In Silico Structural Protein Evaluation of the Phenylalanine Hydroxylase p.(Tyr77His) Variant Associated with Benign Hyperphenylalaninemia as Identified through Mexican Newborn Screening
Abstract
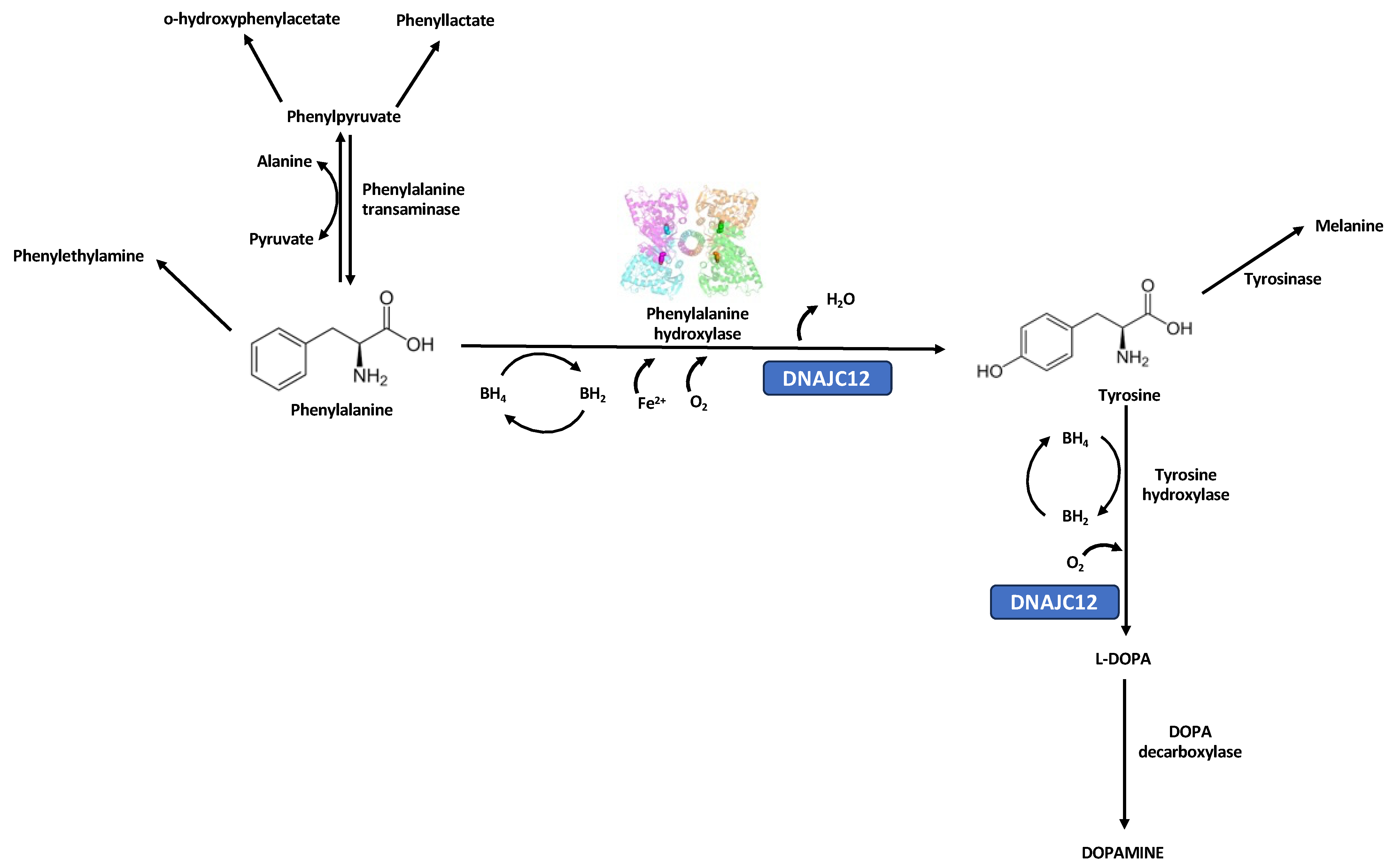
1. Introduction
2. Materials and Methods
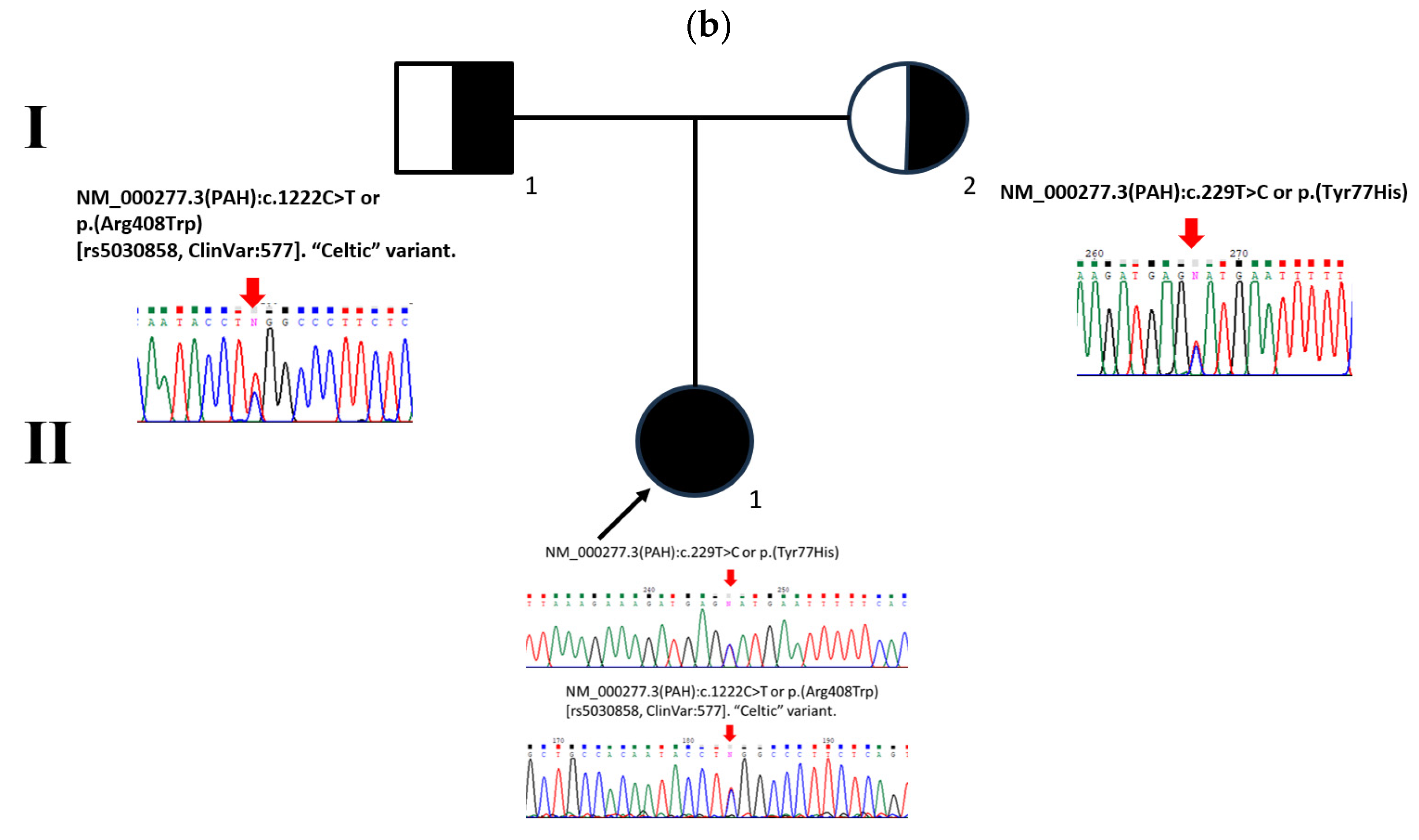
2.1. Patient Description
2.2. Biochemical Confirmatory Analysis
2.3. PAH Genotype Analysis
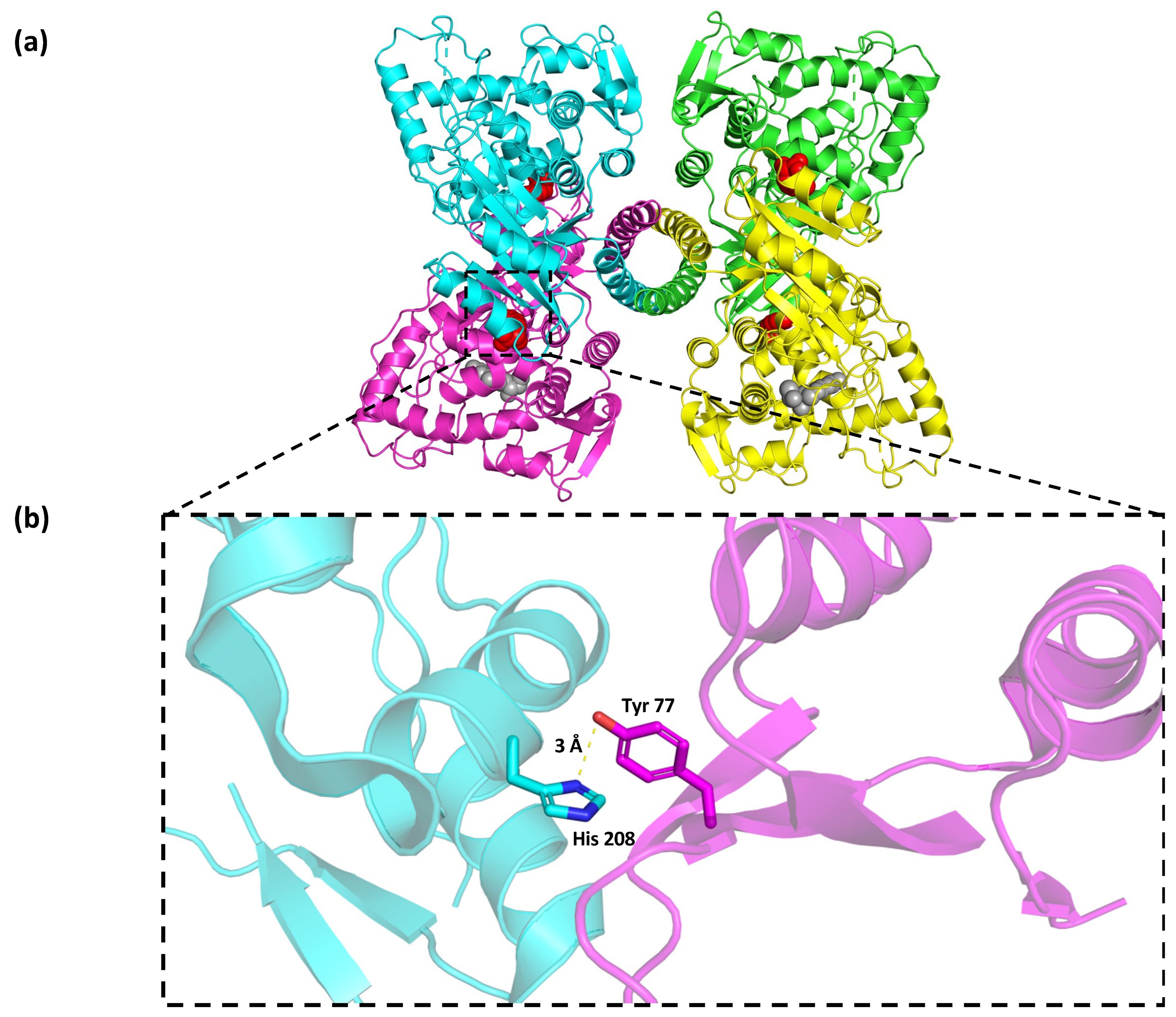
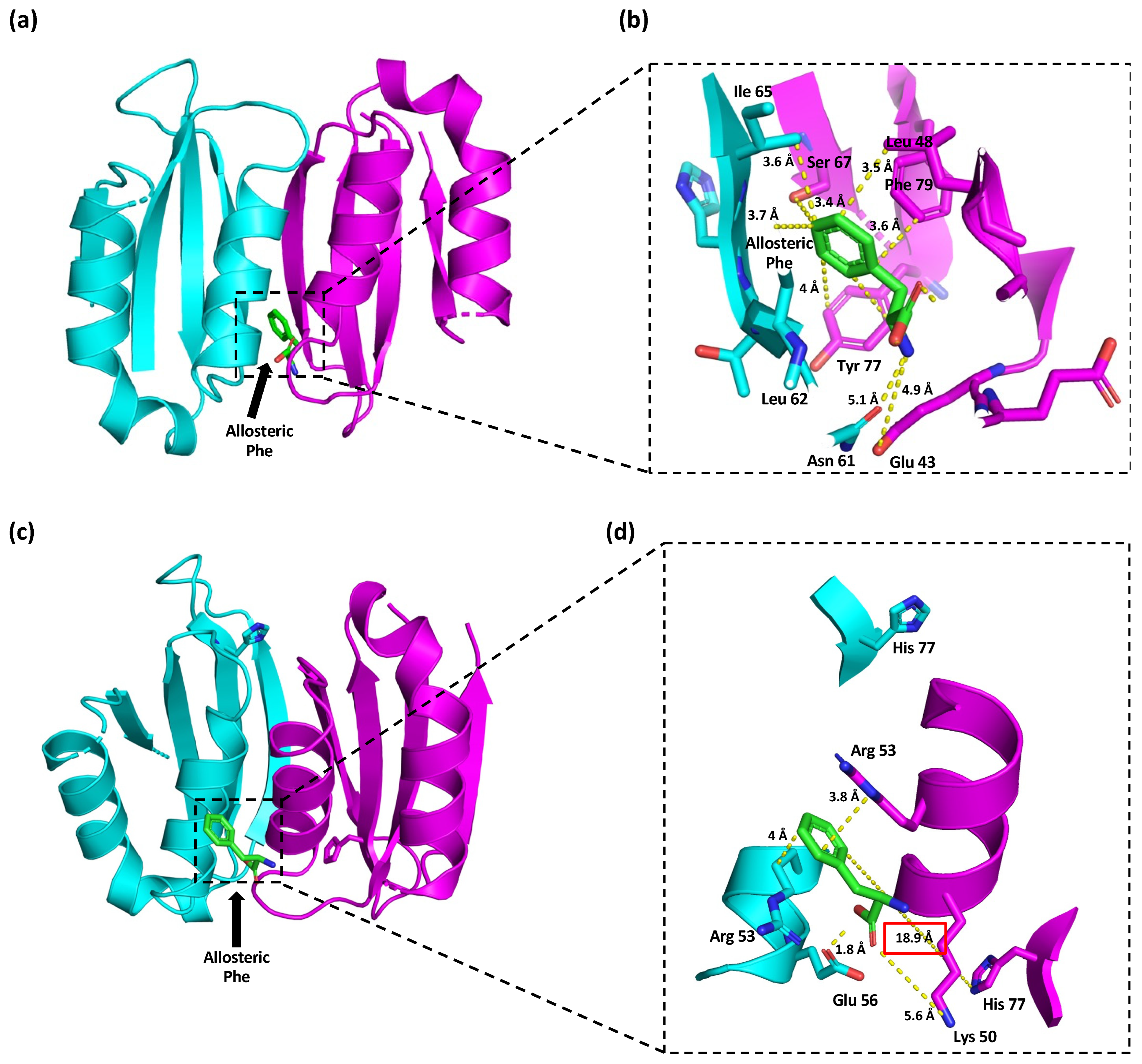
2.4. In Silico Protein Modeling, Docking, and Mutational Analyses of p.(Tyr77His) PAH Variant
