Abstract
This scoping review explores the potential of electronic health records (EHR)-based studies to characterize long COVID. We screened all peer-reviewed publications in the English language from PubMed/MEDLINE, Scopus, and Web of Science databases until 14 September 2023, to identify the studies that defined or characterized long COVID based on data sources that utilized EHR in the United States, regardless of study design. We identified only 17 articles meeting the inclusion criteria. Respiratory conditions were consistently significant in all studies, followed by poor well-being features (n = 14, 82%) and cardiovascular conditions (n = 12, 71%). Some articles (n = 7, 41%) used a long COVID-specific marker to define the study population, relying mainly on ICD-10 codes and clinical visits for post-COVID-19 conditions. Among studies exploring plausible long COVID (n = 10, 59%), the most common methods were RT-PCR and antigen tests. The time delay for EHR data extraction post-test varied, ranging from four weeks to more than three months; however, most studies considering plausible long COVID used a waiting period of 28 to 31 days. Our findings suggest a limited utilization of EHR-derived data sources in defining long COVID, with only 59% of these studies incorporating a validation step.
1. Introduction
Post-COVID conditions, known as long COVID, refer to symptoms that manifest about four or more weeks after the initial infection by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the virus that causes the coronavirus disease of 2019 (COVID-19) [1,2]. Patients with long COVID experience many symptoms and conditions affecting various organ systems, including the respiratory, circulatory, nervous, endocrine, and digestive systems [3]. Moreover, chronic medical conditions and certain risk factors such as high blood pressure, diabetes, and obesity can exacerbate symptoms [4]. The increase in medical spending in the United States (US) as a result of long COVID is estimated at USD 528 billion total (about USD 1570 per person); this equates to roughly 2.5% of the total USD gross domestic product in 2019 [5]. A historical cohort study in Israel estimated the excess cost of long COVID on healthcare utilization as an additional 7.6% of control patient care [6]. Another study estimated productivity-based losses to the German economy at USD 3.7 billion [7]. Due to the debilitating conditions of long COVID, functional impairment, low work productivity, and long-term health complications are anticipated [8,9].
At present, little is known about the pathophysiology of these multisystem complications. While several studies have attempted to characterize long COVID [10,11,12,13], comparing the findings proves difficult due to the heterogeneity of symptoms and study period. Further, the accepted definition of long COVID keeps evolving [14]. The largest initiative in the US to define long COVID has been through the National Institute of Health (NIH) Researching COVID to Enhance Recovery (RECOVER) program. The program identified the 12 most prevalent symptom patterns among those with long COVID, including post-exercise malaise, fatigue, brain fog, dizziness, gastrointestinal symptoms, heart palpitations, changes in sexual desire or capacity, loss of taste or smell, thirst, chronic cough, chest pain, and abnormal movements [15]. As of November 2023, about 450 studies listed on ClinicalTrials.gov are investigating long COVID, including the response to antivirals, lithium therapy, and nitrate supplements, to discover possible treatments [16]. Several studies identified predictors or risk factors of long COVID [17,18,19] but used survey-based approaches that are prone to question heterogeneity and voluntary response bias. Identified examples included increasing age and body mass index (BMI), female sex, frailty, experiencing more than five symptoms or a hospital or emergency room visit during acute COVID-19 illness, and comorbidities such as asthma and heart disease [17,18,19].
Electronic health records (EHR) can provide a reliable, cost-effective, and comprehensive overview of the medical histories of a large population of patients, including previous infections and pre-existing conditions, thus enabling the tracking of symptoms and conditions [20]. According to the NIH, the National COVID Cohort Collaborative (N3C) data serves as the largest open database of patient EHRs in the US [21]. Within EHR systems, a valuable resource for obtaining disease etiology is the International Classification of Diseases, Tenth Revision (ICD-10), used to code and classify all symptoms, procedures, and diagnoses [22]. Unexplainable symptoms of COVID-19 were classified as B94.8 as a placeholder to signify long COVID until 30 September 2021; the code was subsequently changed to U09 [23].
While previous review studies attempted to define long COVID [24,25,26], these efforts are constrained by their heterogeneous designs and lack of specific diagnostic definitions. For example, a previous meta-analysis aimed at characterizing long COVID included 39 studies of diverse study designs, such as cohort, cross-sectional, and case-control [24]. Among these studies, the majority (n = 34) had either a moderate or high risk of bias. Likewise, Kelly et al. [27] identified a wide spectrum of symptoms and significant heterogeneity across the studies, noting that the population was limited to hospitalized patients. An earlier review by Iqbal et al. [28] incorporated 35 articles until March 2021 and highlighted limitations pertaining to various study designs, a limited number of countries, and questionnaire-based cross-sectional studies that fail to capture the evolution of symptoms over time. However, to our knowledge, no review article has comprehensively explored the potential of EHR-based studies to characterize long COVID. Thus, this scoping review aims to collate the studies that defined long COVID based on phenotypes or identified predictors or risk factors derived from various sources that utilized EHR data in the US. We also identified the analytical methods and summarized the common significant phenotypes, predictors, or risk factors.
2. Materials and Methods
We used the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) guidelines to conduct this scoping review.
2.1. Data Source
We used three search engines, PubMed/MEDLINE, Scopus, and Web of Science, to identify peer-reviewed articles until 14 September 2023, in the US regardless of study design.
2.2. Search Strategy
The initial screening of abstracts and titles included all articles that contained our key terms pertaining to long COVID, and those that contained EHR data or electronic medical records (EMR) data (Table 1). Studies were considered eligible if they were written in English, were peer-reviewed, and defined or characterized long COVID phenotypes or their predictors or risk factors. Review articles, pre-prints, case reports, editorials, abstracts, and articles published in early 2020 and before were excluded. Notably, studies that used non-EHR-based features of long COVID were excluded.

