1. Introduction
The use of information systems in clinical patient care has recently grown, generating a high volume of data, as confirmed by Danciu et al. [
1]. These data come from different sources, such as clinical reports, diagnostic test results, monitoring devices, surveys on clinical processes or the care received, and almost any type of documentation related to a medical act. This information is heterogeneous, ranging from numerical quantification to clinical observations in natural language. As a result, the volume of data is very high and requires the use of technologies that allow its effective exploitation. Potential applications of technology in healthcare include system integration, big data, and artificial intelligence [
2]. The use of artificial intelligence, more specifically the areas of machine learning and deep learning, offers the opportunity to obtain information and draw conclusions from large amounts of data through processes that resemble the way human knowledge and reasoning are constructed [
3]. The volume of this information, together with the high level of detail it provides, contributes to overcoming the barriers and limits of traditional analyses [
4]; technology not only allows information to be analyzed more efficiently than using human resources, but it also permits a more in-depth analysis that allows conclusions to be drawn that would otherwise be impossible [
5].
Among the ways data analysis can provide knowledge that improves multiple areas of the health sector, one of the most important is predicting the evolution of a patient’s clinical condition [
6,
7], i.e., estimating a patient’s condition over time.
Estimating patient progression in critical care, where the number of patients that can be treated is very limited and the cost of care per patient is high, could be a relevant achievement in this type of estimation. On the other hand, the criteria for admission to these units are subjective to a high degree, despite the effort to standardize them, as Kim et al. concluded [
8], and this could also be assisted by AI. Based on the relevant clinical data of the patients, it could objectively estimate the probability of survival and the estimated length of stay in the critical care unit. Models like this have been developed using machine learning techniques, which are capable of establishing patterns and drawing conclusions by processing a large volume of historical data. A model with these characteristics could potentially enhance safety in decision-making processes, thereby helping to reduce subjectivity, always as a form of support for clinicians in decision making.
However, the greatest weakness of these systems lies in the interpretation of the information obtained. The predictive models obtained using machine learning techniques are considered a black box because, in practice, it is impossible to determine with reasonable effort the rationale for a given prediction [
9]; this is because it is based on the analysis of multiple parameters from thousands of previous records. Thus, in these models, there is no certainty that the conclusions it provides are reliable since there is no traceability justifying the conclusion, nor is it possible to carry out a strict demonstration of the goodness of the model [
10]. Therefore, validation of the model is essential, since some of the responsibility for these important decisions is assumed by the doctors in the system. Due to this, it is essential to apply a methodology that establishes the steps to follow in the development of an intelligent model, validation against a traditional model, and establishment of the acceptable thresholds of deviation in the results. Similar analysis methodologies have been proposed and successfully applied in virtual screening [
11].
Another aspect to be considered is the effectiveness of these AI-based systems. If we compare their results with linear regression, considered as traditional here, we will obtain results that are inferior [
12]. However, reviews of the literature indicate that the median sample size used is 1250 records [
13]. The use of a larger sample size is expected to provide better results that justify the use of machine learning to make predictions, as has been demonstrated in specific predictions concerning critically ill patients with greater volumes of data [
14]. The possibility of using machine learning to obtain better results than scoring systems in the healthcare field has been recognized [
15]. It has been used in specific applications such as the prediction of upper gastrointestinal bleeding [
16], so we believe it is feasible to investigate it for predicting the evolution of critical patients.
In the following sections of the paper, we detail the objectives to be covered, describe the methodology used, analyze the results, and present the conclusions.
4. Results
In this section, we report on the different tasks involved in the proposed methodology, providing details of its application to the historical and current data.
4.4. Artificial Intelligence
This section contains the application of the AI techniques. Due to its complexity, we present it divided into subsections.
4.4.1. Application of Learning Techniques
The learning process was conducted using the following technologies: Python 3.6.9, with the Anaconda Navigator 2.5.2 distribution and the machine learning libraries FastAI 0.7.0, scikit-learn 0.24.2, and imbalanced-learn 0.8.1.
Two learning processes were carried out to obtain two different models: one to predict patient survival during their stay in the ICU and the other to predict the length of stay in the ICU.
For the estimation of the length of stay, we defined the separation into two types: those of short stay (less than or equal to three days) and those of long stay (more than three days).
