy (5/9, 55.56%), and Tumor diagnosis (13/23, 56.52%) (see Figure 5b). All clinical features of the logical areas Biomarker (4), Case (3), Melanoma patient (3), and Other medication (4) remained completely uncovered.
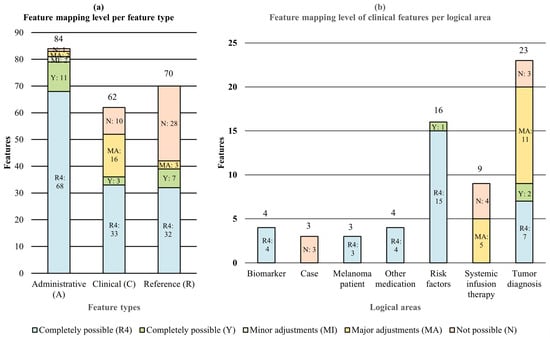
Figure 5.
Mapping level, total of all mapping types (a) per feature types and (b) of clinical features per logical area.
At the feature level, out of 216 features, 21 (9.72%) features could be mapped to the Basisprofil Onkologie [Basic Profile Oncology] without adjustments, 2 (0.93%) could be mapped with minor adjustments, 21 (9.72%) could be mapped with major adjustments, 39 (18.06%) could not be mapped, and 133 (61.75%) could instead be mapped with the standard FHIR specification. With or without adjustments, a total of 177 out of 216 (81.94%) features could be mapped with a combination of the reference FHIR profile and the standard FHIR specification. As a result, a total of 39 (18.06%) features cannot currently be represented in their given forms without using self-defined extensions, which means that the interoperability of this information is limited. Figure 5a cumulatively illustrates, both in proportional and absolute for each feature type, how many features must be adjusted to map our data set with a combination of the Basisprofil Onkologie [Basic Profile Oncology] and the standard FHIR specification.
For the administrative features, a total of 11 (13.10%) features can be mapped completely, 2 (2.38%) can be mapped with (minor) adjustments, 2 (2.38%) can be mapped with major adjustments, 1 (1.19%) cannot be mapped, and the majority with 68 features (80.95%) does not need any mapping, as there is no specific profile or extension in the reference oncological FHIR profile and a mapping to the standard FHIR specification is completely given. Of the clinical features, 3 (4.76%) can be mapped completely, 16 (25.40%) can be mapped with major adjustments, 10 (15.87%) cannot be mapped, and 33 (52.38%) were not mapped to the reference oncological FHIR profile but to the standard FHIR specification. A total of 7 (10.00%) reference features can be mapped completely, 3 (4.29%) can be mapped with major adjustments, and 28 (40.00%) cannot be mapped by the Basisprofil Onkologie [Basic Profile Oncology], whereby an additional 32 (40.00%) features were instead mapped to the standard FHIR specification.

Table 3.
Mapping level of real-world data features to Basisprofil Onkologie [Basic Profile Oncology] FHIR profile [22] or, if not possible, to standard FHIR specification R4 for different feature categories (relative and absolute values with percentage values).
Table 3.
Mapping level of real-world data features to Basisprofil Onkologie [Basic Profile Oncology] FHIR profile [22] or, if not possible, to standard FHIR specification R4 for different feature categories (relative and absolute values with percentage values).
| Feature Type | Mapping Level | Total (n = 212) | ||||
|---|---|---|---|---|---|---|
| To Basisprofil Onkologie [Basic Profile Oncology] | To Standard R4 | To Both | ||||
| Completely Possible (Y) | Minor Adjustments (MI) | Major Adjustments (MA) | Completely Possible (R4) | Not Possible (N) | ||
| Administrative (A) | 11/13.10% | 2/2.38% | 2/2.38% | 68/80.95% | 1/1.19% | 84/38.89% |
| Clinical (C) | 3/4.76% | 0/0.00% | 16/25.40% | 33/52.38% | 10/15.87% | 63/28.70% |
| Reference (R) | 7/10.00% | 0/0.00% | 3/4.29% | 32/45.71% | 28/40.00% | 70/32.41% |
| Total (n = 212) | 21/9.72% | 2/0.93% | 21/9.72% | 129/61.57% | 39/18.06% | 216/100.00% |
3.4. New Features
As shown in Table 3, Figure 5a, and Table S1, the non-mappable features of our data set (39/18.06%), from both the oncological FHIR profile and the standard FHIR specification R4, can be divided into 1 (0.46%) administrative feature, 10 (4.63%) clinical features, and 28 (12.96%) reference features.
