Rodent Models of Audiogenic Epilepsy: Genetic Aspects, Advantages, Current Problems and Perspectives
Abstract
1. Introduction: The Relevance of the Problem
2. Classification and Etiology of Epilepsy
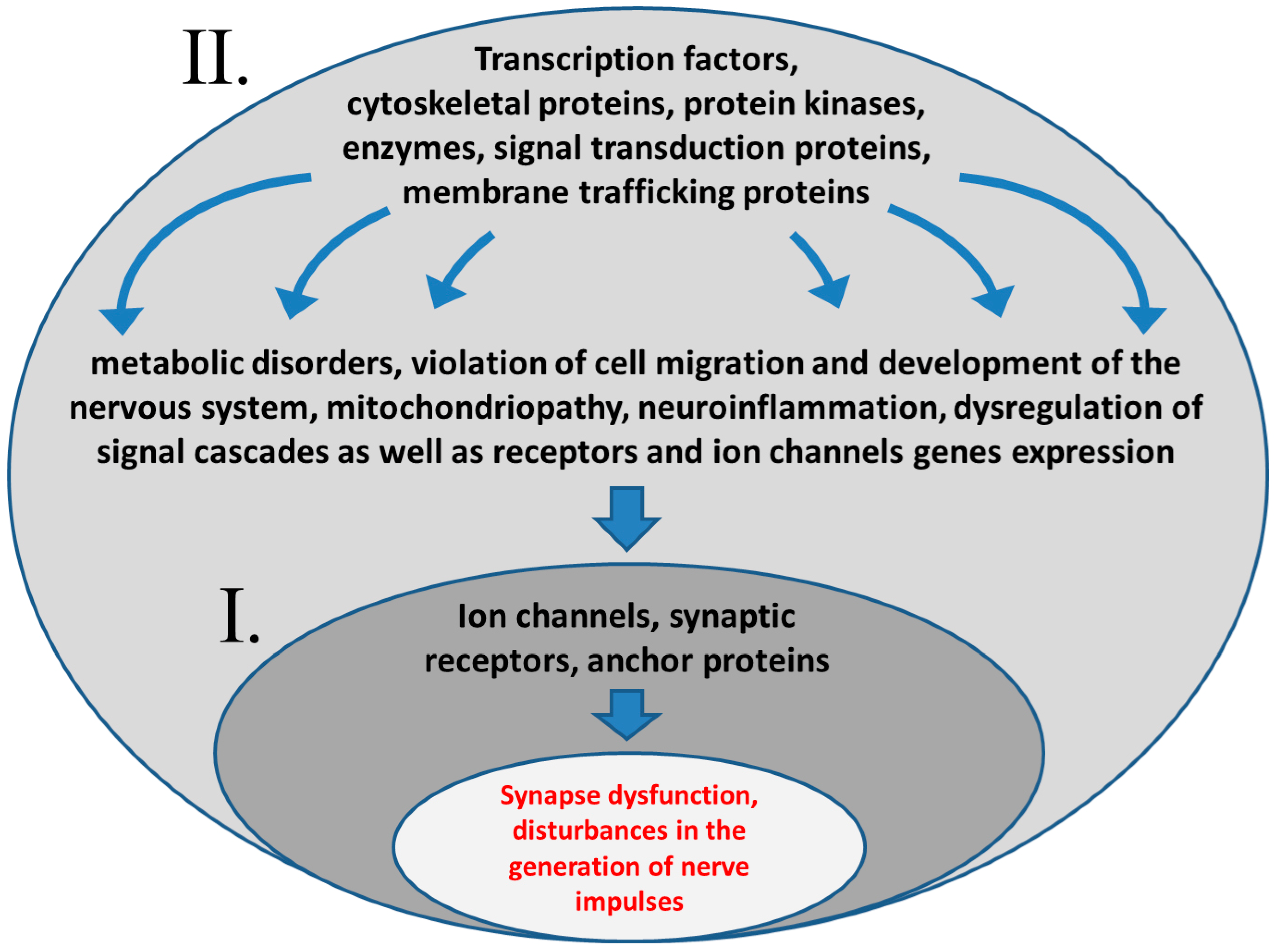
3. Pathophysiological Mechanisms of Epilepsy
4. Genetic Aspects of Human Epilepsy: Brief Review
5. Animal Models of Epilepsy
6. Pathophysiology of Audiogenic Seizures
7. Different Models of Audiogenic Epilepsy in Rodents
7.1. Overview of AE Strains Type
7.2. Monogenic Models
7.3. Models of Pyridoxine-Dependent Epilepsy and Angelman Syndrome
7.4. Models with Polygenic or Unknown Inheritance
| № | Species | Strain | Gene | Protein Function | References |
|---|---|---|---|---|---|
| Monogenic inheritance | |||||
| 1 | Mouse | Fmr1 knockout, model of Martin-Bell (fragile X chromosome) syndrome | Fmr1 | FMRP (fragile X mental retardation protein), mRNA transport | [161,162,163,164] |
| 2 | mouse | Frings | Vlgr1 | Very large G-protein coupled receptor 1, cellular adhesion | [22,23,24,165,166,167] |
| 3 | rat | LDE, lethal dwarfism with epilepsy rats | Wwox | WW domain-containing oxidoreductase | [172] |
| 4 | mouse | CNS-specific Wwox knockout mice | Wwox | WW domain-containing oxidoreductase | [173] |
| 5 | mouse | 5-HT2c receptor mutant mice | Htr2c | Serotonin receptor | [175] |
| 6 | mouse | Gabrb2-/- knockout | Gabrb2 | β2-subunit of the gamma-aminobutyric acid receptor | [184] |
| 7 | rat | Lgi1-mutant rats | Lgi1 | Secreted protein, modulator of disintegrin and metalloproteinase domain-containing proteins and K-channel activity | [191] |
| 8 | mouse | Lgi1-mutant mice | Lgi1 | Secreted protein, modulator of disintegrin and metalloproteinase domain-containing proteins and K-channel activity | [191] |
| 9 | rat | Kcnj16-knockout | Kcnj16 | Kir5.1 potassium channel | [192,193] |
| 10 | mouse | Black Swiss * | Gipc3 | GAIP-interacting protein C terminus, anchoring of the receptor proteins to the cytoskeleton | [194,195,196] |
| 11 | mouse | Tremor | Egr3 | Early growth response protein 3, transcription factor | [197] |
| 12 | mouse | Knockout of eukaryotic elongation factor 1Bδ (eEF1Bδ) long isoform | Eef1d | Eukaryotic elongation factor 1Bδ (eEF1Bδ) long isoform | [199] |
| 13 | mouse | Tef knockout mouse strain | Tef | Proline and acidic amino acid-rich basic leucine zipper (PAR bZip) transcription factor | [202] |
| 14 | mouse | Ube3a mutated mice, model of Angelman syndrome | Ube3a | Ubiquitin–protein ligase E3A | [204,207] |
| Polygenic or putatively polygenic inheritance | |||||
| 15 | mouse | DBA/2J | Kcnj10 | Kir4.1 potassium channel | [209,210] |
| 16 | mouse | 101/HY | unknown | Unknown | [213] |
| 17 | rat | GEPR, genetically epilepsy-prone rats | unknown | Unknown | [137,214,215] |
| 18 | rat | WAR, Wistar audiogenic rats | Hypothetically: Vlgr1 Chrna4 Grin2a Grin2b Kcnq3 Egr3 Ttr | Very large G-protein coupled receptor 1, cellular adhesion Nicotinic acetylcholine receptor Glutamate (NMDA) receptor subunit Glutamate (NMDA) receptor subunit Voltage-gated potassium channel Early growth response protein, transcription factor Transthyretin, transport protein | [83,168,198,217,218,222] |
| 19 | rat | KM, Krushinsky-Molodkina | Hypothetically: Ttr Msh3 Gstm1 | Transthyretin, transport protein MutS Homolog 3, DNA mismatch repair Glutathione S-transferase Mu 1, sulfur metabolism, detoxification | [115] |
| 20 | hamster | GASH/Sal, genetic audiogenic seizure hamster, Salamanca | Hypothetically: Cacna1a Cacna2d3 Grik1 Grin2c Zeb2 Egr3 Ttr Msh3 | Calcium voltage-gated channel subunit Calcium voltage-gated channel subunit Glutamate ionotropic receptor kainate type subunit 1 Glutamate (NMDA) receptor subunit ε-3 Zinc finger E-box-binding homeobox 2, transcription factor Early growth response protein, transcription factor Transthyretin, transport protein MutS Homolog 3, DNA mismatch repair | [217,219,221] |
| 21 | mouse | Ube3a-deleted mice, model of Angelman syndrome | Ube3a Atp10a Gabrb3 | Ubiquitin-protein ligase E3A Phospholipid-transporting ATPase VA (aminophospholipid translocase VA) β3-subunit of the gamma-aminobutyric acid receptor | [204] |
| 22 | mouse | Del(7Gabrb3-Ube3A), model of Angelman syndrome | Ube3a Atp10a Gabrb3 Gabra5 Gabrg3 Oca2 Herc2 | Ubiquitin-protein ligase E3A Phospholipid-transporting ATPase VA (aminophospholipid translocase VA) β3-subunit of the gamma-aminobutyric acid receptor α5-subunit of the gamma-aminobutyric acid receptor γ3-subunit of the gamma-aminobutyric acid receptor Melanocyte-specific transporter protein, membrane transport Giant E3 ubiquitin protein ligase | [204] |
8. The Use of Rodent Strains with AE in AEDs Screening
9. Near-Term Prospects and Conclusions
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| AE | audiogenic epilepsy |
| AEDs | antiepileptic drugs |
| AMPA | α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid |
| AS | Angelman syndrome |
| CAE | childhood absence epilepsy |
| CNS | central nervous system |
| DEEs | developmental epileptic encephalopathies |
| DEGs | differential-expressing genes |
| EAST | epilepsy, ataxia, sensorineural deafness, and tubulopathy |
| EEG | electroencephalography |
| EIEE | early infantile epileptic encephalopathy |
| ERK | extracellular signal-regulated kinase |
| GABA | gamma-aminobutyric acid |
| GASH/Sal | genetic audiogenic seizure hamster from Salamanca |
| GEPR | genetically epilepsy-prone rat |
| IC | inferior colliculi |
| ILAE | The International League Against Epilepsy |
| JAE | including juvenile absence epilepsy |
| JME | juvenile myoclonic epilepsy |
| KM | Krushinsky–Molodkina |
| MAG | myelin-associated glycoprotein |
| MAPK | mitogen-activated protein kinase |
| MES | maximal electroshock test |
| Msh3 | MutS Homolog 3 |
| mtDNA | mitochondrial DNA |
| mTOR | mammalian target of rapamycin |
| NMDA receptors | N-methyl-D-aspartate receptor |
| PAR bZip | proline and acidic amino acid-rich basic leucine zipper |
| PTZ | pentylentetrazole |
| SC | superior colliculi |
| TGFβ | transforming growth factor β |
| tRNA | transfer RNA |
| Ttr | transthyretin |
| WAR | Wistar audiogenic rat |
| WHO | World Health Organization |
| WOREE | WWOX-related epileptic encephalopathy |
| WWOX | WW-domain-containing oxidoreductase |
References
- Betjemann, J.P.; Lowenstein, D.H. Status epilepticus in adults. Lancet Neurol. 2015, 14, 615–624. [Google Scholar] [CrossRef]
- Zimmern, V.; Korff, C. Status Epilepticus in Children. J. Clin. Neurophysiol. 2020, 37, 429–433. [Google Scholar] [CrossRef] [PubMed]
- Bolton, P.F.; Carcani-Rathwell, I.; Hutton, J.; Goode, S.; Howlin, P.; Rutter, M. Epilepsy in autism: Features and correlates. Br. J. Psychiatry 2011, 198, 289–294. [Google Scholar] [CrossRef] [PubMed]
- Strasser, L.; Downes, M.; Kung, J.; Cross, J.H.; De Haan, M. Prevalence and risk factors for autism spectrum disorder in epilepsy: A systematic review and meta-analysis. Dev. Med. Child Neurol. 2018, 60, 19–29. [Google Scholar] [CrossRef]