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Scriver, C.R.; Clow, C.L. Phenylketonuria: Epitome of human biochemical genetics. N. Engl. J. Med. 1980, 303, 1336–1342. [Google Scholar] [CrossRef]
- van Spronsen, F.J.; Blau, N.; Harding, C.; Burlina, A.; Longo, N.; Bosch, A.M. Phenylketonuria. Nat. Rev. Dis. Primers 2021, 7, 36. [Google Scholar] [CrossRef] [PubMed]
- Blau, N.; Van Spronsen, F.J.; Levy, H.L. Phenylketonuria. Lancet 2010, 376, 1417–1427. [Google Scholar] [CrossRef] [PubMed]
- Cleary, M.A.; Skeath, R. Phenylketonuria. Paediatr. Child Health 2019, 29, 111–115. [Google Scholar] [CrossRef]
- Rausell, D.; García-Blanco, A.; Correcher, P.; Vitoria, I.; Vento, M.; Cháfer-Pericás, C. Newly validated biomarkers of brain damage may shed light into the role of oxidative stress in the pathophysiology of neurocognitive impairment in dietary restricted phenylketonuria patients. Pediatr. Res. 2019, 85, 242–250. [Google Scholar] [CrossRef] [PubMed]
- Borrajo, G.J. Newborn screening in Latin America: A brief overview of the state of the art. Proc. Am. J. Med. Genet. Part C Semin. Med. Genet. 2021, 187, 322–328. [Google Scholar] [CrossRef] [PubMed]