Administrative Feature: The non-mappable, administrative feature is the status of an irAE (Observation.status), which is not provided when using the Nebenwirkung [Side Effect] profile of the Basisprofil Onkologie [Basic Profile Oncology]. The Nebenwirkung [Side Effect] profile is based on the FHIR R4 resource AdverseEvent, which does not provide a status, whereas the irAEs in our real-world data set are based on resource Observation, which offers a status.
Clinical Features: The ten non-mappable clinical features can be assigned to the cTNM stage (n = 1), case (n = 3), primary tumor diagnosis (n = 2), and systemic infusion therapy (n = 4) of our data model. These features are represented by FHIR extensions. In our data set, the cTNM stage can include additional information as to whether it is a first diagnosis (Observation.extension.valueBoolean). The case can also redundantly include the same first diagnosis information (EpisodeOfCare.extension.valueBoolean). In addition, the case of our data model can contain the primary case year (EpisodeOfCare.extension.valueString) and reveal whether it is the primary case (EpisodeOfCare.extension.valueBoolean). The primary tumor diagnosis of our data model may also include a structured indication of whether there is a family and/or personal history of neoplasm (both Condition.extension.valueBoolean). The systemic infusion therapy of our data model can also contain information on whether it is an off-label therapy, study therapy, or maintenance therapy (all MedicationAdministration.extension.valueBoolean) and which therapy line (MedicationAdministration.extension.valueString) the treatment is.
Reference Features: Although there are 28 new reference features detected, all of them are an additional reference added to the case (Condition|MedicationAdministration|Observation.extension.valueReference(EpisodeOfCare)). Thus, the two biomarkers comorbidity and tumor marker, all risk factors, all tumor diagnosis classes, and the immune-related adverse event can be expanded to include this reference.
3.5. Mandatory Elements
To implement the oncological FHIR profile correctly, all mandatory elements required by it must be present. The FHIR element EpisodeOfCare.type would have to be added to the real-world data of our data model to implement the Tumorerkrankung [Tumor Disease] profile, the Condition.clinicalStatus element for the Krebsdiagnose [Cancer Diagnosis] profile, the AdverseEvent.actuality and AdverseEvent.category elements for the Nebenwirkung [Side Effect] profile, and the Observation.category:survey element for the Observation-Profil ECOG Performance Status [Observation Profile ECOG Performance Status] profile.
4. Discussion
Although the national oncological FHIR profile also maps (tumor)-specific content, we have shown that this FHIR profile is not sufficient for mapping our specific melanoma use case and is therefore also expected to not be sufficient for prediction models. The developed clinical data model was essential to show these differences in detail and can also be used to provide a basis for mapping real-world data. During feature mapping, it was also shown that minor adjustments were required much less frequently than major adjustments due to terminology inconsistencies.
Below, we discuss the findings and provide interpretations of our results in detail (see Section 4.1, Section 4.2, Section 4.3 and Section 4.4) and address the strengths (see Section 4.5), limitations (see Section 4.6), as well as implications and future work (see Section 4.7).
4.1. Data Model Creation
We determined that the initial creation of a data model is essential to determine which characteristics can be mapped with which FHIR specifications/profiles and which require separate treatments so that they do not become lost in the subsequent processing steps. This finding fits with the finding of Vision Zero GOLD that, in addition to a parameter list, a smart data model is also needed to represent different aspects of the data, e.g., multiplicities [15]. Like related work using object-oriented representations with classes/tables to visualize their data models [12,13,14,19,20,21], we used a UML class diagram. We also showed that it is beneficial to already consider the exact data structure, data types, terminologies, and values for the subsequent mapping process.
4.2. Granularity of Mapping Level
In contrast to the study by Peng et al. [18], we decided to divide the mapping level with adjustments into the categories of minor adjustments and major adjustments, as we assumed that minor adjustments would be needed more frequently due to the use of the SNOMED CT and LOINC terminologies in our real-world data set. Contrary to this expectation, our results show that only 2 out of 23 (8.70%) mapping cases required minor adjustments rather than major adjustments. An explanation could be that although the same terminology or sub-value sets were used in some cases, the structural design or the path within the resource often differed, which means that more complex mapping of the feature is to be expected. In addition, the results confirm Vision Zero’s observation that there is often a lack of terminological consistency when merging existing oncological data sets [15].