- Alsaadi, T.; El Hammasi, K.; Shahrour, T.M.; Shakra, M.; Turkawi, L.; Almaskari, B.; Diab, L.; Raoof, M. Prevalence of depression and anxiety among patients with epilepsy attending the epilepsy clinic at Sheikh Khalifa Medical City, UAE: A cross-sectional study. Epilepsy Behav. 2015, 52, 194–199. [Google Scholar] [CrossRef]
- Josephson, C.B.; Jetté, N. Psychiatric comorbidities in epilepsy. Int. Rev. Psychiatry 2017, 29, 409–424. [Google Scholar] [CrossRef]
- Hassani, M.; Cooray, G.; Sveinsson, O.; Cooray, C. Post-stroke epilepsy in an ischemic stroke cohort-Incidence and diagnosis. Acta Neurol. Scand. 2020, 141, 141–147. [Google Scholar] [CrossRef]
- Fordington, S.; Manford, M. A review of seizures and epilepsy following traumatic brain injury. J. Neurol. 2020, 267, 3105–3111. [Google Scholar] [CrossRef]
- Wang, J.; Lin, Z.J.; Liu, L.; Xu, H.Q.; Shi, Y.W.; Yi, Y.H.; He, N.; Liao, W.P. Epilepsy-associated genes. Seizure 2017, 44, 11–20. [Google Scholar] [CrossRef]
- Perucca, P.; Bahlo, M.; Berkovic, S.F. The Genetics of Epilepsy. Annu. Rev. Genom. Hum. Genet. 2020, 21, 205–230. [Google Scholar] [CrossRef]
- Löscher, W.; Klitgaard, H.; Twyman, R.E.; Schmidt, D. New avenues for anti-epileptic drug discovery and development. Nat. Rev. Drug Discov. 2013, 12, 757–776. [Google Scholar] [CrossRef] [PubMed]
- Vermeulen, J.; Aldenkamp, A.P. Cognitive side-effects of chronic antiepileptic drug treatment: A review of 25 years of research. Epilepsy Res. 1995, 22, 65–95. [Google Scholar] [CrossRef]
- Chen, B.; Choi, H.; Hirsch, L.J.; Katz, A.; Legge, A.; Buchsbaum, R.; Detyniecki, K. Psychiatric and behavioral side effects of antiepileptic drugs in adults with epilepsy. Epilepsy Behav. 2017, 76, 24–31. [Google Scholar] [CrossRef] [PubMed]
- Carreno, M. Levetiracetam. Drugs Today 2007, 43, 769–794. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.L.; Zeng, L.N.; Li, Y.P. Side effects of phenobarbital in epilepsy: A systematic review. Epileptic Disord. 2011, 13, 349–365. [Google Scholar] [CrossRef]
- Shimada, T.; Yamagata, K. Pentylenetetrazole-Induced Kindling Mouse Model. J. Vis. Exp. 2018, 136, e56573. [Google Scholar] [CrossRef]
- Williamson, J.; Singh, T.; Kapur, J. Neurobiology of organophosphate-induced seizures. Epilepsy Behav. 2019, 101, 106426. [Google Scholar] [CrossRef]
- Patra, P.H.; Barker-Haliski, M.; White, H.S.; Whalley, B.J.; Glyn, S.; Sandhu, H.; Jones, N.; Bazelot, M.; Williams, C.M.; McNeish, A.J. Cannabidiol reduces seizures and associated behavioral comorbidities in a range of animal seizure and epilepsy models. Epilepsia 2019, 60, 303–314. [Google Scholar] [CrossRef]
- Banach, M.; Borowicz, K.K. Effects of Chronic Lamotrigine Administration on Maximal Electroshock- Induced Seizures in Mice. CNS Neurol. Disord. Drug Targets 2015, 14, 855–862. [Google Scholar] [CrossRef]
- Gonzalez, D.; Tomasek, M.; Hays, S.; Sridhar, V.; Ammanuel, S.; Chang, C.W.; Pawlowski, K.; Huber, K.M.; Gibson, J.R. Audiogenic Seizures in the. J. NeuroSci. 2019, 39, 9852–9863. [Google Scholar] [CrossRef]
- Ross, K.C.; Coleman, J.R. Developmental and genetic audiogenic seizure models: Behavior and biological substrates. NeuroSci. Biobehav. Rev. 2000, 24, 639–653. [Google Scholar] [CrossRef]
- Skradski, S.L.; Clark, A.M.; Jiang, H.; White, H.S.; Fu, Y.H.; Ptácek, L.J. A novel gene causing a mendelian audiogenic mouse epilepsy. Neuron 2001, 31, 537–544. [Google Scholar] [CrossRef]
- Yagi, H.; Takamura, Y.; Yoneda, T.; Konno, D.; Akagi, Y.; Yoshida, K.; Sato, M. Vlgr1 knockout mice show audiogenic seizure susceptibility. J. Neurochem. 2005, 92, 191–202. [Google Scholar] [CrossRef] [PubMed]
- Myers, K.A.; Nasioulas, S.; Boys, A.; McMahon, J.M.; Slater, H.; Lockhart, P.; Sart, D.D.; Scheffer, I.E. ADGRV1 is implicated in myoclonic epilepsy. Epilepsia 2018, 59, 381–388. [Google Scholar] [CrossRef]
- Doretto, M.C.; Fonseca, C.G.; Lôbo, R.B.; Terra, V.C.; Oliveira, J.A.; Garcia-Cairasco, N. Quantitative study of the response to genetic selection of the Wistar audiogenic rat strain (WAR). Behav. Genet. 2003, 33, 33–42. [Google Scholar] [CrossRef]
- Italiano, D.; Striano, P.; Russo, E.; Leo, A.; Spina, E.; Zara, F.; Striano, S.; Gambardella, A.; Labate, A.; Gasparini, S.; et al. Genetics of reflex seizures and epilepsies in humans and animals. Epilepsy Res. 2016, 121, 47–54. [Google Scholar] [CrossRef]
- Berg, A.T.; Berkovic, S.F.; Brodie, M.J.; Buchhalter, J.; Cross, J.H.; van Emde Boas, W.; Engel, J.; French, J.; Glauser, T.A.; Mathern, G.W.; et al. Revised terminology and concepts for organization of seizures and epilepsies: Report of the ILAE Commission on Classification and Terminology, 2005–2009. Epilepsia 2010, 51, 676–685. [Google Scholar] [CrossRef]
- Porter, R.J. The absence epilepsies. Epilepsia 1993, 34 (Suppl. S3), S42–S48. [Google Scholar] [CrossRef]
- Spencer, S.S. Neural networks in human epilepsy: Evidence of and implications for treatment. Epilepsia 2002, 43, 219–227. [Google Scholar] [CrossRef]
- Vinogradova, L.V. Interhemispheric difference in susceptibility to epileptogenesis: Evidence from the audiogenic kindling model in Wistar rats. Brain Res. 2010, 1329, 175–181. [Google Scholar] [CrossRef]
- Connolly, M.B. Dravet Syndrome: Diagnosis and Long-Term Course. Can. J. Neurol. Sci. 2016, 43 (Suppl. S3), S3–S8. [Google Scholar] [CrossRef] [PubMed]
- Keezer, M.R.; Sisodiya, S.M.; Sander, J.W. Comorbidities of epilepsy: Current concepts and future perspectives. Lancet Neurol. 2016, 15, 106–115. [Google Scholar] [CrossRef]
- Okudan, Z.V.; Özkara, Ç. Reflex epilepsy: Triggers and management strategies. Neuropsychiatr. Dis. Treat. 2018, 14, 327–337. [Google Scholar] [CrossRef] [PubMed]
- Nolan, D.; Fink, J. Genetics of epilepsy. Handb. Clin. Neurol. 2018, 148, 467–491. [Google Scholar] [CrossRef] [PubMed]
- Oyrer, J.; Maljevic, S.; Scheffer, I.E.; Berkovic, S.F.; Petrou, S.; Reid, C.A. Ion Channels in Genetic Epilepsy: From Genes and Mechanisms to Disease-Targeted Therapies. Pharmacol. Rev. 2018, 70, 142–173. [Google Scholar] [CrossRef] [PubMed]
- Lin Lin Lee, V.; Kar Meng Choo, B.; Chung, Y.S.; P Kundap, U.; Kumari, Y.; Shaikh, M.F. Treatment, Therapy and Management of Metabolic Epilepsy: A Systematic Review. Int. J. Mol. Sci. 2018, 19, 871. [Google Scholar] [CrossRef]
- Reddy, C.; Saini, A.G. Metabolic Epilepsy. Indian J. Pediatr. 2021, 88, 1025–1032. [Google Scholar] [CrossRef]
- Alizadeh Khatir, A.; Moghaddam, S.A.; Almukhtar, M.; Ghorbani, H.; Babazadeh, A.; Mehravar, S.; Rostami, A. Toxoplasma infection and risk of epilepsy: A case-control study of incident patients. Microb. Pathog. 2021, 161, 105302. [Google Scholar] [CrossRef]
- Nikbakht, F.; Mohammadkhanizadeh, A.; Mohammadi, E. How does the COVID-19 cause seizure and epilepsy in patients? The potential mechanisms. Mult. Scler. Relat. Disord. 2020, 46, 102535. [Google Scholar] [CrossRef]
- Cattaneo, D.; Giacomelli, A.; MiniSci, D.; Astuti, N.; Meraviglia, P.; Gervasoni, C. Association of HIV Infection with Epilepsy and Other Comorbid Conditions. AIDS Behav. 2020, 24, 1051–1055. [Google Scholar] [CrossRef]
- Gripper, L.B.; Welburn, S.C. The causal relationship between neurocysticercosis infection and the development of epilepsy—A systematic review. Infect. Dis. Poverty 2017, 6, 31. [Google Scholar] [CrossRef] [PubMed]