- García-Flores, E.P.; Herrera-Maldonado, N.; Hinojosa-Trejo, M.A.; Vergara-Vázquez, M.; Halley-Castillo, M.E. Avances y logros del programa de tamiz metabólico neonatal (2012–2018). Acta Pediátr. Méx. 2018, 39, 57–65. [Google Scholar] [CrossRef]
- Hillert, A.; Anikster, Y.; Belanger-Quintana, A.; Burlina, A.; Burton, B.K.; Carducci, C.; Chiesa, A.E.; Christodoulou, J.; Đorđević, M.; Desviat, L.R. The genetic landscape and epidemiology of phenylketonuria. Am. J. Hum. Genet. 2020, 107, 234–250. [Google Scholar] [CrossRef]
- Remec, Z.I.; Trebusak Podkrajsek, K.; Repic Lampret, B.; Kovac, J.; Groselj, U.; Tesovnik, T.; Battelino, T.; Debeljak, M. Next-generation sequencing in newborn screening: A review of current state. Front. Genet. 2021, 12, 662254. [Google Scholar] [CrossRef]
- Pollitt, R. Commentary: What degree of hyperphenylalaninaemia requires treatment? J. Inherit. Metab. Dis. 2012, 35, 927–930. [Google Scholar] [CrossRef]
- van Spronsen, F.J. Mild hyperphenylalaninemia: To treat or not to treat. J. Inherit. Metab. Dis. 2011, 34, 651–656. [Google Scholar] [CrossRef] [PubMed]
- Blau, N. BIOPKU Data Base. Available online: http://www.biopku.org (accessed on 3 October 2023).