4.3. The Effects of the Further Development of Fast Healthcare Interoperability Resources (FHIR)
Since FHIR was introduced in 2011 [1], the standard has continued to evolve. While it consisted of 49 resources at the time the first official version was released (DSTU 1 (Official version), 24 October 2015), the latest official version (FHIR R4, 30 October 2019) consists of 146 resources [35]. For instance, the Observation resource initially had 16 elements [36], and it now consists of 24 elements [37] and was updated nine times within about four years. It can be assumed that both the standard FHIR specification and the oncological FHIR profiles that have already been and will be developed in the future will continue to evolve. Continuous updates of the internal data management models within healthcare institutions will therefore be necessary in the future to maintain a high level of interoperability.
4.4. Applicability of Oncology Standard for Tumor-Specific Melanoma
Our mapping results show that nearly one-third of the clinical features can be mapped (completely or with minor/major adjustments) with the oncological reference FHIR profile. The more generic logical areas of our model, namely Biomarker, Case, Melanoma patient, and Other Medication, and most of the rather generic structured Risk factors cannot be mapped with the oncological reference FHIR profile, but they can be mapped with the standard FHIR specification. As it is not the aim of FHIR profiles to replace all standard resources, we expected that these more generic data would be covered by the standard FHIR specification instead of the oncological FHIR profile. Also, as expected, of the tumor-specific logical areas Systemic infusion therapy and Tumor diagnosis, a little more than half of the clinical features could be mapped. Applying the general 80/20 approach of FHIR [3] to the FHIR profile coverage of (especially clinical features of) dermato-oncology using general oncology shows that the oncological resources/extensions would not be defined with sufficient specificity for our case, although it can be assumed that these clinical features would also be useful for other oncological diseases. Since Vision Zero GOLD has already stated that there is no comprehensive oncology data set [15], we were not surprised that no complete feature coverage was found in our specific area of dermato-oncology as well. Nevertheless, it was shown that the oncological FHIR profile not only represents general oncological information but also provides more (tumor-)specific FHIR profiles and extensions, for example, for the TNM classification used for malignant melanoma.
It met our expectation that very few administrative features would require customization or would not be mappable, as these are mostly not tumor-specific. We expected the same for the reference features, but there is a larger number of non-mappable features, as this relates to a structural specialty of our real-world data set, where most resources are additionally linked to the case. Regarding the clinical features, our results show that 10 out of 62 (16.13%) features cannot be mapped and are therefore potentially new and not (yet) considered in the reference FHIR profile nor the standard FHIR specification R4. Since most of these features are an explicit attribute, e.g., whether there is a family history of neoplasm (Boolean: true/false) or a personal history of neoplasm (Boolean: true/false), it must be discussed whether the advantages of this explicit information outweigh the disadvantages. In an optimal world, all EHR data (from both the patient and their relatives) would be FHIR-based and available in the treating healthcare institution, so this information could be extracted simply by querying whether a related person or the patient themself has already received a neoplasm diagnosis. This querying would make the additional and explicit provision of the same information redundant. However, in the real world, only part of the EHR data (e.g., the past two years of a patient’s EHR, and no EHR data from relatives) are available in a healthcare institution, which means that such a query would rely on incomplete data. Therefore, this explicit information can provide important additional value in the real world.
4.5. Strengths
One of the strengths of our work is that we used a real-world, rich clinical data set from a large university hospital data platform to implement our methodology pipeline. This data platform has been used and further developed for many years for both patient care and research. The mapping of our data set is transparent and granular at the element level (see Table S1) and therefore fully reproducible. In addition, we not only analyze the mappability of our numerous data features according to the plain measure of mappability, but also differentiate between feature types and logical data model areas, as well as different mapping levels with both the reference FHIR profile and the standard FHIR specification.
4.6. Limitations
Although we used a specific data example from only one institution within a single-center study, and one reference FHIR profile to answer our research question, the methodological pipeline of our study remains valid. However, the specific metrics of our use case cannot be generalized, so a general statement about the necessity of FHIR profiles for various oncological diseases cannot be made. As González-Castro et al. pointed out, the flexibility of FHIR also comes with challenges, as there are cases where an aspect can be mapped in different ways using FHIR [12]. To some extent, this flexibility was also observed in the data set and profiles we used, which is why the mapping we present is limited to one way of potentially different possibilities.