- Husari, K.S.; Dubey, D. Autoimmune Epilepsy. Neurotherapeutics 2019, 16, 685–702. [Google Scholar] [CrossRef] [PubMed]
- Geis, C.; Planagumà, J.; Carreño, M.; Graus, F.; Dalmau, J. Autoimmune seizures and epilepsy. J. Clin. Investig. 2019, 129, 926–940. [Google Scholar] [CrossRef] [PubMed]
- Spatola, M.; Dalmau, J. Seizures and risk of epilepsy in autoimmune and other inflammatory encephalitis. Curr. Opin. Neurol. 2017, 30, 345–353. [Google Scholar] [CrossRef] [PubMed]
- Younas, A.; Saim, M.; Maqsood, H.; Younus, S.; Hassan Raza, M. Dyke-Davidoff-Masson Syndrome: A Case Report and Review of Literature. Cureus 2020, 12, e11919. [Google Scholar] [CrossRef] [PubMed]
- Specchio, N.; Pietrafusa, N. New-onset refractory status epilepticus and febrile infection-related epilepsy syndrome. Dev. Med. Child Neurol. 2020, 62, 897–905. [Google Scholar] [CrossRef] [PubMed]
- Mikaeloff, Y.; Jambaqué, I.; Hertz-Pannier, L.; Zamfirescu, A.; Adamsbaum, C.; Plouin, P.; Dulac, O.; Chiron, C. Devastating epileptic encephalopathy in school-aged children (DESC): A pseudo encephalitis. Epilepsy Res. 2006, 69, 67–79. [Google Scholar] [CrossRef]
- Naylor, D.E.; Liu, H.; Niquet, J.; Wasterlain, C.G. Rapid surface accumulation of NMDA receptors increases glutamatergic excitation during status epilepticus. Neurobiol. Dis. 2013, 54, 225–238. [Google Scholar] [CrossRef]
- Kapur, J. Role of NMDA receptors in the pathophysiology and treatment of status epilepticus. Epilepsia Open 2018, 3, 165–168. [Google Scholar] [CrossRef]
- Garrido-Sanabria, E.R.; Otalora, L.F.; Arshadmansab, M.F.; Herrera, B.; Francisco, S.; Ermolinsky, B.S. Impaired expression and function of group II metabotropic glutamate receptors in pilocarpine-treated chronically epileptic rats. Brain Res. 2008, 1240, 165–176. [Google Scholar] [CrossRef]
- Watanabe, Y.; Kaida, Y.; Fukuhara, S.; Takechi, K.; Uehara, T.; Kamei, C. Participation of metabotropic glutamate receptors in pentetrazol-induced kindled seizure. Epilepsia 2011, 52, 140–150. [Google Scholar] [CrossRef] [PubMed]
- Bocchio, M.; Lukacs, I.P.; Stacey, R.; Plaha, P.; Apostolopoulos, V.; Livermore, L.; Sen, A.; Ansorge, O.; Gillies, M.J.; Somogyi, P.; et al. Group II Metabotropic Glutamate Receptors Mediate Presynaptic Inhibition of Excitatory Transmission in Pyramidal Neurons of the Human Cerebral Cortex. Front. Cell. NeuroSci. 2018, 12, 508. [Google Scholar] [CrossRef] [PubMed]
- Doyle, S.; Pyndiah, S.; De Gois, S.; Erickson, J.D. Excitation-transcription coupling via calcium/calmodulin-dependent protein kinase/ERK1/2 signaling mediates the coordinate induction of VGLUT2 and Narp triggered by a prolonged increase in glutamatergic synaptic activity. J. Biol. Chem. 2010, 285, 14366–14376. [Google Scholar] [CrossRef] [PubMed]
- Gangarossa, G.; Di Benedetto, M.; O’Sullivan, G.J.; Dunleavy, M.; Alcacer, C.; Bonito-Oliva, A.; Henshall, D.C.; Waddington, J.L.; Valjent, E.; Fisone, G. Convulsant doses of a dopamine D1 receptor agonist result in Erk-dependent increases in Zif268 and Arc/Arg3.1 expression in mouse dentate gyrus. PLoS ONE 2011, 6, e19415. [Google Scholar] [CrossRef]
- Gangarossa, G.; Castell, L.; Castro, L.; Tarot, P.; Veyrunes, F.; Vincent, P.; Bertaso, F.; Valjent, E. Contrasting patterns of ERK activation in the tail of the striatum in response to aversive and rewarding signals. J. NeuroChem. 2019, 151, 204–226. [Google Scholar] [CrossRef]
- Nateri, A.S.; Raivich, G.; Gebhardt, C.; Da Costa, C.; Naumann, H.; Vreugdenhil, M.; Makwana, M.; Brandner, S.; Adams, R.H.; Jefferys, J.G.; et al. ERK activation causes epilepsy by stimulating NMDA receptor activity. EMBO J. 2007, 26, 4891–4901. [Google Scholar] [CrossRef]
- Pernice, H.F.; Schieweck, R.; Kiebler, M.A.; Popper, B. mTOR and MAPK: From localized translation control to epilepsy. BMC NeuroSci. 2016, 17, 73. [Google Scholar] [CrossRef]
- Lin, T.Y.; Lu, C.W.; Wang, C.C.; Lu, J.F.; Wang, S.J. Hispidulin inhibits the release of glutamate in rat cerebrocortical nerve terminals. Toxicol. Appl. Pharmacol. 2012, 263, 233–243. [Google Scholar] [CrossRef]
- Glazova, M.V.; Nikitina, L.S.; Hudik, K.A.; Kirillova, O.D.; Dorofeeva, N.A.; Korotkov, A.A.; Chernigovskaya, E.V. Inhibition of ERK1/2 signaling prevents epileptiform behavior in rats prone to audiogenic seizures. J. NeuroChem. 2015, 132, 218–229. [Google Scholar] [CrossRef]
- Bozzi, Y.; Borrelli, E. The role of dopamine signaling in epileptogenesis. Front. Cell NeuroSci. 2013, 7, 157. [Google Scholar] [CrossRef]
- Beaulieu, J.M.; Gainetdinov, R.R. The physiology, signaling, and pharmacology of dopamine receptors. Pharmacol. Rev. 2011, 63, 182–217. [Google Scholar] [CrossRef] [PubMed]
- Yakushev, I.Y.; Dupont, E.; Buchholz, H.G.; Tillmanns, J.; Debus, F.; Cumming, P.; Heimann, A.; Fellgiebel, A.; Luhmann, H.J.; Landvogt, C.; et al. In vivo imaging of dopamine receptors in a model of temporal lobe epilepsy. Epilepsia 2010, 51, 415–422. [Google Scholar] [CrossRef] [PubMed]
- Bernedo Paredes, V.E.; Buchholz, H.G.; Gartenschläger, M.; Breimhorst, M.; Schreckenberger, M.; Werhahn, K.J. Reduced D2/D3 Receptor Binding of Extrastriatal and Striatal Regions in Temporal Lobe Epilepsy. PLoS ONE 2015, 10, e0141098. [Google Scholar] [CrossRef] [PubMed]
- Dorofeeva, N.A.; Grigorieva, Y.S.; Nikitina, L.S.; Lavrova, E.A.; Nasluzova, E.V.; Glazova, M.V.; Chernigovskaya, E.V. Effects of ERK1/2 kinases inactivation on the nigrostriatal system of Krushinsky-Molodkina rats genetically prone to audiogenic seizures. Neurol. Res. 2017, 39, 918–925. [Google Scholar] [CrossRef]
- Svob Strac, D.; Pivac, N.; Smolders, I.J.; Fogel, W.A.; De Deurwaerdere, P.; Di Giovanni, G. Monoaminergic Mechanisms in Epilepsy May Offer Innovative Therapeutic Opportunity for Monoaminergic Multi-Target Drugs. Front. NeuroSci. 2016, 10, 492. [Google Scholar] [CrossRef]
- Medvedev, A.E.; Rajgorodskaya, D.I.; Gorkin, V.Z.; Fedotova, I.B.; Semiokhina, A.F. The role of lipid peroxidation in the possible involvement of membrane-bound monoamine oxidases in gamma-aminobutyric acid and glucosamine deamination in rat brain. Focus on chemical pathogenesis of experimental audiogenic epilepsy. Mol. Chem. Neuropathol. 1992, 16, 187–201. [Google Scholar] [CrossRef]
- Dailey, J.W.; Yan, Q.S.; Adams-Curtis, L.E.; Ryu, J.R.; Ko, K.H.; Mishra, P.K.; Jobe, P.C. Neurochemical correlates of antiepileptic drugs in the genetically epilepsy-prone rat (GEPR). Life Sci. 1996, 58, 259–266. [Google Scholar] [CrossRef]
- Chauvel, P.; Trottier, S. Role of noradrenergic ascending system in extinction of epileptic phenomena. Adv. Neurol. 1986, 44, 475–487. [Google Scholar]
- Chauvel, P.; Trottier, S.; Nassif, S.; Dedek, J. Is an alteration of noradrenergic afferents involved in focal epilepsies? Rev. Electroencephalogr. Neuro. Physiol. Clin. 1982, 12, 1–7. [Google Scholar] [CrossRef]
- Sourbron, J.; Lagae, L. Serotonin receptors in epilepsy: Novel treatment targets? Epilepsia Open 2022, 7, 231–246. [Google Scholar] [CrossRef]
- Jerlicz, M.; Kostowski, W.; Bidziński, A.; Hauptman, M.; Dymecki, J. Audiogenic seizures in rats: Relation to noradrenergic neurons of the locus coeruleus. Acta Physiol. Pol. 1978, 29, 409–412. [Google Scholar] [PubMed]