Table 1.
Search terms input into PubMed, Scopus, and Web of Science.
2.3. Study Selection
We imported all the retrieved abstracts and titles into the Covidence online tool https://covidence.org (accessed on 1 August 2023), where duplicates are excluded. Two reviewers (AM and GSC) independently screened abstracts and titles and excluded the articles not about long COVID and those not based on US data. Next, two different reviewers (RAL and GSJ) screened the full texts of the selected articles. In this step, articles were excluded if they did not have a clear definition of long COVID or did not identify risk factors, phenotypes, or predictors. Conflicts were resolved through follow-up discussions to reach a consensus on the final selection of articles. We also conducted a reference check of the selected articles for possible inclusion. Figure 1 shows the process of study selection.
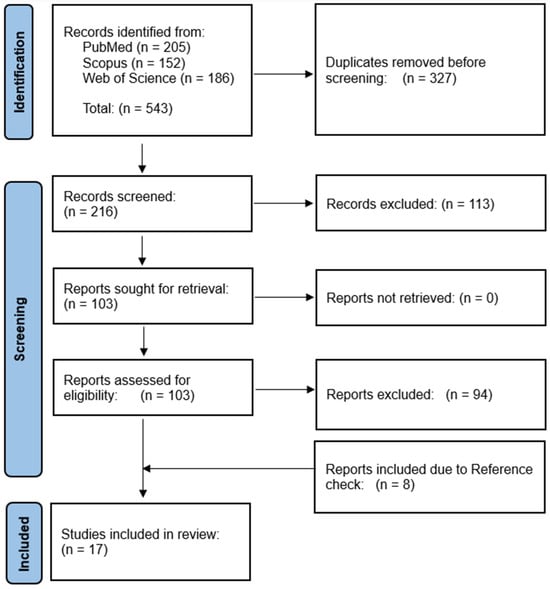
Figure 1.
PRISMA flowchart of EHR-based characterization of long COVID.
2.4. Data Extraction
We extracted information from the selected articles, including author names, study dates, sample size, types of data collected (qualitative and quantitative), study population inclusion criteria, identified long COVID features, methods used to identify such features, methods used to validate results, and study limitations.
2.5. Narrative Synthesis
A summary of long COVID labels assigned to samples in each study was created, and data were categorized based on clinical phenotypes and databases used in each of these papers. We synthesized various methods used to identify the important characteristics of long COVID phenotypes, predictors, and risk factors.
3. Results
Initially, we identified 543 articles from PubMed, Web of Science, and Scopus databases. Of these, 327 studies were duplicates and were thus removed, resulting in 216 unique studies for the initial screening. After the abstract and title screening of those articles, another 113 studies were excluded. We conducted the full-text review of the remaining 103 studies and excluded 94 studies due to a lack of a clear definition of long COVID and/or missing identification of predictors or risk factors, or the studies not based on data from the US. We identified eight additional studies via a reference check of the articles. Therefore, the final selection comprised 17 studies. Table 2 provides a summary of the selected studies.
The total sample size of long COVID study and control populations ranged from 11,209 to 5,213,885. The Veterans Affairs Electronic Healthcare database (n = 5,808,018) and INSIGHT Network, New York City Health System (n = 5,346,357) provided the largest databases of potential patients. Additionally, some datasets used inpatient and outpatient records to identify positive cases of COVID. Figure 2 shows the methods for identifying training data patient labels, classified as long COVID (n = 7/17) or plausibly long COVID (n = 10/17). Among articles that used a long COVID-specific marker (Figure 2a), most (5/7, 71%) chose an ICD-10 code for post COVID-19 condition, unspecified, or sequelae of other specified infectious diseases. Among the studies that used acute COVID-19 illness to identify plausibly long COVID patient training data (n = 10/17), reverse transcription-polymerase chain reaction (RT-PCR) test was the most common (Figure 2b). These ten studies extracted features to study from the EHR after a waiting period following a recorded acute COVID-19 positive test. The waiting period shown in Figure 2b varied from 28 days to more than three months [29], but most used a one-month delay.


Table 2.
Summary of the included studies.
Table 2.
Summary of the included studies.