To obtain the model, an initial approach was made to the data using the following machine learning algorithms that can be used for classification [
29]: Logistic Regression [
30], Random Forest [
31], Support Vector Machines [
32], Gradient Boosting [
33], Neural Network [
34] and K-Nearest Neighbors [
35].
Optimizations were performed on the different models, mainly using the class-balancing (SMOTE) and hyperparameter optimization (Grid Search) techniques.
Optimizations were then performed on the application of each of the machine learning techniques. The metrics were obtained and are indicated in
Table 5 and
Table 6.
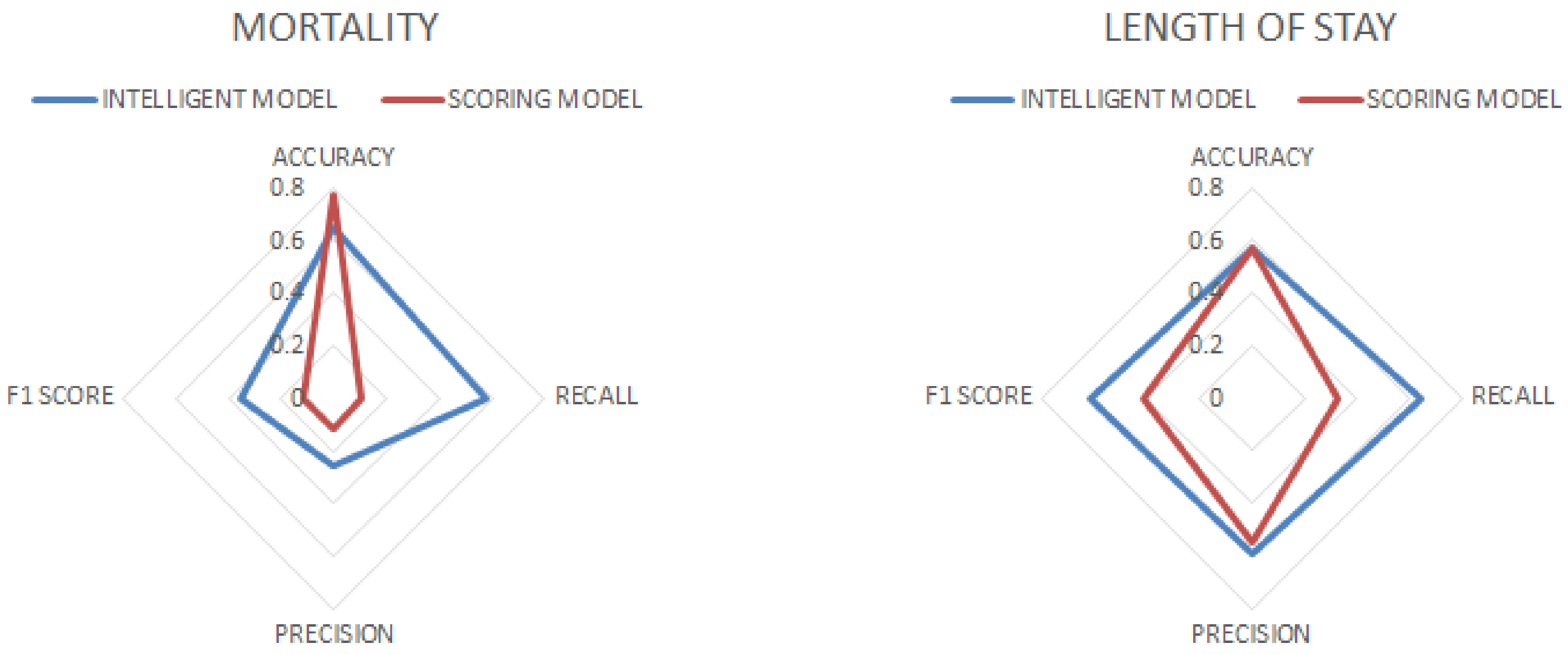
When analyzing the results, it could be seen that with respect to mortality, the K-Nearest Neighbors algorithm had the highest accuracy but a low recall, indicating that it tended to be accurate, but with a low efficiency in identifying positive cases. Gradient Boosting, Random Forest, and Neural Network showed balanced performances. Support Vector Machines had high recall, minimizing false negatives, but with low precision. Logistic Regression was the least effective. With respect to the stay, Logistic Regression had the highest F1 score, with a good balance between precision and recall, although with unremarkable accuracy and ROC AUC. Random Forest and Gradient Boosting showed balanced results for all metrics. Neural Network had high precision but medium recall, and K-Nearest Neighbors has low accuracy, with no outstanding results in any metric.