- Fritschy, J.M. Epilepsy, E/I Balance and GABA (A) Receptor Plasticity. Front. Mol. NeuroSci. 2008, 1, 5. [Google Scholar] [CrossRef] [PubMed]
- Morimoto, K.; Sato, H.; Yamamoto, Y.; Watanabe, T.; Suwaki, H. Antiepileptic effects of tiagabine, a selective GABA uptake inhibitor, in the rat kindling model of temporal lobe epilepsy. Epilepsia 1997, 38, 966–974. [Google Scholar] [CrossRef] [PubMed]
- Fueta, Y.; Vasilets, L.A.; Takeda, K.; Kawamura, M.; Schwarz, W. Down-regulation of GABA-transporter function by hippocampal translation products: Its possible role in epilepsy. NeuroScience 2003, 118, 371–378. [Google Scholar] [CrossRef]
- Cai, K.; Wang, J.; Eissman, J.; Nwosu, G.; Shen, W.; Liang, H.C.; Li, X.J.; Zhu, H.X.; Yi, Y.H.; Song, J.; et al. A missense mutation in SLC6A1 associated with Lennox-Gastaut syndrome impairs GABA transporter 1 protein trafficking and function. Exp. Neurol. 2019, 320, 112973. [Google Scholar] [CrossRef]
- Shaye, H.; Stauch, B.; Gati, C.; Cherezov, V. Molecular mechanisms of metabotropic GABA. Sci. Adv. 2021, 7, eabg3362. [Google Scholar] [CrossRef]
- Zafar, S.; Jabeen, I. Structure, Function, and Modulation of γ-Aminobutyric Acid Transporter 1 (GAT1) in Neurological Disorders: A Pharmacoinformatic Prospective. Front. Chem. 2018, 6, 397. [Google Scholar] [CrossRef]
- Quilichini, P.P.; Chiron, C.; Ben-Ari, Y.; Gozlan, H. Stiripentol, a putative antiepileptic drug, enhances the duration of opening of GABA-A receptor channels. Epilepsia 2006, 47, 704–716. [Google Scholar] [CrossRef]
- Catterall, W.A.; Kalume, F.; Oakley, J.C. NaV1.1 channels and epilepsy. J. Physiol. 2010, 588, 1849–1859. [Google Scholar] [CrossRef]
- Kardos, J.; Szabó, Z.; Héja, L. Framing Neuro-Glia Coupling in Antiepileptic Drug Design. J. Med. Chem. 2016, 59, 777–787. [Google Scholar] [CrossRef]
- Héja, L.; Nyitrai, G.; Kékesi, O.; Dobolyi, A.; Szabó, P.; Fiáth, R.; Ulbert, I.; Pál-Szenthe, B.; Palkovits, M.; Kardos, J. Astrocytes convert network excitation to tonic inhibition of neurons. BMC Biol. 2012, 10, 26. [Google Scholar] [CrossRef] [PubMed]
- Löscher, W.; Gillard, M.; Sands, Z.A.; Kaminski, R.M.; Klitgaard, H. Synaptic Vesicle Glycoprotein 2A Ligands in the Treatment of Epilepsy and Beyond. CNS Drugs 2016, 30, 1055–1077. [Google Scholar] [CrossRef] [PubMed]
- Seifert, G.; Carmignoto, G.; Steinhäuser, C. Astrocyte dysfunction in epilepsy. Brain Res Rev. 2010, 63, 212–221. [Google Scholar] [CrossRef] [PubMed]
- Patel, D.C.; Tewari, B.P.; Chaunsali, L.; Sontheimer, H. Neuron-glia interactions in the pathophysiology of epilepsy. Nat. Rev. NeuroSci. 2019, 20, 282–297. [Google Scholar] [CrossRef]
- Heuser, K.; Szokol, K.; Taubøll, E. The role of glial cells in epilepsy. Tidsskr. Nor. Laegeforen. 2014, 134, 37–41. [Google Scholar] [CrossRef]
- Kaila, K.; Ruusuvuori, E.; Seja, P.; Voipio, J.; Puskarjov, M. GABA actions and ionic plasticity in epilepsy. Curr. Opin. Neurobiol. 2014, 26, 34–41. [Google Scholar] [CrossRef]
- Rana, A.; Musto, A.E. The role of inflammation in the development of epilepsy. J. Neuroinflamm. 2018, 15, 144. [Google Scholar] [CrossRef]
- Vezzani, A.; French, J.; Bartfai, T.; Baram, T.Z. The role of inflammation in epilepsy. Nat. Rev. Neurol. 2011, 7, 31–40. [Google Scholar] [CrossRef]
- Avakyan, G.; Avakyan, G. Transformation of the epileptic system. Current status of the problem and possible ways to solve it. Epilepsy Paroxysmal Cond. 2017, 9, 6–19. [Google Scholar] [CrossRef]
- Verma, I.C.; Bhatia, S.; Arora, V. Genetic Testing in Pediatric Epilepsy. Indian J. Pediatr. 2021, 88, 1017–1024. [Google Scholar] [CrossRef]
- Steinlein, O.K. Genes and mutations in human idiopathic epilepsy. Brain Dev. 2004, 26, 213–218. [Google Scholar] [CrossRef]
- Kjeldsen, M.J.; Corey, L.A.; Christensen, K.; Friis, M.L. Epileptic seizures and syndromes in twins: The importance of genetic factors. Epilepsy Res. 2003, 55, 137–146. [Google Scholar] [CrossRef]
- Berkovic, S.F.; Howell, R.A.; Hay, D.A.; Hopper, J.L. Epilepsies in twins: Genetics of the major epilepsy syndromes. Ann. Neurol. 1998, 43, 435–445. [Google Scholar] [CrossRef]
- Kjeldsen, M.J.; Corey, L.A.; Solaas, M.H.; Friis, M.L.; Harris, J.R.; Kyvik, K.O.; Christensen, K.; Pellock, J.M. Genetic factors in seizures: A population-based study of 47,626 US, Norwegian and Danish twin pairs. Twin Res. Hum. Genet. 2005, 8, 138–147. [Google Scholar] [CrossRef] [PubMed]
- Sugawara, T.; Tsurubuchi, Y.; Agarwala, K.L.; Ito, M.; Fukuma, G.; Mazaki-Miyazaki, E.; Nagafuji, H.; Noda, M.; Imoto, K.; Wada, K.; et al. A missense mutation of the Na+ channel alpha II subunit gene Na(v)1.2 in a patient with febrile and afebrile seizures causes channel dysfunction. Proc. Natl. Acad. Sci. USA 2001, 98, 6384–6389. [Google Scholar] [CrossRef]
- Falace, A.; Filipello, F.; La Padula, V.; Vanni, N.; Madia, F.; De Pietri Tonelli, D.; de Falco, F.A.; Striano, P.; Dagna Bricarelli, F.; Minetti, C.; et al. TBC1D24, an ARF6-interacting protein, is mutated in familial infantile myoclonic epilepsy. Am. J. Hum. Genet. 2010, 87, 365–370. [Google Scholar] [CrossRef]
- Fehr, S.; Wilson, M.; Downs, J.; Williams, S.; Murgia, A.; Sartori, S.; Vecchi, M.; Ho, G.; Polli, R.; Psoni, S.; et al. The CDKL5 disorder is an independent clinical entity associated with early-onset encephalopathy. Eur. J. Hum. Genet. 2013, 21, 266–273. [Google Scholar] [CrossRef]
- Zhou, Q.; Wang, J.; Xia, L.; Li, R.; Zhang, Q.; Pan, S. SYN1 Mutation Causes X-Linked Toothbrushing Epilepsy in a Chinese Family. Front. Neurol. 2021, 12, 736977. [Google Scholar] [CrossRef]
- Garcia, C.C.; Blair, H.J.; Seager, M.; Coulthard, A.; Tennant, S.; Buddles, M.; Curtis, A.; Goodship, J.A. Identification of a mutation in synapsin I, a synaptic vesicle protein, in a family with epilepsy. J. Med. Genet. 2004, 41, 183–186. [Google Scholar] [CrossRef]
- Thakran, S.; Guin, D.; Singh, P.; Kukal, S.; Rawat, C.; Yadav, S.; Kushwaha, S.S.; Srivastava, A.K.; Hasija, Y.; Saso, L.; et al. Genetic Landscape of Common Epilepsies: Advancing towards Precision in Treatment. Int. J. Mol. Sci. 2020, 21, 7784. [Google Scholar] [CrossRef]
- Ferraro, T.N.; Buono, R.J. Polygenic epilepsy. Adv. Neurol. 2006, 97, 389–398. [Google Scholar] [PubMed]
- Koeleman, B.P.C. What do genetic studies tell us about the heritable basis of common epilepsy? Polygenic or complex epilepsy? NeuroSci. Lett 2018, 667, 10–16. [Google Scholar] [CrossRef] [PubMed]
- Leu, C.; Richardson, T.G.; Kaufmann, T.; van der Meer, D.; Andreassen, O.A.; Westlye, L.T.; Busch, R.M.; Davey Smith, G.; Lal, D. Pleiotropy of polygenic factors associated with focal and generalized epilepsy in the general population. PLoS ONE 2020, 15, e0232292. [Google Scholar] [CrossRef] [PubMed]
- Farnaes, L.; Nahas, S.A.; Chowdhury, S.; Nelson, J.; Batalov, S.; Dimmock, D.M.; Kingsmore, S.F.; Investigators, R. Rapid whole-genome sequencing identifies a novel. Mol. Case Stud. 2017, 3, a001776. [Google Scholar] [CrossRef] [PubMed]
- Kodera, H.; Ohba, C.; Kato, M.; Maeda, T.; Araki, K.; Tajima, D.; Matsuo, M.; Hino-Fukuyo, N.; Kohashi, K.; Ishiyama, A.; et al. De novo GABRA1 mutations in Ohtahara and West syndromes. Epilepsia 2016, 57, 566–573. [Google Scholar] [CrossRef] [PubMed]