- Himmelreich, N.; Shen, N.; Okun, J.G.; Thiel, C.; Hoffmann, G.F.; Blau, N. Relationship between genotype, phenylalanine hydroxylase expression and in vitro activity and metabolic phenotype in phenylketonuria. Mol. Genet. Metab. 2018, 125, 86–95. [Google Scholar] [CrossRef]
- Vela-Amieva, M.; Alcántara-Ortigoza, M.A.; Ibarra-González, I.; González-del Angel, A.; Fernández-Hernández, L.; Guillén-López, S.; López-Mejía, L.; Carrillo-Nieto, R.I.; Belmont-Martínez, L.; Fernández-Lainez, C. An Updated PAH Mutational Spectrum of Phenylketonuria in Mexican Patients Attending a Single Center: Biochemical, Clinical-Genotyping Correlations. Genes 2021, 12, 1676. [Google Scholar] [CrossRef] [PubMed]
- Vela-Amieva, M.; Abreu-Gonzalez, M.; Gonzalez-del Angel, A.; Ibarra-Gonzalez, I.; Fernandez-Lainez, C.; Barrientos-Rios, R.; Monroy-Santoyo, S.; Guillén-López, S.; Alcántara-Ortigoza, M. Phenylalanine hydroxylase deficiency in Mexico: Genotype–phenotype correlations, BH4 responsiveness and evidence of a founder effect. Clin. Genet. 2015, 88, 62–67. [Google Scholar] [CrossRef] [PubMed]
- Ohlsson, A.; Bruhn, H.; Nordenström, A.; Zetterström, R.H.; Wedell, A.; von Döbeln, U. The Spectrum of PAH Mutations and Increase of Milder Forms of Phenylketonuria in Sweden during 1965–2014. In JIMD Reports; Springer: Cham, Switzerland, 2016; Volume 34, pp. 19–26. [Google Scholar]
- Hill, D.; Burnworth, L.; Skea, W.; Pfeifer, R. Quantitative HPLC analysis of plasma amino acids as orthophthaldialdehyde/ethanethiol derivatives. J. Liq. Chromatogr. 1982, 5, 2369–2393. [Google Scholar] [CrossRef]
- Flydal, M.I.; Alcorlo-Pagés, M.; Johannessen, F.G.; Martínez-Caballero, S.; Skjærven, L.; Fernandez-Leiro, R.; Martinez, A.; Hermoso, J.A. Structure of full-length human phenylalanine hydroxylase in complex with tetrahydrobiopterin. Proc. Natl. Acad. Sci. USA 2019, 116, 11229–11234. [Google Scholar] [CrossRef] [PubMed]
- Patel, D.; Kopec, J.; Fitzpatrick, F.; McCorvie, T.J.; Yue, W.W. Structural basis for ligand-dependent dimerization of phenylalanine hydroxylase regulatory domain. Sci. Rep. 2016, 6, 23748. [Google Scholar] [CrossRef]
- DeLano, W.L. Pymol: An open-source molecular graphics tool. CCP4 Newsl. Protein Crystallogr. 2002, 40, 82–92. [Google Scholar]
- Laskowski, R.A.; Jabłońska, J.; Pravda, L.; Vařeková, R.S.; Thornton, J.M. PDBsum: Structural summaries of PDB entries. Protein Sci. 2018, 27, 129–134. [Google Scholar] [CrossRef]
- Krieger, E.; Joo, K.; Lee, J.; Lee, J.; Raman, S.; Thompson, J.; Tyka, M.; Baker, D.; Karplus, K. Improving physical realism, stereochemistry, and side-chain accuracy in homology modeling: Four approaches that performed well in CASP8. Proteins Struct. Funct. Bioinform. 2009, 77, 114–122. [Google Scholar] [CrossRef]
- Grosdidier, A.; Zoete, V.; Michielin, O. SwissDock, a protein-small molecule docking web service based on EADock DSS. Nucleic Acids Res. 2011, 39, W270–W277. [Google Scholar] [CrossRef] [PubMed]
- Genome Aggregation Database (gnomAD). Available online: https://gnomad.broadinstitute.org/ (accessed on 3 October 2023).