| Study | Year | Study Design | Total Sample Size | Total Participants (n = Study Population) | EHR Source | Main Methods | Method of Validation |
|---|---|---|---|---|---|---|---|
| Al-Aly et al. [30] | 2021 | Observational Study | 5,213,885 | 73,435 | US Department of Veterans Affairs electronic healthcare databases | Cox regression | N/A |
| Baskett et al. [31] | 2022 | Retrospective Cohort Study | N/A | 17,487 | Cerner Real-World data set | Logistic regression; Propensity score matching | N/A |
| Estiri et al. [29] | 2021 | Retrospective Cohort Study | 96,025 | 22,475 | Mass General Brigham Hospital | MLHO; Multivariate time series analysis | Clinical expert review |
| Fritsche et al. [32] | 2023 | Case-Control | 63,675 | 1724 | Michigan Medicine EHR data | Logistic regression | Sensitivity analysis; Logistic regression; AAUC |
| Haupert et al. [33] | 2022 | Case-Crossover Design | 204,597 | 44,198 | Michigan Medicine Health System | Logistic regression | N/A |
| Jiang et al. [34] | 2022 | Retrospective Cohort Study | 85,196 | 28,558 | N3C | Deep neural networks; Extreme gradient boosting (decision tree); PCA | AUC; F1 score |
| Khullar et al. [35] | 2023 | Retrospective Cohort Study | 310,220 | 62,339 | INSIGHT Network; New York City Health Systems | Logistic regression | Sensitivity analysis |
| Lorman et al. [36] | 2023 | Observational Study | 14,399 | 1309 | RECOVER PEDSnet EHR | Propensity score matching; Decision trees | Sensitivity analysis |
| Nasir et al. [37] | 2023 | Observational Study | 11,209 | 4091 | Health Choice Network | Bayesian structural time series modeling | N/A |
| Pfaff et al. [38] | 2023 | Retrospective Cohort Study | 36,880 | 33,782 | N3C | Clustering; Network analysis | N/A |
| Pfaff et al. [23] | 2022 | Retrospective Cohort Study | 1,793,604 | 73,972 | N3C | Extreme gradient boosting (decision tree) | Cross-validation; AUC; Precision; Recall; F-score |
| Rao et al. [39] | 2022 | Exploratory, Retrospective Cohort | 659,286 | 59,893 | PEDSnet | Cox regression; Logistic regression | N/A |
| Reese et al. [40] | 2023 | Retrospective Cohort | 5,434,528 | 20,532 | N3C | NLP; k-means clustering | N/A |
| Sengupta et al. [41] | 2022 | Retrospective Cohort | 49,950 | 7511 | N3C | Convolutional and LSTM neural networks | AUC |
| Wang et al. [42] | 2022 | Observational Study | 51,485 | 26,117 | Mass General Brigham Hospital | Rule-based NLP | Precision |
| Zang et al. [43] | 2023 | Observational Study | 361,401 (INSIGHT) 199,351 (OneFlorida+) | 35,275 (INSIGHT) 22,341 (OneFlorida+) | INSIGHT CRN; OneFlorida+ CRN | Propensity score matching | Sensitivity analysis |
| H Zhang et al. [44] | 2022 | Retrospective Cohort Study | 3460 | 20,881 | INSIGHT CRN; OneFlorida+ CRN | Clustering; NLP via topic modeling | Topic Coherence; Sensitivity analysis |
Abbreviations: AAUC: area under the covariate-adjusted receiver operating characteristic curve; AUC: area under the receiver operating characteristic curve; CRN: clinical research network MLHO: Machine Learns Health Outcomes; PEDSnet: a National Pediatric Learning Health System; N3C: National COVID Cohort Collaborative; NLP: natural language processing; LSTM: long short-term memory; PCA: principal component analysis.
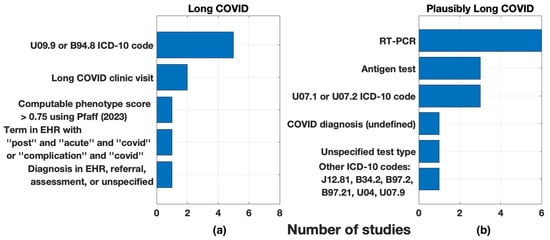
Figure 2.
Markers used for training data patient labels: (a) long COVID and (b) plausibly long COVID. Plausible long COVID markers were paired with a time duration post-test.
Figure 3 depicts the candidate features for the definition, predictors, or risk factors of long COVID. All studies used diagnostic categories as candidates. Many studies organized their diagnostic category candidates by grouping ICD-10 codes into diagnostic clusters using the Clinical Classifications Software Refined. In contrast, some used the R package PheWAS to map ICD-10 codes to unique PheCodes. Notably, five (29%) studies used a combination of social determinants of health (SDOH) and patient demographics, and four (24%) studies included medications. Only one study [34] used quantitative data based on biophysical metrics recorded during acute COVID-19 hospitalizations.
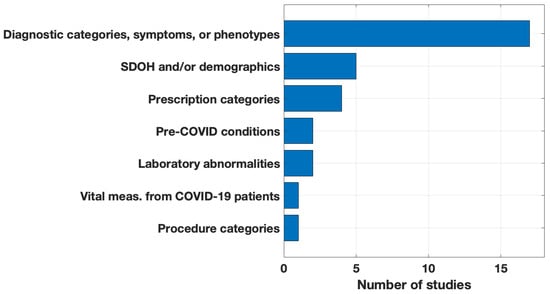
Figure 3.
Candidate features for the definition, predictors, or risk factors of long COVID.
The main data analysis methods used by the studies are presented in Figure 4. In this figure, we depicted the methods used by at least two studies. See Table 2 for methods for each study. Due to the relatively large sample sizes, most studies used data science techniques. Eight (47%) articles discussed the implementation of machine learning for feature down-selection or identification of significant features.
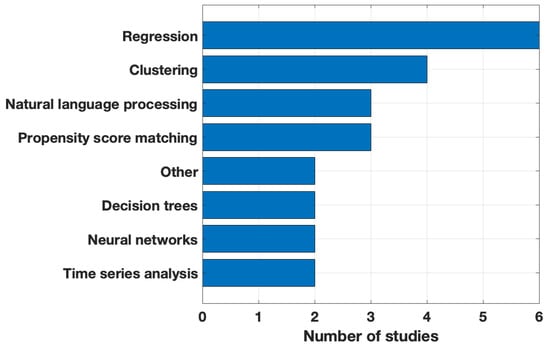
Figure 4.
Main data analysis method used. Methods in the Other category: network analysis and MLHO (Machine Learns Health Outcomes).