We then implemented a staking model using Gradient Boosting and SVC as base models. We used Gradient Boosting for mortality because it had high accuracy, a relatively high ROC, and could distinguish effectively between patient types, while for length of stay, it presented a balanced performance with reasonable accuracy and F1 score values. We used SVC for mortality because it had high recall, which is critical to predict deaths correctly and minimize negative biases.
We used Logistic Regression as the meta-classifier because it is computationally simple and allows for easy interpretation of the results.
Once the stacking model was implemented, class balancing and hyperparameter optimization techniques were utilized. The metrics were obtained and are indicated in
Table 7.
The metrics obtained were superior to those of the base models, especially for length of stay.
4.4.2. Model Validation
The results achieved in the first approximations already suggested the need to make adjustments to obtain more robust models with more conclusive metrics.
For the optimization, the poor performances of the recall and f-beta indicators were analyzed. There were imbalance problems in the data set used. This was the case for the ICU survival data (survival during hospital admission in the critical unit); from a total of 24,876 patients, the number of patients who survived (20,731) was much higher than the number of patients who died (4145).
It was necessary to consider that, in clinical contexts, it is very common for data to be unbalanced. That is, there is an unequal number of elements belonging to one class or another.
To improve the obtained results, resampling techniques and the generation of synthetic samples were carried out [
36], which are recommended to compensate for the imbalance in the data set [
37]—in this case, the disproportion in the survival rate.
Specifically, we obtained the best results by applying oversampling techniques, namely, the minority class synthetic oversampling technique [
38], and undersampling techniques, namely, Tomek links (optimization of the Condensed Nearest Neighbors technique [
39]).
In addition, a hyperparameter optimization process was carried out, specifically using Grid Search, to build and evaluate the models with different hyperparameter configurations until the most accurate model was obtained.
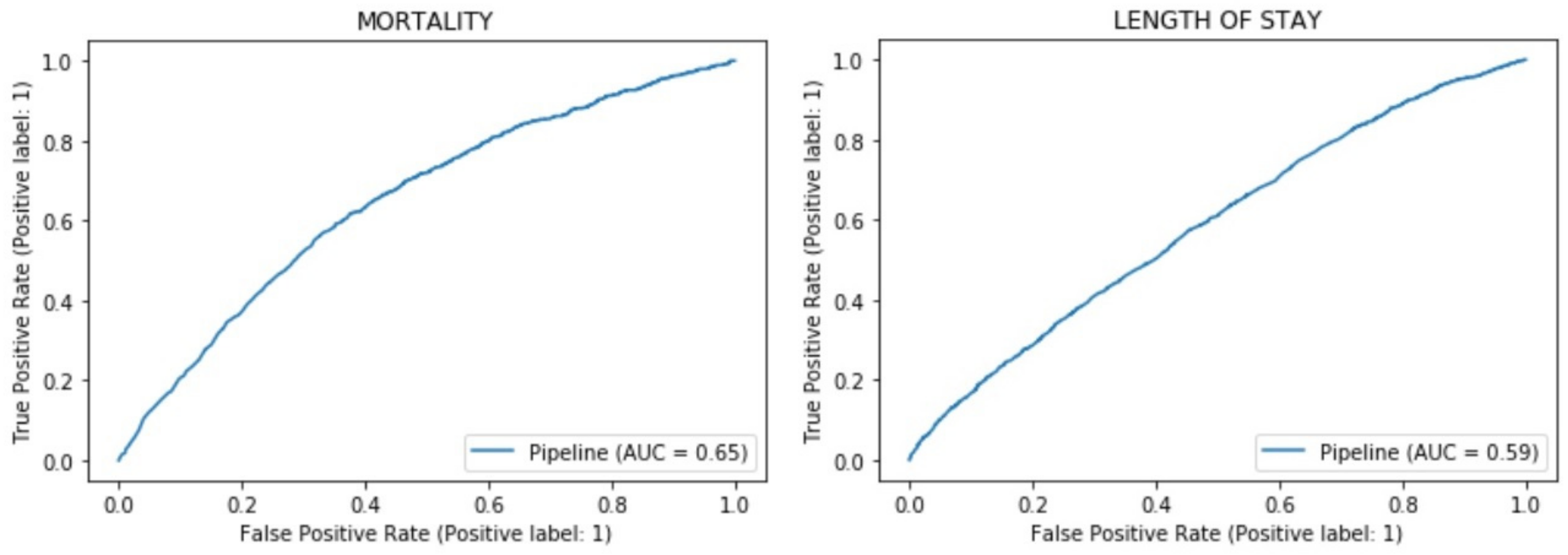
We calculated the Receiver Operating Characteristic (ROC) values, and the curve is displayed in
Figure 5.