Even though both our real-world data and our data model, as well as the Basisprofil Onkologie [Basic Profile Oncology], are based on the standard FHIR specification R4, the newer FHIR specification R5 already exists. There have been changes from R4 to R5 where, for example, the FHIR resource AdverseEvent now also has a status and would therefore no longer be considered as a new (administrative) feature.
4.7. Implications and Future Work
Our results should be considered when estimating the effort required to (automatically) implement new or updated FHIR profiles for real-world data on the one hand, and for the (further) development of (dermato-)oncological profiles/extensions on the other. For the continuous process of data mapping in a hospital, this means that not only must updates to the standard resources be implemented, but also oncological FHIR profiles and their updates should be implemented for a high degree of interoperability. At the moment, both should be expanded to prevent the loss of specific data (e.g., labeling of a first diagnosis and first case; the year of the first case; labeling of family and personal history of neoplasm; labeling of off-label, study, and maintenance therapy; and therapy line), as these are essential for a correct and comprehensive melanoma- and patient-specific data model and research based on it, for example, in the fields of Artificial Intelligence and prediction. Future research should further consider more recent oncological reference FHIR profiles, like GOLD, and the MII basic and oncology modules once they have been released. It should also be examined whether explicit information, which must be assumed to be incomplete and/or in an unsuitable format in the real world, would offer added value in a majority of the use cases and should therefore be taken into account in the (further) development of (new) oncological FHIR profiles. Since we conducted a single-center study, an extension to a multicenter study is a reasonable next step to compare the results of multiple institutions.
5. Conclusions
To ensure interoperability, an oncological FHIR profile can be used to a certain extent for a tumor-specific use case if it is used in combination with the standard FHIR specification. However, our study of one selected use case showed that some features, including clinical ones, are either lost or must be stored and exchanged in self-defined extensions when implementing such a combination, resulting in limited interoperability. As it can be assumed that the existing standard FHIR specifications and existing oncological FHIR profiles will continue to evolve, new oncological FHIR profiles will continue to be developed. At the same time, the internal system infrastructures and thus the internal data models of healthcare institutions will continue to evolve. Healthcare institutions will therefore need to make a certain effort and stay up to date to ensure a high level of interoperability.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/informatics11030042/s1, Table S1: Real-world melanoma data classes that are (partially) mapped to the HL7 Basisprofil Onkologie [Basic Profile Oncology] and data classes that are otherwise representable by the standard FHIR specification R4.
Author Contributions
Conceptualization, J.S. and B.B.; methodology, J.S. and B.B.; software, J.S.; validation, J.S.; formal analysis, J.S.; investigation, J.S.; resources, J.S., D.S., E.L., F.N., G.C.L., M.A. and C.L.B.; data curation, J.S., G.C.L. and M.A.; writing—original draft preparation, J.S.; writing—review and editing, B.B., E.L., F.N., G.C.L. and C.L.B.; visualization, J.S.; supervision, B.B.; project administration, J.S.; funding acquisition, B.B., E.L., F.N., and D.S. All authors have read and agreed to the published version of the manuscript.
Funding
This work was funded by a Ph. D. grant from the DFG Research Training Group 2535, Knowledge- and data-based personalization of medicine at the point of care (WisPerMed), University of Duisburg-Essen, Germany. We additionally acknowledge the support of the Open Access Publication Fund of the University of Duisburg-Essen, Germany.
Institutional Review Board Statement
This study was approved by the Ethics Committee of the Medical Faculty of the University of Duisburg-Essen, Germany (approval number 21-10393-BO, 3 August 2022).
Informed Consent Statement
Not applicable.
Data Availability Statement
Restrictions apply to the availability of the patient data, which were used within a research agreement for the current work and are therefore not publicly available.
Acknowledgments
We thank the Smart Hospital Information Platform (SHIP) team of the Institute for Artificial Intelligence in Medicine (IKIM), especially Katarzyna Borys (University Hospital Essen), for providing the EHR data and technical support in accessing them. We also thank the Department of Dermatology, Venereology and Allergology (University Hospital Essen) for their cooperation and for providing the detailed EHR data. We thank Wolfgang Galetzka (University Hospital Essen) and Sylvia Nürnberg (University Hospital Essen) for providing very helpful feedback on this study.
Conflicts of Interest
The authors declare no conflicts of interest.