All articles incorporated statistical tests, but approaches varied. Nearly all articles (n = 15, 88%) reported results using hazard ratios, risk ratios, odds ratios, or p-values. Hazard ratios were often found using Cox survival models, and reported p-values often underwent the Bonferroni correction. Some studies reduced their initial pool of candidates to a final set based on feature importance criteria such as hazard ratios greater than 1 or p-values less than 0.05. In contrast, others merely discussed the top 20 or 30 features.
We categorized the phenotypes, predictors, and risk factors of long COVID identified by the articles (Figure 5). As some articles used hundreds of candidates in their analyses, Figure 5 only shows the categories of most significant findings. The studies varied significantly in how they clustered and summarized their results; we grouped these by broad categories to facilitate comparison. All articles found respiratory conditions significant, nearly all studies (n = 14, 82%) reported poor well-being features as significant, and most (n = 12, 71%) included cardiovascular conditions. Examples of these top three categories were: respiratory conditions (e.g., respiratory failure, asthma), poor general well-being (e.g., fatigue, pain), and cardiovascular conditions and diseases (e.g., chest pain, coronary disease). Uncommon findings included the circumstances of the acute or post-COVID-19 infection (e.g., extended hospital stay, SARS-CoV-2 variant, outpatient utilization) and the presence of certain medications (e.g., corticosteroids, cough preparations, anticoagulant drugs).
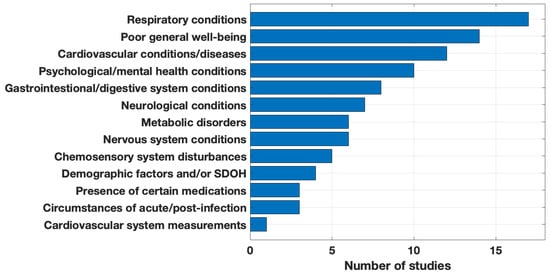
Figure 5.
Significant phenotypes, predictors, and risk factors of long COVID.
After identifying long COVID features based on training data (Figure 5), ten of the studies (59%) included a validation step to test their purported long COVID phenotype, predictors, or risk factors on test data. Roughly one-fourth used their entire suite of candidate features in a long COVID prediction task and measured their success with the area under the receiver operating characteristic curve (AUC) or F scores. About 30% of the studies used sensitivity analysis to test the robustness of their results. As two examples of validation methods, Wang et al. [42] used natural language processing (NLP) to validate their reduced symptom lexicon on a test dataset and achieved an average precision and estimated recall of 0.94 and 0.84. Estiri et al. [29] employed clinical experts to review the phenotypes identified by their method.
4. Discussion
To our knowledge, this is the first scoping review for identifying long COVID phenotypes, predictors, and risk factors based on various sources that utilized EHR data in the US. We found 17 articles that met the eligibility criteria. The articles were classified based on how long COVID was defined, methodologies, and the identification of significant risk factors or phenotypes. ICD-10 codes (U09.9 or B 94.8) were the most common markers. A large majority of the studies reported poor general well-being and respiratory and cardiovascular conditions as significant features.
The studies collectively suggest the heterogeneous nature of long COVID, implying that it does not manifest as a uniform condition. Instead, it suggests diverse phenotypic clusters, which include respiratory, neuropsychiatric, cardiovascular, and pain and fatigue subtypes [40]. These subtypes are characterized by distinct clinical features, patient demographics, and associations with various organ systems. Ten of the twelve symptom patterns identified by the NIH RECOVER initiative were considered as features by the studies and fell under broad, significant categories (Figure 5). Both thirst and changes in sexual desire were absent as individual features but may fall under broad symptom characterizations [40]. Phenotypic clustering utilizing machine learning methods may provide a unique approach to examining phenotype commonalities and drastic differences that may enhance our understanding of long COVID’s heterogeneity. While several of the phenotypes found by the studies are common in understanding long COVID, latent phenotypes like substance use disorders (e.g., opioid use) present unique ways to define the disease.
Beyond the symptoms of long COVID, some studies reported predictors and risk factors for developing the condition. We noted that SDOH and demographic information cannot plausibly be used to form a clinical definition of long COVID but may indicate the likelihood of developing the condition. Such studies included patients who recovered quickly from their acute infection in their test population, therefore diluting the conclusions that can be drawn from observed symptoms, diagnoses, and medicines.
While our review focused on US data, significant international studies, particularly those from the UK, offer valuable insights. A comprehensive study in the UK, including 58 million individuals, aimed to estimate long COVID prevalence based on diagnostic codes. However, it lacked phenotype or risk factors identification and relied heavily on clinician-entered or referral codes [45]. Mayor et al. (2022) in the UK developed an EHR-based phenotype for long COVID, to facilitate a standardized method for defining and identifying cases from routine data. Using this phenotype, they compared pre-pandemic and pandemic symptoms, and found that this method allows the identification of long COVID cases in routine data sets, though validation against other UK reports was necessary [46]. In Scotland, Jeffrey et al. (2024) analyzed EHR data from 4.6 million participants, estimating a 1.7% long COVID prevalence using various measures including clinical codes, free text, sick notes, and an operational definition with the latter identifying the most cases [47]. Additionally, international EHR-based studies from Germany [48], France, Italy, and Singapore by [49,50] reported diverse long COVID symptoms, consistent with our findings. However, US and international studies have been unable to define long COVID by obtaining globally aggregated data due to challenges in privacy regulations, standardization, quality, and interoperability that complicates data integration.