The obtained metrics allowed for analysis of the model. In relation to mortality, the accuracy was reasonable, although the precision and recall were mediocre. Regarding length of stay, the accuracy was similar to that of mortality, and the recall and precision were much better. In both cases, the ROC AUC indicated that the predictions were valid, but it did not provide a definitive degree of confidence.
Author Contributions
Conceptualization, S.O.-T. and E.M.B.; methodology, S.O.-T. and J.-M.G.-M.; software, E.M.B.; validation, E.M.B. and S.O.-T.; formal analysis, E.M.B. and A.C.-M.; investigation, E.M.B. and J.-M.G.-M.; resources, E.M.B.; data curation, E.M.B.; writing—original draft preparation, E.M.B.; writing—review and editing, J.-M.G.-M.; visualization, A.C.-M. and J.-M.G.-M.; supervision, S.O.-T. and J.-M.G.-M.; project administration, E.M.B. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Due to the sensitive nature of the data, the clinical center that provided the data has not authorized public distribution, so supporting data are not available.
Conflicts of Interest
The authors declare no conflicts of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
References
- Danciu, I.; Cowan, J.D.; Basford, M.; Wang, X.; Saip, A.; Osgood, S.; Shirey-Rice, J.; Kirby, J.; Harris, P.A. Secondary use of clinical data: The Vanderbilt approach. J. Biomed. Inform. 2014, 52, 28–35. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.C.; Islam, S.U.; Noor, A.; Khan, S.; Afsar, W.; Nazir, S. Influential Usage of Big Data and Artificial Intelligence in Healthcare. Comput. Math. Methods Med. 2021, 2021, e5812499. [Google Scholar] [CrossRef] [PubMed]
- PKFA What is Artificial Intelligence? “Success is no accident It is hard work, perseverance, learning, studying, sacrifice and most of all, love of what you are doing or learning to do”. 1984; p. 65.
- Jiang, F.; Jiang, Y.; Zhi, H.; Dong, Y.; Li, H.; Ma, S.; Wang, Y.; Dong, Q.; Shen, H.; Wang, Y. Artificial intelligence in healthcare: Past, present and future. Stroke Vasc. Neurol. 2017, 2, 230–243. [Google Scholar] [CrossRef] [PubMed]
- Hayes-Roth, F. Artificial Intelligence: What Works and What Doesn’t? AI Mag. 1997, 18, 99. [Google Scholar]
- Xu, Y.; Liu, X.; Cao, X.; Huang, C.; Liu, E.; Qian, S.; Liu, X.; Wu, Y.; Dong, F.; Qiu, C.-W.; et al. Artificial intelligence: A powerful paradigm for scientific research. Innovation 2021, 2, 100179. [Google Scholar] [CrossRef]