Abbreviations
The following abbreviations are used in this manuscript:
| ADT | Arbeitsgemeinschaft Deutscher Tumorzentren [Association of German Tumor Centers] |
| CDM | Common Data Model |
| CDS | Core Data Set |
| cTNM | Clinical TNM |
| DiGA | Digitale Gesundheitsanwendungen [Digital Health Applications] |
| EHR | Electronic Health Record |
| ePA | elektronische Patientenakte [Personal Health Record] |
| FHIR | Fast Healthcare Interoperability Resources |
| GEKID | Gesellschaft der epidemiologischen Krebsregister in Deutschland [Association of Population-Based Cancer Registries in Germany] |
| gematik | Nationale Agentur für Digitale Medizin [National Agency for Digital Medicine] |
| GOLD | German OncoLogical Data Standard |
| HL7 | Health Level 7 |
| irAE | Immune-Related Adverse Event |
| ISiK | Informationstechnische Systeme in Krankenhäusern [Information Technology in Hospital] |
| LOINC | Logical Observation Identifiers Names and Codes |
| MII | Medizininformatik Initiative [Medical Informatics Initiative] |
| MIO | Medizinische Informationsobjekte [Medical Information Object] |
| NCIm | National Cancer Institute meta thesaurus |
| nNGM | Nationales Netzwerk Genomische Medizin Lungenkrebs [German National Network Genomic Medicine Lung Cancer] |
| oBDS | Bundeseinheitlicher Onkologischer Basisdatensatz [German Uniform Basic Oncology Data Set] |
| OMOP | Observational Medical Outcomes Partnership |
| pTNM | Pathological TNM |
| SHIP | Smart Hospital Information Platform |
| SNOMED CT | SNOMED Clinical Terms |
| UML | Unified Modeling Language |
| URL | Uniform Resource Locator |
References
- Vorisek, C.N.; Lehne, M.; Klopfenstein, S.A.I.; Mayer, P.J.; Bartschke, A.; Haese, T.; Thun, S. Fast Healthcare Interoperability Resources (FHIR) for Interoperability in Health Research: Systematic Review. JMIR Med. Inform. 2022, 10, e35724. [Google Scholar] [CrossRef] [PubMed]
- Bosca, D.; Moner, D.; Maldonado, J.A.; Robles, M. Combining Archetypes with Fast Health Interoperability Resources in Future-proof Health Information Systems. In Digital Healthcare Empowering Europeans; IOS Press: Amsterdam, Netherlands, 2015; Volume 210, pp. 180–184. [Google Scholar]
- FHIR Overview. Architects. Available online: https://hl7.org/fhir/R4/overview-arch.html (accessed on 6 May 2024).
- Medizinische Informationsobjekte. Available online: https://mio.kbv.de/site/mio#tab-Rund+um+die+MIOs (accessed on 6 May 2024).
- Weber, S.; Heitmann, K.U. Interoperabilität im Gesundheitswesen: Auch für digitale Gesundheitsanwendungen (DiGA) verordnet. Bundesgesundheitsbl 2021, 64, 1262–1268. [Google Scholar] [CrossRef] [PubMed]
- Interoperabilität dank ISiK. Available online: https://fachportal.gematik.de/informationen-fuer/isik (accessed on 6 May 2024).