- ClinVar NCBI. Available online: https://www.ncbi.nlm.nih.gov/clinvar/ (accessed on 3 October 2023).
- Richards, S.; Aziz, N.; Bale, S.; Bick, D.; Das, S.; Gastier-Foster, J.; Grody, W.W.; Hegde, M.; Lyon, E.; Spector, E. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015, 17, 405–423. [Google Scholar] [CrossRef] [PubMed]
- Kleinberger, J.; Maloney, K.A.; Pollin, T.I.; Jeng, L.J.B. An openly available online tool for implementing the ACMG/AMP standards and guidelines for the interpretation of sequence variants. Genet. Med. 2016, 18, 1165. [Google Scholar] [CrossRef] [PubMed]
- DiLella, A.G.; Marvi, J.; Brayton, K.; Woo, S.L. An amino-acid substitution involved in phenylketonuria is in linkage disequilibrium with DNA haplotype 2. Nature 1987, 327, 333–336. [Google Scholar] [CrossRef] [PubMed]
- Desviat, L.; Pérez, B.; Gutierrez, E.; Sanchez, A.; Barrios, B.; Ugarte, M. Molecular basis of phenylketonuria in Cuba. Hum. Mutat. 2001, 18, 252. [Google Scholar] [CrossRef] [PubMed]
- Acosta, P.; Matalon, K.M. Nutrition Management of Patients with Inherited Disorders of Aromatic Amino Acid Metabolism. In Nutrition Management of Patients with Inherited Metabolic Disorders, 1st ed.; Phyllis, B.A., Ed.; Jones and Bartlett Publishers: Sudbury, MA, USA, 2010; pp. 119–153. [Google Scholar]
- Kobe, B.; Jennings, I.G.; House, C.M.; Michell, B.J.; Goodwill, K.E.; Santarsiero, B.D.; Stevens, R.C.; Cotton, R.G.; Kemp, B.E. Structural basis of autoregulation of phenylalanine hydroxylase. Nat. Struct. Biol. 1999, 6, 442–448. [Google Scholar] [CrossRef]
- Gersting, S.W.; Kemter, K.F.; Staudigl, M.; Messing, D.D.; Danecka, M.K.; Lagler, F.B.; Sommerhoff, C.P.; Roscher, A.A.; Muntau, A.C. Loss of function in phenylketonuria is caused by impaired molecular motions and conformational instability. Am. J. Hum. Genet. 2008, 83, 5–17. [Google Scholar] [CrossRef]
- Flydal, M.I.; Martinez, A. Phenylalanine hydroxylase: Function, structure, and regulation. IUBMB Life 2013, 65, 341–349. [Google Scholar] [CrossRef]
- Guldberg, P.; Rey, F.; Zschocke, J.; Romano, V.; François, B.; Michiels, L.; Ullrich, K.; Hoffmann, G.F.; Burgard, P.; Schmidt, H. A European multicenter study of phenylalanine hydroxylase deficiency: Classification of 105 mutations and a general system for genotype-based prediction of metabolic phenotype. Am. J. Hum. Genet. 1998, 63, 71–79. [Google Scholar] [CrossRef]
- Regier DS, G.C. Phenylalanine Hydroxylase Deficiency. Available online: https://www.ncbi.nlm.nih.gov/books/NBK1504/ (accessed on 12 September 2023).
- Lorey, F. Asian immigration and public health in California: Thalassemia in newborns in California. J. Pediatr. Hematol. Oncol. 2000, 22, 564–566. [Google Scholar] [CrossRef]
- Reich, S.; Hennermann, J.; Vetter, B.; Neumann, L.M.; Shin, Y.S.; Söling, A.; MÖnch, E.; Kulozik, A.E. An unexpectedly high frequency of hypergalactosemia in an immigrant Bosnian population revealed by newborn screening. Pediatr. Res. 2002, 51, 598–601. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Perfil Migratorio de México Boletín Anual 2022. Organización Internacional para las Migraciónes (OIM). Organización de las Naciones Unidas (ONU) Migración. Available online: https://mexico.iom.int/sites/g/files/tmzbdl1686/files/documents/2023-03/Perfil%20Migratorio-%20Boletin%20Anual%202022%20%283%29.pdf (accessed on 3 October 2023).