The studies identified for this review a broad range of symptoms, complications, and clinical conditions significantly associated with long COVID. However, only one study [37] examined causality, using Bayesian structural time series models. This complexity underscores the need for a more comprehensive understanding of clinical conditions to elucidate the causal relationships. Moreover, while the studies primarily relied on ICD-10 codes to define long COVID, only three studies leveraged the potential of NLP for extracting information from unstructured clinical notes in EHR data. Combined with longitudinal EHR data, this approach can enhance our understanding of patterns, progression, treatment, and management of long COVID. However, it is crucial to acknowledge the challenges associated with NLP, such as its time-consuming nature and the potential for inconsistencies, which should be carefully considered in future research endeavors. While computer-assisted coding is efficient, it lacks human interactions, increasing the risk of inadvertently including irrelevant or erroneous information in the dataset.
EHR, EMR, and electronic patient records (EPR) systems are often used interchangeably, but they differ in scope and usage. EHRs were the most frequently encountered systems in this review. They serve as digital repositories of a patient’s complete medical history from all healthcare providers and are intended for sharing with other healthcare entities, practices, and hospitals. EMRs focus on a single practice or hospital. Some EMR systems offer integrated care, possibly within larger hospital corporations. Lastly, EPR usage is primarily limited to Europe, which was not used in the articles. The localized usage of the term EPR may indicate the use of other terminologies globally that we have not accounted for in our literature search strategy, as our search was confined to specific health information systems. For future studies, it would be beneficial to include broader research terms, such as data hubs, data lakes, registries, claims, and repositories to encompass a more comprehensive scope of relevant literature.
The included studies shared some common limitations. There is no widely accepted definition for long COVID to date, and the disease was defined based on various arbitrary time intervals. Additionally, the symptoms and conditions of long COVID were examined mostly in the initial months. Most studies failed to differentiate between incident and prevalent symptoms, contributing to ambiguity in characterizing long COVID. To address these challenges, a standardized research protocol that facilitates capturing systematic tracking of symptoms seems imperative. Leveraging longitudinal EHR-derived data may provide a more comprehensive understanding of the emerging symptoms and monitor longer-term trends and effects.
Nearly all studies acknowledged data-related limitations. The use of diagnosis codes is a limitation as they may not capture all signs, symptoms, or laboratory results found in clinical notes. Reliance on diagnostic codes could result in missing information and biases. Additionally, the intensity of the pandemic and misinformation might introduce confirmatory bias between healthcare providers and patients. Some articles [33,43] acknowledged the need for replication studies in other cohorts. Several articles excluded hospitalized COVID-19 patients, which might not reflect the complete spectrum of long COVID. This limitation highlights the importance of including more hospitalized patients in future studies.
Temporal bias is an issue because the choice of time windows for analysis varies among studies. Some studies expressed concerns about not accounting for within-person time-varying confounders, such as changes in health-seeking behavior during the pandemic. The studies’ requirement of pre- and post-COVID-19 visits is acknowledged as potentially biased toward patients with more complex health histories. The cohort case window ratio is a subjective parameter, indicating variability in the study design. Additionally, some studies mentioned potential timing biases related to developing and implementing specific diagnosis codes. Some touched on the issue of data representation concerning various population groups and the inability to generalize the data. The absence of medication information used for COVID-19 therapy, particularly in severe cases, is noted as a potential limitation, as is the lack of data on viral variants for individual patients.
Further to the previously mentioned limitations, our study also presents several constraints. First, the exclusion of non-English articles may introduce language bias. Due to the evolving nature of the topic, the omission of pre-prints and conference articles could result in the loss of crucial information. Additionally, it is possible that some articles were missed during the screening process, although we mitigated this by conducting reference checks and backward searches. Another recurring limitation was the lack of comprehensive insight into the underlying mechanisms that lead to long COVID. Most studies did not explore the indirect effects of long COVID, such as the social, economic, and behavioral changes that may arise due to the condition. This knowledge gap poses a significant challenge in fully understanding and addressing long COVID.
5. Conclusions
In conclusion, this scoping review has provided an overview of state-of-the-art research on long COVID, drawing from various sources that utilized EHR data in the US for defining and characterizing this condition. The findings suggest that while a consensus on the definition of long COVID remains elusive, and understanding long COVID remains evolving and uncertain, ICD-10 codes are commonly used for identification, and poor general well-being, respiratory conditions, and cardiovascular conditions are consistently associated with long COVID. Moreover, while data science techniques are widely employed, the lack of validation and causality assessments is evident, highlighting the need for more robust methodologies. The complex nature of long COVID, encompassing various symptoms and clinical conditions, underscores the need for more in-depth studies, including those leveraging longitudinal EHR data. Despite these uncertainties and gaps, this review lays the groundwork for future research endeavors aimed at harnessing the potential of health information systems to better understand the epidemiology of long COVID.
Author Contributions
Conceptualization, A.M.; methodology, all authors; software, all authors; formal analysis, all authors; writing—original draft preparation, all authors; writing—review and editing, all authors. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Crook, H.; Raza, S.; Nowell, J.; Young, M.; Edison, P. Long COVID—Mechanisms, risk factors, and management. BMJ 2021, 374, n1648. [Google Scholar] [CrossRef] [PubMed]
- Devi, K.P.; Pourkarim, M.R.; Thijssen, M.; Sureda, A.; Khayatkashani, M.; Cismaru, C.A.; Neagoe, I.B.; Habtemariam, S.; Razmjouei, S.; Khayat Kashani, H.R. A perspective on the applications of furin inhibitors for the treatment of SARS-CoV-2. Pharmacol. Rep. 2022, 74, 425–430. [Google Scholar] [CrossRef] [PubMed]
- Garg, M.; Maralakunte, M.; Garg, S.; Dhooria, S.; Sehgal, I.; Bhalla, A.S.; Vijayvergiya, R.; Grover, S.; Bhatia, V.; Jagia, P. The conundrum of ‘long-COVID-19: A narrative review. Int. J. Gen. Med. 2021, 14, 2491–2506. [Google Scholar] [CrossRef]
- Makhoul, E.; Aklinski, J.L.; Miller, J.; Leonard, C.; Backer, S.; Kahar, P.; Parmar, M.S.; Khanna, D. A review of COVID-19 in relation to metabolic syndrome: Obesity, hypertension, diabetes, and dyslipidemia. Cureus 2022, 14, e27438. [Google Scholar] [CrossRef]
- Cutler, D.M. The Economic Cost of Long COVID: An Update. Publish Online July 2022. Available online: https://scholar.harvard.edu/cutler/news/long-covid (accessed on 9 June 2024).