- Shamout, F.; Zhu, T.; Clifton, D.A. Machine Learning for Clinical Outcome Prediction. IEEE Rev. Biomed. Eng. 2021, 14, 116–126. [Google Scholar] [CrossRef]
- Kim, S.H.; Chan, C.W.; Olivares, M.; Escobar, G. ICU Admission Control: An Empirical Study of Capacity Allocation and Its Implication for Patient Outcomes. Manag. Sci. 2015, 61, 19–38. [Google Scholar] [CrossRef]
- Castelvecchi, D. Can we open the black box of AI? Nat. News 2016, 538, 20. [Google Scholar] [CrossRef]
- Adadi, A.; Berrada, M. Peeking Inside the Black-Box: A Survey on Explainable Artificial Intelligence (XAI). IEEE Access 2018, 6, 52138–52160. [Google Scholar] [CrossRef]
- Wójcikowski, M.; Ballester, P.J.; Siedlecki, P. Performance of machine-learning scoring functions in structure-based virtual screening. Sci. Rep. 2017, 7, 46710. [Google Scholar] [CrossRef]
- Keuning, B.E.; Kaufmann, T.; Wiersema, R.; Granholm, A.; Pettilä, V.; Møller, M.H.; Christiansen, C.F.; Forte, J.C.; Snieder, H.; Keus, F.; et al. Mortality prediction models in the adult critically ill: A scoping review. Acta Anaesthesiol. Scand. 2020, 64, 424–442. [Google Scholar] [CrossRef] [PubMed]
- Christodoulou, E.; Ma, J.; Collins, G.S.; Steyerberg, E.W.; Verbakel, J.Y.; Van Calster, B. A systematic review shows no performance benefit of machine learning over logistic regression for clinical prediction models. J. Clin. Epidemiol. 2019, 110, 12–22. [Google Scholar] [CrossRef] [PubMed]
- Nemati, S.; Holder, A.; Razmi, F.; Stanley, M.D.; Clifford, G.D.; Buchman, T.G. An Interpretable Machine Learning Model for Accurate Prediction of Sepsis in the ICU. Crit. Care Med. 2018, 46, 547–553. [Google Scholar] [CrossRef] [PubMed]
- Shen, C.; Hu, Y.; Wang, Z.; Zhang, X.; Zhong, H.; Wang, G.; Yao, X.; Xu, L.; Cao, D.; Hou, T. Can machine learning consistently improve the scoring power of classical scoring functions? Insights into the role of machine learning in scoring functions. Brief. Bioinform. 2021, 22, 497–514. [Google Scholar] [CrossRef] [PubMed]
- Shung, D.L.; Au, B.; Taylor, R.A.; Tay, J.K.; Laursen, S.B.; Stanley, A.J.; Dalton, H.R.; Ngu, J.; Schultz, M.; Laine, L. Validation of a Machine Learning Model That Outperforms Clinical Risk Scoring Systems for Upper Gastrointestinal Bleeding. Gastroenterology 2020, 158, 160–167. [Google Scholar] [CrossRef]