- Cossette, P.; Lortie, A.; Vanasse, M.; Saint-Hilaire, J.M.; Rouleau, G.A. Autosomal dominant juvenile myoclonic epilepsy and GABRA1. Adv. Neurol. 2005, 95, 255–263. [Google Scholar]
- Belousova, E.D.; Zavadenko, N.N.; Kholin, A.A.; Sharkov, A.A. New classifications of epilepsies and seizure types created by the International League against Epilepsy. Zhurnal Nevrol. Psikhiatrii Im. SS Korsakova 2017, 117, 99–106. [Google Scholar] [CrossRef]
- Nakayama, T.; Ishii, A.; Yoshida, T.; Nasu, H.; Shimojima, K.; Yamamoto, T.; Kure, S.; Hirose, S. Somatic mosaic deletions involving SCN1A cause Dravet syndrome. Am. J. Med. Genet. A 2018, 176, 657–662. [Google Scholar] [CrossRef]
- Guerrini, R.; Marini, C.; Mantegazza, M. Genetic epilepsy syndromes without structural brain abnormalities: Clinical features and experimental models. Neurotherapeutics 2014, 11, 269–285. [Google Scholar] [CrossRef]
- Liu, X.-X.; Yang, L.; Shao, L.-X.; He, Y.; Wu, G.; Bao, Y.-H.; Lu, N.-N.; Gong, D.-M.; Lu, Y.-P.; Cui, T.-T.; et al. Endothelial Cdk5 deficit leads to the development of spontaneous epilepsy through CXCL1/CXCR2-mediated reactive astrogliosis. J. Exp. Med. 2019, 217, e20180992. [Google Scholar] [CrossRef]
- Breuss, M.W.; Sultan, T.; James, K.N.; Rosti, R.O.; Scott, E.; Musaev, D.; Furia, B.; Reis, A.; Sticht, H.; Al-Owain, M.; et al. Autosomal-Recessive Mutations in the tRNA Splicing Endonuclease Subunit TSEN15 Cause Pontocerebellar Hypoplasia and Progressive Microcephaly. Am. J. Hum. Genet. 2016, 99, 228–235. [Google Scholar] [CrossRef] [PubMed]
- Poulton, C.; Oegema, R.; Heijsman, D.; Hoogeboom, J.; Schot, R.; Stroink, H.; Willemsen, M.A.; Verheijen, F.W.; Van De Spek, P.; Kremer, A.; et al. Progressive cerebellar atrophy and polyneuropathy: Expanding the spectrum of PNKP mutations. Neurogenetics 2012, 14, 43–51. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.S. Molecular Genetics of Neuronal Migration Disorders. Curr. Neurol. Neurosci. Rep. 2011, 11, 171–178. [Google Scholar] [CrossRef]
- Finsterer, J. Mitochondriopathies. Eur. J. Neurol. 2004, 11, 163–186. [Google Scholar] [CrossRef]
- Lim, A.; Thomas, R.H. The mitochondrial epilepsies. Eur. J. Paediatr. Neurol. 2020, 24, 47–52. [Google Scholar] [CrossRef] [PubMed]
- Musto, E.; Gardella, E.; Møller, R.S. Recent advances in treatment of epilepsy-related sodium channelopathies. Eur. J. Paediatr. Neurol. 2020, 24, 123–128. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Han, X.; Blendy, J.A.; Porter, B.E. Decreased CREB levels suppress epilepsy. Neurobiol. Dis. 2012, 45, 253–263. [Google Scholar] [CrossRef]
- Switon, K.; Kotulska, K.; Janusz-Kaminska, A.; Zmorzynska, J.; Jaworski, J. Molecular neurobiology of mTOR. NeuroScience 2017, 341, 112–153. [Google Scholar] [CrossRef]
- Robison, A.J. Emerging role of CaMKII in neuropsychiatric disease. Trends NeuroSci. 2014, 37, 653–662. [Google Scholar] [CrossRef]
- Liu, J.; Reeves, C.; Michalak, Z.; Coppola, A.; Diehl, B.; Sisodiya, S.M.; Thom, M. Evidence for mTOR pathway activation in a spectrum of epilepsy-associated pathologies. Acta Neuropathol. Commun. 2014, 2, 71. [Google Scholar] [CrossRef]
- Chuvakova, L.N.; Funikov, S.Y.; Rezvykh, A.P.; Davletshin, A.I.; Evgen’ev, M.B.; Litvinova, S.A.; Fedotova, I.B.; Poletaeva, I.I.; Garbuz, D.G. Transcriptome of the Krushinsky-Molodkina Audiogenic Rat Strain and Identification of Possible Audiogenic Epilepsy-Associated Genes. Front. Mol. NeuroSci. 2021, 14, 738930. [Google Scholar] [CrossRef] [PubMed]
- Venediktova, N.I.; Gorbacheva, O.S.; Belosludtseva, N.V.; Fedotova, I.B.; Surina, N.M.; Poletaeva, I.I.; Kolomytkin, O.V.; Mironova, G.D. Energetic, oxidative and ionic exchange in rat brain and liver mitochondria at experimental audiogenic epilepsy (Krushinsky–Molodkina model). J. Bioenerg. Biomembr. 2017, 49, 149–158. [Google Scholar] [CrossRef] [PubMed]
- Zavala-Tecuapetla, C.; Cuellar-Herrera, M.; Luna-Munguia, H. Insights into Potential Targets for Therapeutic Intervention in Epilepsy. Int. J. Mol. Sci. 2020, 21, 8573. [Google Scholar] [CrossRef] [PubMed]
- Sanz, P.; Garcia-Gimeno, M.A. Reactive Glia Inflammatory Signaling Pathways and Epilepsy. Int. J. Mol. Sci. 2020, 21, 4096. [Google Scholar] [CrossRef]
- Dixit, A.B.; Banerjee, J.; Tripathi, M.; Sarkar, C.; Chandra, P.S. Synaptic roles of cyclin-dependent kinase 5 & its implications in epilepsy. Indian J. Med. Res. 2017, 145, 179–188. [Google Scholar] [CrossRef]
- Crino, P.B. mTOR Signaling in Epilepsy: Insights from Malformations of Cortical Development. Cold Spring Harb. Perspect. Med. 2015, 5, a022442. [Google Scholar] [CrossRef]
- Kandratavicius, L.; Balista, P.A.; Lopes-Aguiar, C.; Ruggiero, R.N.; Umeoka, E.H.; Garcia-Cairasco, N.; Bueno-Junior, L.S.; Leite, J.P. Animal models of epilepsy: Use and limitations. Neuropsychiatr. Dis. Treat. 2014, 10, 1693–1705. [Google Scholar] [CrossRef]
- McNamara, J.O.; Bonhaus, D.W.; Shin, C.; Crain, B.J.; Gellman, R.L.; Giacchino, J.L. The kindling model of epilepsy: A critical review. CRC Crit. Rev. Clin. Neurobiol. 1985, 1, 341–391. [Google Scholar]
- Ferlazzo, E.; Gasparini, S.; Beghi, E.; Sueri, C.; Russo, E.; Leo, A.; Labate, A.; Gambardella, A.; Belcastro, V.; Striano, P.; et al. Epilepsy in cerebrovascular diseases: Review of experimental and clinical data with meta-analysis of risk factors. Epilepsia 2016, 57, 1205–1214. [Google Scholar] [CrossRef]
- Bonvallet, M.; Dell, P.; Hugelin, A. Olfactory, gustatory, visceral, vagal, visual and auditory projections in the grey formations of the forebrain of the cat. J. Physiol. 1952, 44, 222–224. [Google Scholar]
- Kopeloff, L.M. Experimental epilepsy in the mouse. Proc. Soc. Exp. Biol. Med. 1960, 104, 500–504. [Google Scholar] [CrossRef] [PubMed]
- Gorter, J.A.; van Vliet, E.A.; Lopes da Silva, F.H. Which insights have we gained from the kindling and post-status epilepticus models? J. NeuroSci. Methods 2016, 260, 96–108. [Google Scholar] [CrossRef] [PubMed]
- Castel-Branco, M.M.; Alves, G.L.; Figueiredo, I.V.; Falcão, A.C.; Caramona, M.M. The maximal electroshock seizure (MES) model in the preclinical assessment of potential new antiepileptic drugs. Methods Find. Exp. Clin. Pharmacol. 2009, 31, 101–106. [Google Scholar] [CrossRef]
- Yuen, E.S.; Trocóniz, I.F. Can pentylenetetrazole and maximal electroshock rodent seizure models quantitatively predict antiepileptic efficacy in humans? Seizure 2015, 24, 21–27. [Google Scholar] [CrossRef] [PubMed]
- De Sarro, G.; Russo, E.; Citraro, R.; Meldrum, B.S. Genetically epilepsy-prone rats (GEPRs) and DBA/2 mice: Two animal models of audiogenic reflex epilepsy for the evaluation of new generation AEDs. Epilepsy Behav. 2017, 71, 165–173. [Google Scholar] [CrossRef]
- Sarkisova, K.; van Luijtelaar, G. The WAG/Rij strain: A genetic animal model of absence epilepsy with comorbidity of depression [corrected]. Prog. Neuro-Psychopharmacol. Biol. Psychiatry 2010, 35, 854–876. [Google Scholar] [CrossRef]
- Seyfried, T.N. Audiogenic seizures in mice. Fed. Proc. 1979, 38, 2399–2404. [Google Scholar]