- Sagy, Y.W.; Feldhamer, I.; Brammli-Greenberg, S.; Lavie, G. Estimating the economic burden of long-COVID: The additive cost of healthcare utilisation among COVID-19 recoverees in Israel. BMJ Glob. Health 2023, 8, e012588. [Google Scholar] [CrossRef] [PubMed]
- Gandjour, A. Long COVID: Costs for the German economy and health care and pension system. BMC Health Serv. Res. 2023, 23, 641. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Ramirez, D.C.; Normand, K.; Zhaoyun, Y.; Torres-Castro, R. Long-term impact of COVID-19: A systematic review of the literature and meta-analysis. Biomedicines 2021, 9, 900. [Google Scholar] [CrossRef]
- Ham, D.I. Long-Haulers and Labor Market Outcomes; Federal Reserve Bank of Minneapolis: Minneapolis, MN, USA, 2022; Available online: https://www.minneapolisfed.org/institute/working-papers-institute/iwp60.pdf (accessed on 9 June 2024).
- Amin-Chowdhury, Z.; Ladhani, S.N. Causation or confounding: Why controls are critical for characterizing long COVID. Nat. Med. 2021, 27, 1129–1130. [Google Scholar] [CrossRef]
- Barizien, N.; Le Guen, M.; Russel, S.; Touche, P.; Huang, F.; Vallée, A. Clinical characterization of dysautonomia in long COVID-19 patients. Sci. Rep. 2021, 11, 14042. [Google Scholar] [CrossRef] [PubMed]
- Davis, H.E.; Assaf, G.S.; McCorkell, L.; Wei, H.; Low, R.J.; Re’em, Y.; Redfield, S.; Austin, J.P.; Akrami, A. Characterizing long COVID in an international cohort: 7 months of symptoms and their impact. EClinicalMedicine 2021, 38, 101019. [Google Scholar] [CrossRef]
- Deer, R.R.; Rock, M.A.; Vasilevsky, N.; Carmody, L.; Rando, H.; Anzalone, A.J.; Basson, M.D.; Bennett, T.D.; Bergquist, T.; Boudreau, E.A. Characterizing long COVID: Deep phenotype of a complex condition. EBioMedicine 2021, 74, 103722. [Google Scholar] [CrossRef]
- Taquet, M.; Dercon, Q.; Luciano, S.; Geddes, J.R.; Husain, M.; Harrison, P.J. Incidence, co-occurrence, and evolution of long-COVID features: A 6-month retrospective cohort study of 273,618 survivors of COVID-19. PLoS Med. 2021, 18, e1003773. [Google Scholar] [CrossRef] [PubMed]
- Thaweethai, T.; Jolley, S.E.; Karlson, E.W.; Levitan, E.B.; Levy, B.; McComsey, G.A.; McCorkell, L.; Nadkarni, G.N.; Parthasarathy, S.; Singh, U. Development of a definition of postacute sequelae of SARS-CoV-2 infection. JAMA 2023, 329, 1934–1946. [Google Scholar] [CrossRef] [PubMed]
- Bonilla, H.; Peluso, M.J.; Rodgers, K.; Aberg, J.A.; Patterson, T.F.; Tamburro, R.; Baizer, L.; Goldman, J.D.; Rouphael, N.; Deitchman, A.; et al. Therapeutic trials for long COVID-19: A call to action from the interventions taskforce of the RECOVER initiative. Front. Immunol. 2023, 14, 1129459. [Google Scholar] [CrossRef] [PubMed]
- Jones, R.; Davis, A.; Stanley, B.; Julious, S.; Ryan, D.; Jackson, D.J.; Halpin, D.M.; Hickman, K.; Pinnock, H.; Quint, J.K. Risk predictors and symptom features of long COVID within a broad primary care patient population including both tested and untested patients. Pragmatic Obs. Res. 2021, 12, 93–104. [Google Scholar] [CrossRef] [PubMed]
- Knight, D.R.; Munipalli, B.; Logvinov, I.I.; Halkar, M.G.; Mitri, G.; Hines, S.L. Perception, prevalence, and prediction of severe infection and post-acute sequelae of COVID-19. Am. J. Med. Sci. 2022, 363, 295–304. [Google Scholar] [CrossRef] [PubMed]
- Sudre, C.H.; Murray, B.; Varsavsky, T.; Graham, M.S.; Penfold, R.S.; Bowyer, R.C.; Pujol, J.C.; Klaser, K.; Antonelli, M.; Canas, L.S. Attributes and predictors of long COVID. Nat. Med. 2021, 27, 626–631. [Google Scholar] [CrossRef] [PubMed]
- Mollalo, A.; Hamidi, B.; Lenert, L.; Alekseyenko, A.V. Characterizing Patient Phenotypes and Emerging Trends in Application of Spatial Analysis in Individual-Level Health Data. Res. Square 2023. [Google Scholar] [CrossRef]
- NIH. N3C: Translating Health Data into Health Solutions. Available online: https://ncats.nih.gov/sites/default/files/NCATS-N3C-One-Pager-508.pdf (accessed on 1 December 2023).