- Ciortuz, G.; Wiedekopf, J.; Fudickar, S. Integration von Wearables und Nutzung von digitalen Biomarkern zur Diagnostik und Therapie im Gesundheitswesen. In Health Data Management; Henke, V., Hülsken, G., Schneider, H., Varghese, J., Eds.; Springer: Wiesbaden, Germany, 2024; pp. 323–336. ISBN 978-3-658-43235-5. [Google Scholar]
- Semler, S.C.; Wissing, F.; Heyder, R. German Medical Informatics Initiative. Methods Inf. Med. 2018, 57, e50–e56. [Google Scholar] [CrossRef] [PubMed]
- Smith, A.D.; Roda, D.; Yap, T.A. Strategies for modern biomarker and drug development in oncology. J. Hematol. Oncol. 2014, 7, 70. [Google Scholar] [CrossRef] [PubMed]
- Pflugfelder, A.; Kochs, C.; Blum, A.; Capellaro, M.; Czeschik, C.; Dettenborn, T.; Dill, D.; Dippel, E.; Eigentler, T.; Feyer, P.; et al. Malignant melanoma S3-guideline “diagnosis, therapy and follow-up of melanoma”. J. Dtsch. Dermatol. Ges. 2013, 11 (Suppl. S6), 1–116. [Google Scholar] [CrossRef] [PubMed]
- Deppenwiese, N.; Delpy, P.; Lambarki, M.; Lablans, M. ADT2FHIR—A Tool for Converting ADT/GEKID Oncology Data to HL7 FHIR Resources. In German Medical Data Sciences 2021: Digital Medicine: Recognize–Understand–Heal; IOS Press: Amsterdam, The Netherlands, 2021; Volume 283, pp. 104–110. [Google Scholar] [CrossRef]
- González-Castro, L.; Cal-González, V.M.; Del Fiol, G.; López-Nores, M. CASIDE: A data model for interoperable cancer survivorship information based on FHIR. J. Biomed. Inform. 2021, 124, 103953. [Google Scholar] [CrossRef] [PubMed]
- Oeppert, L.; Hartz, T.; Wehner, K.; Schrader, T.; Meier, J. FHIR-Datenmodell zur Übermittlung von Tumordaten zwischen Krebsregistern und IQTIG. GMS Med. Inform. Biom. Und Epidemiol. 2021, 17, Doc17. [Google Scholar] [CrossRef]
- Lambarki, M.; Kern, J.; Croft, D.; Engels, C.; Deppenwiese, N.; Kerscher, A.; Kiel, A.; Palm, S.; Lablans, M. Oncology on FHIR: A Data Model for Distributed Cancer Research. In German Medical Data Sciences: Bringing Data to Life; IOS Press: Amsterdam, The Netherlands, 2021; Volume 278, pp. 203–210. [Google Scholar] [CrossRef]
- Digitalisierung: Mit Einem Konsentierten Datenformat gegen den Krebs. Available online: https://vision-zero-oncology.de/projekte-arbeitsgruppe-digitalisierung.php (accessed on 6 May 2024).
- Project NCCN Chemotherapy Order Templates. Available online: https://simplifier.net/NCCNChemotherapyOrde (accessed on 6 May 2024).
- Peng, Y.; Henke, E.; Reinecke, I.; Zoch, M.; Sedlmayr, M.; Bathelt, F. An ETL-process design for data harmonization to participate in international research with German real-world data based on FHIR and OMOP CDM. Int. J. Med. Inform. 2023, 169, 104925. [Google Scholar] [CrossRef]
- Peng, Y.; Nassirian, A.; Ahmadi, N.; Sedlmayr, M.; Bathelt, F. Towards the Representation of Genomic Data in HL7 FHIR and OMOP CDM. In German Medical Data Sciences 2021: Digital Medicine: Recognize—Understand—Heal; IOS Press: Amsterdam, The Netherlands, 2021; Volume 283, pp. 86–94. [Google Scholar] [CrossRef]
- Stellmach, C.; Sass, J.; Auber, B.; Boeker, M.; Wienker, T.; Heidel, A.J.; Benary, M.; Schumacher, S.; Ossowski, S.; Klauschen, F.; et al. Creation of a structured molecular genomics report for Germany as a local adaption of HL7’s Genomic Reporting Implementation Guide. J. Am. Med. Inform. Assoc. 2023, 30, 1179–1189. [Google Scholar] [CrossRef] [PubMed]
- Holweg, F.; Achenbach, S.; Deppenwiese, N.; Gaede, L.; Prokosch, H.-U. Towards a FHIR-Based Data Model for Coronary Angiography Observations. In Healthcare of the Future 2022; IOS Press: Amsterdam, The Netherlands, 2022; Volume 292, pp. 96–99. [Google Scholar] [CrossRef]
- Gundler, C.; Zhu, Q.R.; Trübe, L.; Dadkhah, A.; Gutowski, T.; Rosch, M.; Langebrake, C.; Nürnberg, S.; Baehr, M.; Ückert, F. A Unified Data Architecture for Assessing Motor Symptoms in Parkinson’s Disease. In German Medical Data Sciences 2023—Science. Close to People; IOS Press: Amsterdam, The Netherlands, 2023; Volume 307, pp. 22–30. [Google Scholar] [CrossRef]
- Project of HL7 Deutschland e.V.: Basisprofil Onkologie. Available online: https://simplifier.net/BasisprofileOnkologie (accessed on 6 May 2024).
- Project GOLD—German OncoLogical Data Standard. Available online: https://simplifier.net/GOLD---German-OncoLogical-Data-Standard (accessed on 6 May 2024).