- Maxson, S.C. A genetic context for the study of audiogenic seizures. Epilepsy Behav. 2017, 71, 154–159. [Google Scholar] [CrossRef]
- Fedotova, I.B.; Surina, N.M.; Nikolaev, G.M.; Revishchin, A.V.; Poletaeva, I.I. Rodent Brain Pathology, Audiogenic Epilepsy. Biomedicines 2021, 9, 1641. [Google Scholar] [CrossRef]
- A Fless, D.; Salimov, R.M. An analysis of the phase of prespasmodic motor excitation in rats with audiogenic seizures. Bull. Exp. Biol. Med. 1974, 78, 31–33. [Google Scholar] [CrossRef]
- Salimov, R.M.; Fless, D.A. Influence of convulsive and pre-convulsive components of an audiogenic seizure on the process of consolidation of temporary connections. Bull. Exp. Biol. Med. 1977, 83, 649–651. [Google Scholar] [CrossRef]
- Poletaeva, I.I.; Surina, N.M.; Kostina, Z.A.; Perepelkina, O.V.; Fedotova, I.B. The Krushinsky-Molodkina rat strain: The study of audiogenic epilepsy for 65years. Epilepsy Behav. 2017, 71, 130–141. [Google Scholar] [CrossRef] [PubMed]
- Reigel, C.E.; Dailey, J.W.; Jobe, P.C. The genetically epilepsy-prone rat: An overview of seizure-prone characteristics and responsiveness to anticonvulsant drugs. Life Sci. 1986, 39, 763–774. [Google Scholar] [CrossRef]
- Zhao, D.Y.; Wu, X.R.; Pei, Y.Q.; Zuo, Q.H. Kindling phenomenon of hyperthermic seizures in the epilepsy-prone versus the epilepsy-resistant rat. Brain Res. 1985, 358, 390–393. [Google Scholar] [CrossRef]
- Bo, X.; Zhiguo, W.; Xiaosu, Y.; Guoliang, L.; Guangjie, X. Analysis of gene expression in genetic epilepsy-prone rat using a cDNA expression array. Seizure 2002, 11, 418–422. [Google Scholar] [CrossRef]
- Muñoz, L.J.; Carballosa-Gautam, M.M.; Yanowsky, K.; García-Atarés, N.; López, D.E. The genetic audiogenic seizure hamster from Salamanca: The GASH:Sal. Epilepsy Behav. 2017, 71, 181–192. [Google Scholar] [CrossRef]
- Dolina, S.; Peeling, J.; Sutherland, G.; Pillay, N.; Greenberg, A. Effect of sustained pyridoxine treatment on seizure susceptibility and regional brain amino acid levels in genetically epilepsy-prone BALB/c mice. Epilepsia 1993, 34, 33–42. [Google Scholar] [CrossRef]
- Martin, B.; Dieuset, G.; Pawluski, J.L.; Costet, N.; Biraben, A. Audiogenic seizure as a model of sudden death in epilepsy: A comparative study between four inbred mouse strains from early life to adulthood. Epilepsia 2020, 61, 342–349. [Google Scholar] [CrossRef]
- Poletaeva, I.I.; Fedotova, I.B.; Sourina, N.M.; Kostina, Z.A. Audiogenic seizures-biological phenomenon and experimental model of human epilepsies. Clin. Genet. Asp. Epilepsy 2011, 115–148. [Google Scholar] [CrossRef]
- Laird, H.E.; Dailey, J.W.; Jobe, P.C. Neurotransmitter abnormalities in genetically epileptic rodents. Fed. Proc. 1984, 43, 2505–2509. [Google Scholar]
- Mishra, P.K.; Kahle, E.H.; Bettendorf, A.F.; Dailey, J.W.; Jobe, P.C. Anticonvulsant effects of intracerebroventricularly administered norepinephrine are potentiated in the presence of monoamine oxidase inhibition in severe seizure genetically epilepsy-prone rats (GEPR-9s). Life Sci. 1993, 52, 1435–1441. [Google Scholar] [CrossRef]
- Kash, S.F.; Johnson, R.S.; Tecott, L.H.; Noebels, J.L.; Mayfield, R.D.; Hanahan, D.; Baekkeskov, S. Epilepsy in mice deficient in the 65-kDa isoform of glutamic acid decarboxylase. Proc. Natl. Acad. Sci. USA 1997, 94, 14060–14065. [Google Scholar] [CrossRef] [PubMed]
- N’Gouemo, P.; Faingold, C.L. Periaqueductal gray neurons exhibit increased responsiveness associated with audiogenic seizures in the genetically epilepsy-prone rat. NeuroScience 1998, 84, 619–625. [Google Scholar] [CrossRef]
- N’Gouemo, P.; Faingold, C.L. The periaqueductal grey is a critical site in the neuronal network for audiogenic seizures: Modulation by GABA(A), NMDA and opioid receptors. Epilepsy Res. 1999, 35, 39–46. [Google Scholar] [CrossRef]
- Willott, J.F.; Lu, S.M. Midbrain pathways of audiogenic seizures in DBA/2 mice. Exp. Neurol. 1980, 70, 288–299. [Google Scholar] [CrossRef]
- Leite, J.P.; Garcia-Cairasco, N.; Cavalheiro, E.A. New insights from the use of pilocarpine and kainate models. Epilepsy Res. 2002, 50, 93–103. [Google Scholar] [CrossRef]
- Garcia-Cairasco, N.; Sabbatini, R.M. Possible interaction between the inferior colliculus and the substantia nigra in audiogenic seizures in Wistar rats. Physiol. Behav. 1991, 50, 421–427. [Google Scholar] [CrossRef]
- Simler, S.; Hirsch, E.; Danober, L.; Motte, J.; Vergnes, M.; Marescaux, C. C-fos expression after single and kindled audiogenic seizures in Wistar rats. NeuroSci. Lett. 1994, 175, 58–62. [Google Scholar] [CrossRef]
- Browning, R.A.; Wang, C.; Nelson, D.K.; Jobe, P.C. Effect of precollicular transection on audiogenic seizures in genetically epilepsy-prone rats. Exp. Neurol. 1999, 155, 295–301. [Google Scholar] [CrossRef]
- Ribak, C.E.; Manio, A.L.; Navetta, M.S.; Gall, C.M. In situ hybridization for c-fos mRNA reveals the involvement of the superior colliculus in the propagation of seizure activity in genetically epilepsy-prone rats. Epilepsy Res. 1997, 26, 397–406. [Google Scholar] [CrossRef]
- Ribak, C.E.; Morin, C.L. The role of the inferior colliculus in a genetic model of audiogenic seizures. Anat. Embryol. 1995, 191, 279–295. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Benito, D.; Gómez-Nieto, R.; Hernández-Noriega, S.; Murashima, A.A.B.; de Oliveira, J.A.C.; Garcia-Cairasco, N.; López, D.E.; Hyppolito, M.A. Morphofunctional alterations in the olivocochlear efferent system of the genetic audiogenic seizure-prone hamster GASH:Sal. Epilepsy Behav. 2017, 71, 193–206. [Google Scholar] [CrossRef] [PubMed]
- Zivanovic, D.; Stanojlovic, O.; Mirkovic, S.; Susic, V. Ontogenetic study of metaphit-induced audiogenic seizures in rats. Brain Res. Dev. Brain Res. 2005, 155, 42–48. [Google Scholar] [CrossRef] [PubMed]
- Susic, V.; Reith, M.E.; Zlokovic, B.V.; Lajtha, A.; Jacobson, A.E.; Rice, K.C.; Lipovac, M.N. Electroencephalographic characteristics of audiogenic seizures induced in metaphit-treated small rodents. Epilepsia 1991, 32, 783–790. [Google Scholar] [CrossRef]
- 165 Stanojlović, O.; Zivanović, D.; Mirković, S.; Vucević, D. Behavioral and electroencephalographic effects of delta sleep inducing peptide and its analogue on metaphit-induced audiogenic seizures in rats. Srp. Arh. Celok. Lek. 2004, 132, 421–426. [Google Scholar] [CrossRef]
- Kovalzon, V.M.; Strekalova, T.V. Delta sleep-inducing peptide (DSIP): A still unresolved riddle. J. Neurochem. 2006, 97, 303–309. [Google Scholar] [CrossRef]
- Tukhovskaya, E.A.; Ismailova, A.M.; Shaykhutdinova, E.R.; Slashcheva, G.A.; Prudchenko, I.A.; Mikhaleva, I.I.; Khokhlova, O.N.; Murashev, A.N.; Ivanov, V.T. Delta Sleep-Inducing Peptide Recovers Motor Function in SD Rats after Focal Stroke. Molecules 2021, 26, 5173. [Google Scholar] [CrossRef]
- Khvatova, E.M.; Samartzev, V.N.; Zagoskin, P.P.; Prudchenko, I.A.; Mikhaleva, I.I. Delta sleep inducing peptide (DSIP): Effect on respiration activity in rat brain mitochondria and stress protective potency under experimental hypoxia. Peptides 2003, 24, 307–311. [Google Scholar] [CrossRef]