- Aguilar Martinez, P.; Angastiniotis, M.; Eleftheriou, A.; Gulbis, B.; Manu Pereira, M.D.M.; Petrova-Benedict, R.; Corrons, J.-L.V. Haemoglobinopathies in Europe: Health & Migration Policy Perspectives. Orphanet J. Rare Dis. 2014, 9, 97. [Google Scholar] [PubMed]



| Variant | c.229T>C or p.(Tyr77His) | c.1222C>T or p.(Arg408Trp) [rs5030858] |
|---|---|---|
| Variant type | Substitution | Substitution |
| Coding effect | Missense | Missense |
| Gene region | Exon 3 | Exon 12 |
| Protein domain | Regulatory | Catalytic |
| Enzyme activity | Unknown | 2% |
| Allelic phenotype value (APV) | Unknown | 0 |
| Worldwide allele frequency (gnomAD) [24] | Unknown | 0.0008910 |
| ClinVar [25] | Uncertain significance (ID: RCV003316903.1) | Pathogenic (ID: 577) |
| ACMG/AMP criteria for the interpretation of sequence variants [26,27] | Pathogenic IIIb (PS4, PM2, PM3, PP2, PP3, PP4) | Pathogenic IIb (PS3, PS4, PP1-S, PM1, PM2, PM3, PP1-M, PP1, PP2, PP3, PP4, PP5) |
| First reported | Ohlsson, et al., 2016 [16] | DiLella, et al., 1987 [28] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vela-Amieva, M.; Alcántara-Ortigoza, M.A.; González-del Angel, A.; Ibarra-González, I.; Fernández-Hernández, L.; Guillén-López, S.; López-Mejía, L.; Fernández-Lainez, C. In Silico Structural Protein Evaluation of the Phenylalanine Hydroxylase p.(Tyr77His) Variant Associated with Benign Hyperphenylalaninemia as Identified through Mexican Newborn Screening. Children 2023, 10, 1865. https://doi.org/10.3390/children10121865
Vela-Amieva M, Alcántara-Ortigoza MA, González-del Angel A, Ibarra-González I, Fernández-Hernández L, Guillén-López S, López-Mejía L, Fernández-Lainez C. In Silico Structural Protein Evaluation of the Phenylalanine Hydroxylase p.(Tyr77His) Variant Associated with Benign Hyperphenylalaninemia as Identified through Mexican Newborn Screening. Children. 2023; 10(12):1865. https://doi.org/10.3390/children10121865
Chicago/Turabian StyleVela-Amieva, Marcela, Miguel Angel Alcántara-Ortigoza, Ariadna González-del Angel, Isabel Ibarra-González, Liliana Fernández-Hernández, Sara Guillén-López, Lizbeth López-Mejía, and Cynthia Fernández-Lainez. 2023. "In Silico Structural Protein Evaluation of the Phenylalanine Hydroxylase p.(Tyr77His) Variant Associated with Benign Hyperphenylalaninemia as Identified through Mexican Newborn Screening" Children 10, no. 12: 1865. https://doi.org/10.3390/children10121865
APA StyleVela-Amieva, M., Alcántara-Ortigoza, M. A., González-del Angel, A., Ibarra-González, I., Fernández-Hernández, L., Guillén-López, S., López-Mejía, L., & Fernández-Lainez, C. (2023). In Silico Structural Protein Evaluation of the Phenylalanine Hydroxylase p.(Tyr77His) Variant Associated with Benign Hyperphenylalaninemia as Identified through Mexican Newborn Screening. Children, 10(12), 1865. https://doi.org/10.3390/children10121865