- Lombardozzi, K.; Bible, S.; Eckman, J.; Hamrick, A.; Kellett, N.; Burnette, J.; Cox, D.; Justice, L.; Bendyk, H.; Morrow, C. Evaluation of efficiency and accuracy of a streamlined data entry process into an outcomes database. In Critical Care Medicine; Lippincott Williams & Wilkins: Philadelphia, PA, USA, 2009; p. A370. [Google Scholar]
- Juba, B.; Le, H.S. Precision-Recall versus Accuracy and the Role of Large Data Sets. In Proceedings of the AAAI Conference on Artificial Intelligence, Honolulu, HI, USA, 27 January–1 February 2019; Volume 33, pp. 4039–4048. [Google Scholar]
- Kramer, A.A.; Higgins, T.L.; Zimmerman, J.E. Comparing Observed and Predicted Mortality Among ICUs Using Different Prognostic Systems: Why Do Performance Assessments Differ? Crit. Care Med. 2015, 43, 261–269. [Google Scholar] [CrossRef] [PubMed]
- Thompson, D.; Hsu, Y.; Chang, B.; Marsteller, J. Impact of nursing staffing on patient outcomes in intensive care unit. J. Nurs. Care 2013, 2, 128. [Google Scholar]
- Maldonado Belmonte, E.; Otón Tortosa, S. Design and implementation of a predictive system in a clinical intensive care unit. In Aula de Tecnologías de la Información y Comunicaciones Avanzadas (ATICA); Universidad de Alcalá: Madrid, Spain, 2017; pp. 733–739. [Google Scholar]
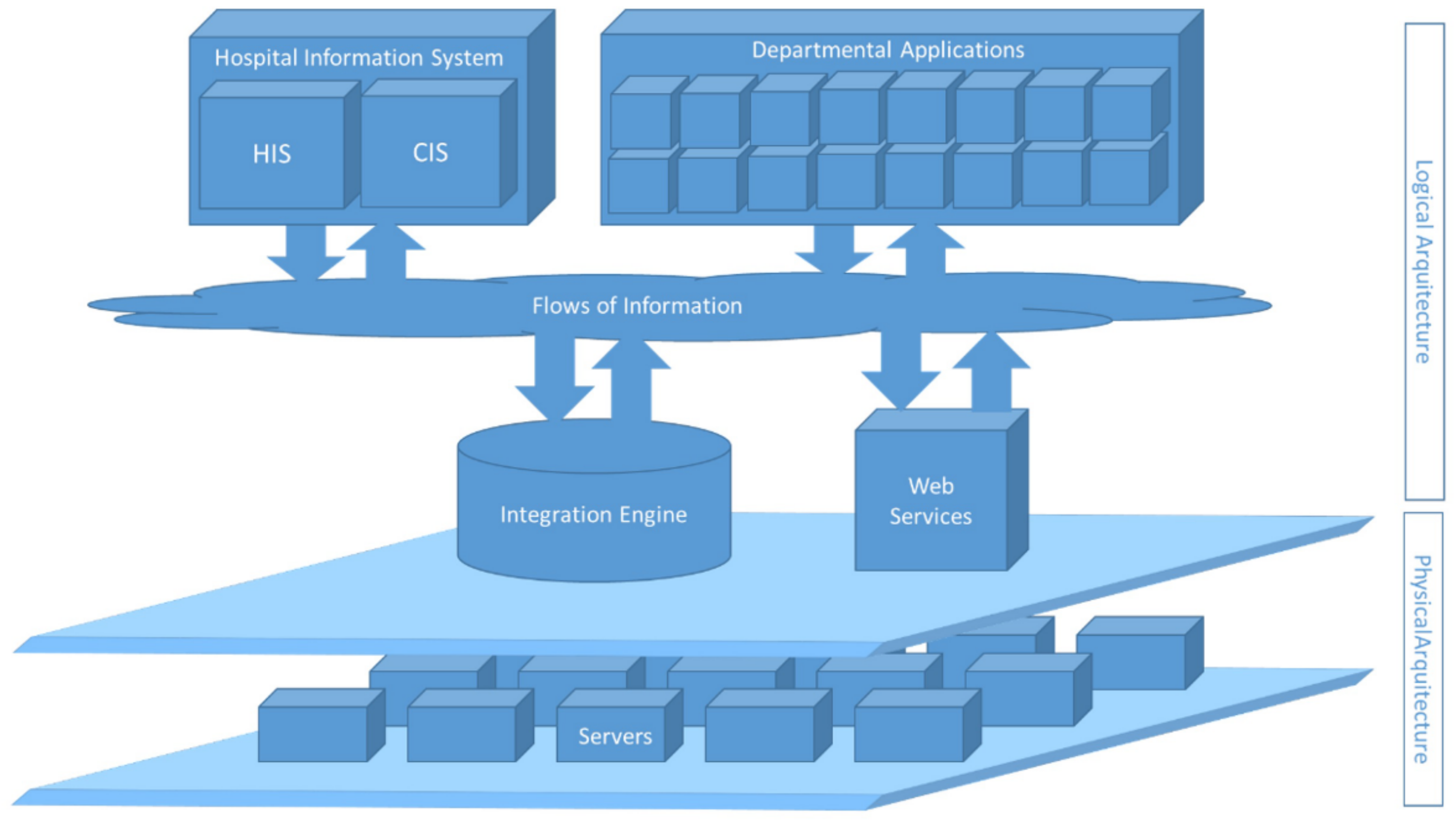
- Maldonado Belmonte, E.; Otón Tortosa, S.; Ruggia Frick, R.J. Proposal for a Standard Architecture for the Integration of Clinical Information Systems in a Complex Hospital Environment. Informatics 2021, 8, 87. [Google Scholar] [CrossRef]
- Hettinger, B.J.; Brazile, R.P. Health Level Seven (HL7): Standard for healthcare electronic data transmissions. Comput. Nurs. 1994, 12, 13–16. [Google Scholar]
- Scope and Field of Application. Available online: https://dicom.nema.org/medical/dicom/current/output/chtml/part01/chapter_1.html#sect_1.1 (accessed on 17 March 2022).
- The Value of SNOMEDCT, S.N.O.M.E.D. Available online: https://www.snomed.org/snomed-ct/why-snomed-ct (accessed on 17 March 2022).
- International Classification of Diseases (ICD). Available online: https://www.who.int/standards/classifications/classification-of-diseases (accessed on 17 March 2022).
- Home, L.O.I.N.C. Available online: https://loinc.org/ (accessed on 17 March 2022).