- Gupta, V.; Awasthi, N.; Wagner, B.J. Specific Activation of the Glucocorticoid Receptor and Modulation of Signal Transduction Pathways in Human Lens Epithelial Cells. Investig. Opthalmol. Vis. Sci. 2007, 48, 1724–1734. [Google Scholar] [CrossRef]
- Fedotova, I.B.; Semiokhina, A.F.; Arkhipova, G.V.; Nikashin, A.V.; Burlakova, E.B. Differences in the lipid composition of the brain of rats of the Krushinskiĭ-Molodkina strain during an audiogenic seizure attack and in myoclonus. Zhurnal Vyss. Nervn. Deiatelnosti Im. IP Pavlov. 1988, 38, 374–377. [Google Scholar]
- Baulac, S.; Ishida, S.; Mashimo, T.; Boillot, M.; Fumoto, N.; Kuwamura, M.; Ohno, Y.; Takizawa, A.; Aoto, T.; Ueda, M.; et al. A rat model for LGI1-related epilepsies. Hum. Mol. Genet. 2012, 21, 3546–3557. [Google Scholar] [CrossRef] [PubMed]
- Bassell, G.J.; Warren, S.T. Fragile X syndrome: Loss of local mRNA regulation alters synaptic development and function. Neuron 2008, 60, 201–214. [Google Scholar] [CrossRef] [PubMed]
- Berry-Kravis, E.; Filipink, R.A.; Frye, R.E.; Golla, S.; Morris, S.M.; Andrews, H.; Choo, T.H.; Kaufmann, W.E.; Consortium, F. Seizures in Fragile X Syndrome: Associations and Longitudinal Analysis of a Large Clinic-Based Cohort. Front. Pediatr. 2021, 9, 736255. [Google Scholar] [CrossRef] [PubMed]
- Musumeci, S.A.; Calabrese, G.; Bonaccorso, C.M.; D’Antoni, S.; Brouwer, J.R.; Bakker, C.E.; Elia, M.; Ferri, R.; Nelson, D.L.; Oostra, B.A.; et al. Audiogenic seizure susceptibility is reduced in fragile X knockout mice after introduction of FMR1 transgenes. Exp. Neurol. 2007, 203, 233–240. [Google Scholar] [CrossRef] [PubMed]
- Musumeci, S.A.; Bosco, P.; Calabrese, G.; Bakker, C.; De Sarro, G.B.; Elia, M.; Ferri, R.; Oostra, B.A. Audiogenic seizures susceptibility in transgenic mice with fragile X syndrome. Epilepsia 2000, 41, 19–23. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Toth, M. Fragile X mice develop sensory hyperreactivity to auditory stimuli. NeuroScience 2001, 103, 1043–1050. [Google Scholar] [CrossRef]
- Pacey, L.K.; Tharmalingam, S.; Hampson, D.R. Subchronic administration and combination metabotropic glutamate and GABAB receptor drug therapy in fragile X syndrome. J. Pharmacol. Exp. Ther. 2011, 338, 897–905. [Google Scholar] [CrossRef]
- McMillan, D.R.; Kayes-Wandover, K.M.; Richardson, J.A.; White, P.C. Very large G protein-coupled receptor-1, the largest known cell surface protein, is highly expressed in the developing central nervous system. J. Biol. Chem. 2002, 277, 785–792. [Google Scholar] [CrossRef]
- Shin, D.; Lin, S.T.; Fu, Y.H.; Ptácek, L.J. Very large G protein-coupled receptor 1 regulates myelin-associated glycoprotein via Gαs/Gαq-mediated protein kinases A/C. Proc. Natl. Acad. Sci. USA 2013, 110, 19101–19106. [Google Scholar] [CrossRef]
- Yagi, H.; Noguchi, Y.; Kitamura, K.; Sato, M. Deficiency of Vlgr1 resulted in deafness and susceptibility to audiogenic seizures while the degree of hearing impairment was not correlated with seizure severity in C57BL/6- and 129-backcrossed lines of Vlgr1 knockout mice. NeuroSci. Lett. 2009, 461, 190–195. [Google Scholar] [CrossRef]
- Damasceno, S.; Fonseca, P.A.S.; Rosse, I.C.; Moraes, M.F.D.; de Oliveira, J.A.C.; Garcia-Cairasco, N.; Brunialti Godard, A.L. Putative Causal Variant on. Front. Neurol. 2021, 12, 647859. [Google Scholar] [CrossRef]
- Bednarek, A.K.; Laflin, K.J.; Daniel, R.L.; Liao, Q.; Hawkins, K.A.; Aldaz, C.M. WWOX, a novel WW domain-containing protein mapping to human chromosome 16q23.3-24.1, a region frequently affected in breast cancer. Cancer Res. 2000, 60, 2140–2145. [Google Scholar] [PubMed]
- Repudi, S.; Steinberg, D.J.; Elazar, N.; Breton, V.L.; Aquilino, M.S.; Saleem, A.; Abu-Swai, S.; Vainshtein, A.; Eshed-Eisenbach, Y.; Vijayaragavan, B.; et al. Neuronal deletion of Wwox, associated with WOREE syndrome, causes epilepsy and myelin defects. Brain 2021, 144, 3061–3077. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Zhou, W.; Shi, J.; Zhang, B.; Wang, H. A Chinese patient with epilepsy and WWOX compound heterozygous mutations. Epileptic Disord. 2020, 22, 120–124. [Google Scholar] [CrossRef]
- Suzuki, H.; Katayama, K.; Takenaka, M.; Amakasu, K.; Saito, K.; Suzuki, K. A spontaneous mutation of the Wwox gene and audiogenic seizures in rats with lethal dwarfism and epilepsy. Genes Brain Behav. 2009, 8, 650–660. [Google Scholar] [CrossRef] [PubMed]
- Knowles, J.K.; Xu, H.; Soane, C.; Batra, A.; Saucedo, T.; Frost, E.; Tam, L.T.; Fraga, D.; Ni, L.; Villar, K.; et al. Maladaptive myelination promotes generalized epilepsy progression. Nat. NeuroSci. 2022, 25, 596–606. [Google Scholar] [CrossRef] [PubMed]
- Repudi, S.; Kustanovich, I.; Abu-Swai, S.; Stern, S.; Aqeilan, R.I. Neonatal neuronal WWOX gene therapy rescues Wwox null phenotypes. EMBO Mol. Med. 2021, 13, e14599. [Google Scholar] [CrossRef]
- Brennan, T.J.; Seeley, W.W.; Kilgard, M.; Schreiner, C.E.; Tecott, L.H. Sound-induced seizures in serotonin 5-HT2c receptor mutant mice. Nat. Genet. 1997, 16, 387–390. [Google Scholar] [CrossRef]
- Celada, P.; Puig, M.V.; Artigas, F. Serotonin modulation of cortical neurons and networks. Front. Integr. NeuroSci. 2013, 7, 25. [Google Scholar] [CrossRef]
- Jiang, X.; Xing, G.; Yang, C.; Verma, A.; Zhang, L.; Li, H. Stress impairs 5-HT2A receptor-mediated serotonergic facilitation of GABA release in juvenile rat basolateral amygdala. Neuropsychopharmacology 2009, 34, 410–423. [Google Scholar] [CrossRef]
- Crunelli, V.; Di Giovanni, G. Differential Control by 5-HT and 5-HT1A, 2A, 2C Receptors of Phasic and Tonic GABAA Inhibition in the Visual Thalamus. CNS NeuroSci. Ther. 2015, 21, 967–970. [Google Scholar] [CrossRef] [PubMed]
- Bagdy, G.; Kecskemeti, V.; Riba, P.; Jakus, R. Serotonin and epilepsy. J. NeuroChem. 2007, 100, 857–873. [Google Scholar] [CrossRef] [PubMed]
- Jakus, R.; Graf, M.; Juhasz, G.; Gerber, K.; Levay, G.; Halasz, P.; Bagdy, G. 5-HT2C receptors inhibit and 5-HT1A receptors activate the generation of spike-wave discharges in a genetic rat model of absence epilepsy. Exp. Neurol. 2003, 184, 964–972. [Google Scholar] [CrossRef]
- Srivastava, S.; Cohen, J.; Pevsner, J.; Aradhya, S.; McKnight, D.; Butler, E.; Johnston, M.; Fatemi, A. A novel variant in GABRB2 associated with intellectual disability and epilepsy. Am. J. Med. Genet. A 2014, 164A, 2914–2921. [Google Scholar] [CrossRef]
- Yang, Y.; Xiangwei, W.; Zhang, X.; Xiao, J.; Chen, J.; Yang, X.; Jia, T.; Yang, Z.; Jiang, Y.; Zhang, Y. Phenotypic spectrum of patients with GABRB2 variants: From mild febrile seizures to severe epileptic encephalopathy. Dev. Med. Child Neurol. 2020, 62, 1213–1220. [Google Scholar] [CrossRef]
- El Achkar, C.M.; Harrer, M.; Smith, L.; Kelly, M.; Iqbal, S.; Maljevic, S.; Niturad, C.E.; Vissers, L.E.L.M.; Poduri, A.; Yang, E.; et al. Characterization of the GABRB2-Associated Neurodevelopmental Disorders. Ann. Neurol. 2021, 89, 573–586. [Google Scholar] [CrossRef]
- Yeung, R.K.; Xiang, Z.H.; Tsang, S.Y.; Li, R.; Ho, T.Y.C.; Li, Q.; Hui, C.K.; Sham, P.C.; Qiao, M.Q.; Xue, H. Gabrb2-knockout mice displayed schizophrenia-like and comorbid phenotypes with interneuron-astrocyte-microglia dysregulation. Transl. Psychiatry 2018, 8, 128. [Google Scholar] [CrossRef]