- Project of Medizininformatik Initiative: Medizininformatik Initiative—Modul Onkologie. Available online: https://simplifier.net/MedizininformatikInitiative-ModulOnkologie (accessed on 6 May 2024).
- Whiteman, D.C.; Green, A.C.; Olsen, C.M. The Growing Burden of Invasive Melanoma: Projections of Incidence Rates and Numbers of New Cases in Six Susceptible Populations through 2031. J. Investig. Dermatol. 2016, 136, 1161–1171. [Google Scholar] [CrossRef] [PubMed]
- Zimmer, L.; Goldinger, S.M.; Hofmann, L.; Loquai, C.; Ugurel, S.; Thomas, I.; Schmidgen, M.I.; Gutzmer, R.; Utikal, J.S.; Göppner, D.; et al. Neurological, respiratory, musculoskeletal, cardiac and ocular side-effects of anti-PD-1 therapy. Eur. J. Cancer 2016, 60, 210–225. [Google Scholar] [CrossRef] [PubMed]
- Kähler, K.C.; Hassel, J.C.; Heinzerling, L.; Loquai, C.; Mössner, R.; Ugurel, S.; Zimmer, L.; Gutzmer, R. Management of side effects of immune checkpoint blockade by anti-CTLA-4 and anti-PD-1 antibodies in metastatic melanoma. JDDG J. Der Dtsch. Dermatol. Ges. 2016, 14, 662–681. [Google Scholar] [CrossRef] [PubMed]
- Leitlinienprogramm Onkologie (Deutsche Krebsgesellschaft, Deutsche Krebshilfe, AWMF): Diagnostik, Therapie und Nachsorge des Melanoms, Langversion 3.3, 2020, AWMF Registernummer: 032/024OL. Available online: http://www.leitlinienprogramm-onkologie.de/leitlinien/melanom/ (accessed on 6 May 2024).
- Kähler, K.C.; Hassel, J.C.; Heinzerling, L.; Loquai, C.; Thoms, K.-M.; Ugurel, S.; Zimmer, L.; Gutzmer, R. Side effect management during immune checkpoint blockade using CTLA-4 and PD-1 antibodies for metastatic melanoma—An update. JDDG J. Der Dtsch. Dermatol. Ges. 2020, 18, 582–609. [Google Scholar] [CrossRef] [PubMed]
- Baldini, G.; Arzideh, K.; Trienes, J.; Schlötterer, J.; Seifert, C.; Nensa, F. Aufbau einer Automatisierten NLP-Pipeline zur De-Identifikation Klinischer Dokumente; German Medical Science GMS Publishing House: Düsseldorf, Germany, 2023. [Google Scholar] [CrossRef]
- Hosch, R.; Baldini, G.; Parmar, V.; Borys, K.; Koitka, S.; Engelke, M.; Arzideh, K.; Ulrich, M.; Nensa, F. FHIR-PYrate: A data science friendly Python package to query FHIR servers. BMC Health Serv. Res. 2023, 23, 734. [Google Scholar] [CrossRef] [PubMed]
- Benson, T.; Grieve, G. FHIR Resources. In Principles of Health Interoperability; Benson, T., Grieve, G., Eds.; Springer International Publishing: Cham, Switzerland, 2016; pp. 361–379. ISBN 978-3-319-30368-0. [Google Scholar]
- Ulrich, H.; Behrend, P.; Wiedekopf, J.; Drenkhahn, C.; Kock-Schoppenhauer, A.-K.; Ingenerf, J. Hands on the Medical Informatics Initiative Core Data Set—Lessons Learned from Converting the MIMIC-IV. In German Medical Data Sciences 2021: Digital Medicine: Recognize—Understand—Heal; IOS Press: Amsterdam, The Netherlands, 2021; Volume 283, pp. 119–126. [Google Scholar] [CrossRef]
- Medical Informatics Initiative Germany. Basic Modules of the MII Core Data Set. Available online: https://www.medizininformatik-initiative.de/en/basic-modules-mii-core-data-set (accessed on 6 May 2024).
- FHIR Specification. Publication (Version) History. Available online: https://hl7.org/fhir/directory.html (accessed on 6 May 2024).
- FHIR Specification DSTU 1—Resource Observation—Content. Available online: https://hl7.org/fhir/DSTU1/observation.html (accessed on 6 May 2024).
- HL7 FHIR Release 4—Resource Observation—Content. Available online: https://hl7.org/fhir/R4/observation.html (accessed on 6 May 2024).
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).