- Maldonado Belmonte, E.; Pérez Vallina, M.; López Martínez, R.; Cuadrado Martínez, V.; Nájera Cano, S. Design and implementation of a clinical information standardization platform: Moving towards semantic interoperability. In Proceedings of the National Congress on Health Informatics; SEIS, Sociedad Española de Informática de la Salud: Madrid, Spain, 2021; Volume 1, pp. 303–305. [Google Scholar]
- Gong, D. Top 6 Machine Learning Algorithms for Classification. Medium 2022. Available online: https://towardsdatascience.com/top-machine-learning-algorithms-for-classification-2197870ff501 (accessed on 4 April 2022).
- Dreiseitl, S.; Ohno-Machado, L. Logistic regression and artificial neural network classification models: A methodology review. J. Biomed. Inform. 2002, 35, 352–359. [Google Scholar] [CrossRef]
- Breiman, L. Random Forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Cervantes, J.; Garcia-Lamont, F.; Rodríguez-Mazahua, L.; Lopez, A. A comprehensive survey on support vector machine classification: Applications, challenges and trends. Neurocomputing 2020, 408, 189–215. [Google Scholar] [CrossRef]
- Bentéjac, C.; Csörgő, A.; Martínez-Muñoz, G. A comparative analysis of gradient boosting algorithms. Artif. Intell. Rev. 2021, 54, 1937–1967. [Google Scholar] [CrossRef]
- Zhang, G.P. Neural networks for classification: A survey. IEEE Trans. Syst. Man Cybern. Part C (Appl. Rev.) 2000, 30, 451–462. [Google Scholar] [CrossRef]
- Mucherino, A.; Papajorgji, P.J.; Pardalos, P.M. (Eds.) k-Nearest Neighbor Classification. In Data Mining in Agriculture; Springer Optimization and Its Applications; Springer: New York, NY, USA, 2009; pp. 83–106. [Google Scholar] [CrossRef]
- More, A. Survey of resampling techniques for improving classification performance in unbalanced datasets. arXiv 2016, arXiv:1608.06048. [Google Scholar]
- Wei, Q.; Dunbrack, R. The Role of Balanced Training and Testing Data Sets for Binary Classifiers in Bioinformatics. PLoS ONE 2013, 8, e67863. [Google Scholar] [CrossRef]
- Chawla, N.V.; Bowyer, K.W.; Hall, L.O.; Kegelmeyer, W.P. SMOTE: Synthetic Minority Over-sampling Technique. J. Artif. Intell. Res. 2002, 16, 321–357. [Google Scholar] [CrossRef]
- Sain, H.; Purnami, S.W. Combine Sampling Support Vector Machine for Imbalanced Data Classification. Procedia Comput. Sci. 2015, 72, 59–66. [Google Scholar] [CrossRef]
- Summary of NQF-Endorsed Intensive Care Outcomes Models for Risk Adjusted Mortality and Length of Stay (ICOMmort and ICOMlos). 3. Philip R. Lee Institute for Health Policy Studies. Available online: https://healthpolicy.ucsf.edu/icu-outcomes (accessed on 10 December 2022).
- Hourmant, Y.; Mailloux, A.; Valade, S.; Lemiale, V.; Azoulay, E.; Darmon, M. Impact of early ICU admission on outcome of critically ill and critically ill cancer patients: A systematic review and meta-analysis. J. Crit. Care 2021, 61, 82–88. [Google Scholar] [CrossRef]
- Pinsky, M.R. Hemodynamic Evaluation and Monitoring in the ICU. Chest 2007, 132, 2020–2029. [Google Scholar] [CrossRef]
- Majumdar, M. Haemodynamic Monitoring in the Intensive Care Unit. In Intensive Care; IntechOpen: London, UK, 2017. [Google Scholar] [CrossRef]
- Keszler, M. State of the art in conventional mechanical ventilation. J. Perinatol. 2009, 29, 262–275. [Google Scholar] [CrossRef] [PubMed]
- Sise, M.J.; Shackford, S.R.; Sise, C.B.; Sack, D.I.; Paci, G.M.; Yale, R.S.; O’Reilly, E.B.; Norton, V.C.; Huebner, B.R.; Peck, K.A. Early Intubation in the Management of Trauma Patients: Indications and Outcomes in 1,000 Consecutive Patients. J. Trauma. Acute Care Surg. 2009, 66, 32. [Google Scholar] [CrossRef] [PubMed]
| Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).