- Kurbasic, I.; Pandza, H.; Masic, I.; Huseinagic, S.; Tandir, S.; Alicajic, F.; Toromanovic, S. The advantages and limitations of international classification of diseases, injuries and causes of death from aspect of existing health care system of Bosnia and Herzegovina. Acta Inform. Med. 2008, 16, 159. [Google Scholar] [CrossRef]
- Pfaff, E.R.; Girvin, A.T.; Bennett, T.D.; Bhatia, A.; Brooks, I.M.; Deer, R.R.; Dekermanjian, J.P.; Jolley, S.E.; Kahn, M.G.; Kostka, K. Identifying who has long COVID in the USA: A machine learning approach using N3C data. Lancet Digit. Health 2022, 4, e532–e541. [Google Scholar] [CrossRef]
- Michelen, M.; Manoharan, L.; Elkheir, N.; Cheng, V.; Dagens, A.; Hastie, C.; O’Hara, M.; Suett, J.; Dahmash, D.; Bugaeva, P. Characterising long COVID: A living systematic review. BMJ Glob. Health 2021, 6, e005427. [Google Scholar] [CrossRef] [PubMed]
- Akbarialiabad, H.; Taghrir, M.H.; Abdollahi, A.; Ghahramani, N.; Kumar, M.; Paydar, S.; Razani, B.; Mwangi, J.; Asadi-Pooya, A.A.; Malekmakan, L. Long COVID, a comprehensive systematic scoping review. Infection 2021, 49, 1163–1186. [Google Scholar] [CrossRef] [PubMed]
- Aiyegbusi, O.L.; Hughes, S.E.; Turner, G.; Rivera, S.C.; McMullan, C.; Chandan, J.S.; Haroon, S.; Price, G.; Davies, E.H.; Nirantharakumar, K. Symptoms, complications and management of long COVID: A review. J. R. Soc. Med. 2021, 114, 428–442. [Google Scholar] [CrossRef] [PubMed]
- Kelly, J.D.; Curteis, T.; Rawal, A.; Murton, M.; Clark, L.J.; Jafry, Z.; Shah-Gupta, R.; Berry, M.; Espinueva, A.; Chen, L. SARS-CoV-2 post-acute sequelae in previously hospitalised patients: Systematic literature review and meta-analysis. Eur. Respir. Rev. 2023, 32, 220254. [Google Scholar] [CrossRef] [PubMed]
- Iqbal, F.M.; Lam, K.; Sounderajah, V.; Clarke, J.M.; Ashrafian, H.; Darzi, A. Characteristics and predictors of acute and chronic post-COVID syndrome: A systematic review and meta-analysis. EClinicalMedicine 2021, 36, 100899. [Google Scholar] [CrossRef] [PubMed]
- Estiri, H.; Strasser, Z.H.; Brat, G.A.; Semenov, Y.R.; Patel, C.J.; Murphy, S.N. Evolving phenotypes of non-hospitalized patients that indicate long COVID. BMC Med. 2021, 19, 249. [Google Scholar] [CrossRef] [PubMed]
- Al-Aly, Z.; Xie, Y.; Bowe, B. High-dimensional characterization of post-acute sequelae of COVID-19. Nature 2021, 594, 259–264. [Google Scholar] [CrossRef] [PubMed]
- Baskett, W.I.; Qureshi, A.I.; Shyu, D.; Armer, J.M.; Shyu, C.-R. COVID-specific long-term sequelae in comparison to common viral respiratory infections: An analysis of 17 487 infected adult patients. Open Forum Infect. Dis. 2023, 10, ofac683. [Google Scholar] [CrossRef]
- Fritsche, L.G.; Jin, W.; Admon, A.J.; Mukherjee, B. Characterizing and predicting post-acute sequelae of SARS-CoV-2 infection (PASC) in a large academic medical center in the US. J. Clin. Med. 2023, 12, 1328. [Google Scholar] [CrossRef]
- Haupert, S.R.; Shi, X.; Chen, C.; Fritsche, L.G.; Mukherjee, B. A Case-Crossover Phenome-wide association study (PheWAS) for understanding Post-COVID-19 diagnosis patterns. J. Biomed. Inform. 2022, 136, 104237. [Google Scholar] [CrossRef] [PubMed]
- Jiang, S.; Loomba, J.; Sharma, S.; Brown, D. Vital Measurements of Hospitalized COVID-19 Patients as a Predictor of Long COVID: An EHR-based Cohort Study from the RECOVER Program in N3C. In Proceedings of the 2022 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), Las Vegas, NV, USA, 6–8 December 2022; pp. 3023–3030. [Google Scholar]
- Khullar, D.; Zhang, Y.; Zang, C.; Xu, Z.; Wang, F.; Weiner, M.G.; Carton, T.W.; Rothman, R.L.; Block, J.P.; Kaushal, R. Racial/ethnic disparities in post-acute sequelae of SARS-CoV-2 infection in New York: An EHR-based cohort study from the RECOVER program. J. Gen. Intern. Med. 2023, 38, 1127–1136. [Google Scholar] [CrossRef] [PubMed]
- Lorman, V.; Rao, S.; Jhaveri, R.; Case, A.; Mejias, A.; Pajor, N.M.; Patel, P.; Thacker, D.; Bose-Brill, S.; Block, J. Understanding pediatric long COVID using a tree-based scan statistic approach: An EHR-based cohort study from the RECOVER Program. JAMIA Open 2023, 6, ooad016. [Google Scholar] [CrossRef] [PubMed]
- Nasir, M.; Cook, N.; Parras, D.; Mukherjee, S.; Miller, G.; Ferres, J.L.; Chung-Bridges, K. Using Data Science and a Health Equity Lens to Identify Long-COVID Sequelae Among Medically Underserved Populations. J. Health Care Poor Underserved 2023, 34, 521–534. [Google Scholar] [CrossRef] [PubMed]
- Pfaff, E.R.; Madlock-Brown, C.; Baratta, J.M.; Bhatia, A.; Davis, H.; Girvin, A.; Hill, E.; Kelly, E.; Kostka, K.; Loomba, J. Coding long COVID: Characterizing a new disease through an ICD-10 lens. BMC Med. 2023, 21, 58. [Google Scholar] [CrossRef] [PubMed]
- Rao, S.; Lee, G.M.; Razzaghi, H.; Lorman, V.; Mejias, A.; Pajor, N.M.; Thacker, D.; Webb, R.; Dickinson, K.; Bailey, L.C. Clinical features and burden of postacute sequelae of SARS-CoV-2 infection in children and adolescents. JAMA Pediatr. 2022, 176, 1000–1009. [Google Scholar] [CrossRef] [PubMed]
- Reese, J.T.; Blau, H.; Casiraghi, E.; Bergquist, T.; Loomba, J.J.; Callahan, T.J.; Laraway, B.; Antonescu, C.; Coleman, B.; Gargano, M. Generalisable long COVID subtypes: Findings from the NIH N3C and RECOVER programmes. EBioMedicine 2023, 87, 104413. [Google Scholar] [CrossRef] [PubMed]
- Sengupta, S.; Loomba, J.; Sharma, S.; Brown, D.E.; Thorpe, L.; Haendel, M.A.; Chute, C.G.; Hong, S. Analyzing historical diagnosis code data from NIH N3C and RECOVER Programs using deep learning to determine risk factors for Long COVID. In Proceedings of the 2022 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), Las Vegas, NV, USA, 6–8 December 2022; pp. 2797–2802. [Google Scholar]
- Wang, L.; Foer, D.; MacPhaul, E.; Lo, Y.-C.; Bates, D.W.; Zhou, L. PASCLex: A comprehensive post-acute sequelae of COVID-19 (PASC) symptom lexicon derived from electronic health record clinical notes. J. Biomed. Inform. 2022, 125, 103951. [Google Scholar] [CrossRef] [PubMed]
- Zang, C.; Zhang, Y.; Xu, J.; Bian, J.; Morozyuk, D.; Schenck, E.J.; Khullar, D.; Nordvig, A.S.; Shenkman, E.A.; Rothman, R.L. Data-driven analysis to understand long COVID using electronic health records from the RECOVER initiative. Nat. Commun. 2023, 14, 1948. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Zang, C.; Xu, Z.; Zhang, Y.; Xu, J.; Bian, J.; Morozyuk, D.; Khullar, D.; Zhang, Y.; Nordvig, A.S. Data-driven identification of post-acute SARS-CoV-2 infection subphenotypes. Nat. Med. 2023, 29, 226–235. [Google Scholar] [CrossRef]
- Walker, A.J.; MacKenna, B.; Inglesby, P.; Tomlinson, L.; Rentsch, C.T.; Curtis, H.J.; Morton, C.E.; Morley, J.; Mehrkar, A.; Bacon, S. Clinical coding of long COVID in English primary care: A federated analysis of 58 million patient records in situ using OpenSAFELY. Br. J. Gen. Pract. 2021, 71, e806–e814. [Google Scholar] [CrossRef] [PubMed]
- Mayor, N.; Meza-Torres, B.; Okusi, C.; Delanerolle, G.; Chapman, M.; Wang, W.; Anand, S.; Feher, M.; Macartney, J.; Byford, R. Developing a long COVID phenotype for postacute COVID-19 in a national primary care sentinel cohort: Observational retrospective database analysis. JMIR Public Health Surveill. 2022, 8, e36989. [Google Scholar] [CrossRef] [PubMed]
- Jeffrey, K.; Woolford, L.; Maini, R.; Basetti, S.; Batchelor, A.; Weatherill, D.; White, C.; Hammersley, V.; Millington, T.; Macdonald, C. Prevalence and risk factors for long COVID among adults in Scotland using electronic health records: A national, retrospective, observational cohort study. EClinicalMedicine 2024, 71, 102590. [Google Scholar] [CrossRef] [PubMed]
- Kessler, R.; Philipp, J.; Wilfer, J.; Kostev, K. Predictive Attributes for Developing Long COVID—A Study Using Machine Learning and Real-World Data from Primary Care Physicians in Germany. J. Clin. Med. 2023, 12, 3511. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.G.; Dagliati, A.; Shakeri Hossein Abad, Z.; Xiong, X.; Bonzel, C.-L.; Xia, Z.; Tan, B.W.; Avillach, P.; Brat, G.A.; Hong, C. International electronic health record-derived post-acute sequelae profiles of COVID-19 patients. NPJ Digit. Med. 2022, 5, 81. [Google Scholar] [CrossRef]
- Dagliati, A.; Strasser, Z.H.; Abad, Z.S.H.; Klann, J.G.; Wagholikar, K.B.; Mesa, R.; Visweswaran, S.; Morris, M.; Luo, Y.; Henderson, D.W. Characterization of long COVID temporal sub-phenotypes by distributed representation learning from electronic health record data: A cohort study. Eclinicalmedicine 2023, 64, 102210. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