- Zhu, Y.H.; Liu, H.; Zhang, L.Y.; Zeng, T.; Song, Y.; Qin, Y.R.; Li, L.; Liu, L.; Li, J.; Zhang, B.; et al. Downregulation of LGI1 promotes tumor metastasis in esophageal squamous cell carcinoma. Carcinogenesis 2014, 35, 1154–1161. [Google Scholar] [CrossRef][Green Version]
- Cowell, J.K. LGI1: From zebrafish to human epilepsy. Prog. Brain Res. 2014, 213, 159–179. [Google Scholar] [CrossRef]
- Kinboshi, M.; Shimizu, S.; Mashimo, T.; Serikawa, T.; Ito, H.; Ikeda, A.; Takahashi, R.; Ohno, Y. Down-Regulation of Astrocytic Kir4.1 Channels during the Audiogenic Epileptogenesis in. Int. J. Mol. Sci. 2019, 20, 1013. [Google Scholar] [CrossRef]
- Kunapuli, P.; Kasyapa, C.S.; Hawthorn, L.; Cowell, J.K. LGI1, a putative tumor metastasis suppressor gene, controls in vitro invasiveness and expression of matrix metalloproteinases in glioma cells through the ERK1/2 pathway. J. Biol. Chem. 2004, 279, 23151–23157. [Google Scholar] [CrossRef] [PubMed]
- Yamagata, A.; Fukai, S. Insights into the mechanisms of epilepsy from structural biology of LGI1-ADAM22. Cell Mol. Life Sci. 2020, 77, 267–274. [Google Scholar] [CrossRef] [PubMed]
- Senechal, K.R.; Thaller, C.; Noebels, J.L. ADPEAF mutations reduce levels of secreted LGI1, a putative tumor suppressor protein linked to epilepsy. Hum. Mol. Genet. 2005, 14, 1613–1620. [Google Scholar] [CrossRef] [PubMed]
- Chabrol, E.; Navarro, V.; Provenzano, G.; Cohen, I.; Dinocourt, C.; Rivaud-Péchoux, S.; Fricker, D.; Baulac, M.; Miles, R.; Leguern, E.; et al. Electroclinical characterization of epileptic seizures in leucine-rich, glioma-inactivated 1-deficient mice. Brain 2010, 133, 2749–2762. [Google Scholar] [CrossRef]
- Manis, A.D.; Palygin, O.; Isaeva, E.; Levchenko, V.; LaViolette, P.S.; Pavlov, T.S.; Hodges, M.R.; Staruschenko, A. Kcnj16 knockout produces audiogenic seizures in the Dahl salt-sensitive rat. JCI Insight 2021, 6, e143251. [Google Scholar] [CrossRef]
- Staruschenko, A.; Hodges, M.R.; Palygin, O. Kir5.1 channels: Potential role in epilepsy and seizure disorders. Am. J. Physiol. Cell Physiol. 2022, 323, C706–C717. [Google Scholar] [CrossRef]
- Misawa, H.; Sherr, E.H.; Lee, D.J.; Chetkovich, D.M.; Tan, A.; Schreiner, C.E.; Bredt, D.S. Identification of a monogenic locus (jams1) causing juvenile audiogenic seizures in mice. J. NeuroSci. 2002, 22, 10088–10093. [Google Scholar] [CrossRef]
- Drayton, M.; Noben-Trauth, K. Mapping quantitative trait loci for hearing loss in Black Swiss mice. Hear. Res. 2006, 212, 128–139. [Google Scholar] [CrossRef]
- Charizopoulou, N.; Lelli, A.; Schraders, M.; Ray, K.; Hildebrand, M.S.; Ramesh, A.; Srisailapathy, C.R.; Oostrik, J.; Admiraal, R.J.; Neely, H.R.; et al. Gipc3 mutations associated with audiogenic seizures and sensorineural hearing loss in mouse and human. Nat. Commun. 2011, 2, 201. [Google Scholar] [CrossRef]
- Garcia-Gomes, M.S.A.; Zanatto, D.A.; Galvis-Alonso, O.Y.; Mejia, J.; Antiorio, A.T.F.B.; Yamamoto, P.K.; Olivato, M.C.M.; Sandini, T.M.; Flório, J.C.; Lebrun, I.; et al. Behavioral and neurochemical characterization of the spontaneous mutation tremor, a new mouse model of audiogenic seizures. Epilepsy Behav. 2020, 105, 106945. [Google Scholar] [CrossRef]
- Kim, J.H.; Roberts, D.S.; Hu, Y.; Lau, G.C.; Brooks-Kayal, A.R.; Farb, D.H.; Russek, S.J. Brain-derived neurotrophic factor uses CREB and Egr3 to regulate NMDA receptor levels in cortical neurons. J. NeuroChem. 2012, 120, 210–219. [Google Scholar] [CrossRef] [PubMed]
- Kaitsuka, T.; Kiyonari, H.; Shiraishi, A.; Tomizawa, K.; Matsushita, M. Deletion of Long Isoform of Eukaryotic Elongation Factor 1Bδ Leads to Audiogenic Seizures and Aversive Stimulus-Induced Long-Lasting Activity Suppression in Mice. Front. Mol. NeuroSci. 2018, 11, 358. [Google Scholar] [CrossRef] [PubMed]
- Kaitsuka, T.; Tomizawa, K.; Matsushita, M. Transformation of eEF1Bδ into heat-shock response transcription factor by alternative splicing. EMBO Rep. 2011, 12, 673–681. [Google Scholar] [CrossRef]
- Reuter, M.S.; Tawamie, H.; Buchert, R.; Hosny Gebril, O.; Froukh, T.; Thiel, C.; Uebe, S.; Ekici, A.B.; Krumbiegel, M.; Zweier, C.; et al. Diagnostic Yield and Novel Candidate Genes by Exome Sequencing in 152 Consanguineous Families With Neurodevelopmental Disorders. JAMA Psychiatry 2017, 74, 293–299. [Google Scholar] [CrossRef] [PubMed]
- Gachon, F.; Fonjallaz, P.; Damiola, F.; Gos, P.; Kodama, T.; Zakany, J.; Duboule, D.; Petit, B.; Tafti, M.; Schibler, U. The loss of circadian PAR bZip transcription factors results in epilepsy. Genes Dev. 2004, 18, 1397–1412. [Google Scholar] [CrossRef] [PubMed]
- Falsaperla, R.; Corsello, G. Pyridoxine dependent epilepsies: New therapeutical poInt of view. Ital. J. Pediatr. 2017, 43, 68. [Google Scholar] [CrossRef]
- Rotaru, D.C.; Mientjes, E.J.; Elgersma, Y. Angelman Syndrome: From Mouse Models to Therapy. NeuroScience 2020, 445, 172–189. [Google Scholar] [CrossRef]
- Gentile, J.K.; Tan, W.H.; Horowitz, L.T.; Bacino, C.A.; Skinner, S.A.; Barbieri-Welge, R.; Bauer-Carlin, A.; Beaudet, A.L.; Bichell, T.J.; Lee, H.S.; et al. A neurodevelopmental survey of Angelman syndrome with genotype-phenotype correlations. J. Dev. Behav. Pediatr. 2010, 31, 592–601. [Google Scholar] [CrossRef]
- Buiting, K.; Williams, C.; Horsthemke, B. Angelman syndrome—Insights into a rare neurogenetic disorder. Nat. Rev. Neurol. 2016, 12, 584–593. [Google Scholar] [CrossRef]
- Jiang, Y.H.; Armstrong, D.; Albrecht, U.; Atkins, C.M.; Noebels, J.L.; Eichele, G.; Sweatt, J.D.; Beaudet, A.L. Mutation of the Angelman ubiquitin ligase in mice causes increased cytoplasmic p53 and deficits of contextual learning and long-term potentiation. Neuron 1998, 21, 799–811. [Google Scholar] [CrossRef]
- Sonzogni, M.; Wallaard, I.; Santos, S.S.; Kingma, J.; du Mee, D.; van Woerden, G.M.; Elgersma, Y. A behavioral test battery for mouse models of Angelman syndrome: A powerful tool for testing drugs and novel. Mol. Autism 2018, 9, 47. [Google Scholar] [CrossRef] [PubMed]
- Neumann, P.E.; Collins, R.L. Confirmation of the influence of a chromosome 7 locus on susceptibility to audiogenic seizures. Mamm. Genome 1992, 3, 250–253. [Google Scholar] [CrossRef] [PubMed]
- Ferraro, T.N.; Golden, G.T.; Smith, G.G.; Martin, J.F.; Lohoff, F.W.; Gieringer, T.A.; Zamboni, D.; Schwebel, C.L.; Press, D.M.; Kratzer, S.O.; et al. Fine mapping of a seizure susceptibility locus on mouse Chromosome 1: Nomination of Kcnj10 as a causative gene. Mamm. Genome 2004, 15, 239–251. [Google Scholar] [CrossRef] [PubMed]
- Jensen, T.P.; Filoteo, A.G.; Knopfel, T.; Empson, R.M. Presynaptic plasma membrane Ca2+ ATPase isoform 2a regulates excitatory synaptic transmission in rat hippocampal CA3. J. Physiol. 2007, 579, 85–99. [Google Scholar] [CrossRef] [PubMed]
- Inyushin, M.; Kucheryavykh, L.Y.; Kucheryavykh, Y.V.; Nichols, C.G.; Buono, R.J.; Ferraro, T.N.; Skatchkov, S.N.; Eaton, M.J. Potassium channel activity and glutamate uptake are impaired in astrocytes of seizure-susceptible DBA/2 mice. Epilepsia 2010, 51, 1707–1713. [Google Scholar] [CrossRef] [PubMed]
- Boiarshinova, O.S.; Malashenko, A.M.; Bizikoeva, F.Z.; Revishchin, A.V.; Lil’p, I.G.; Poletaeva, I.I. The 1xC3 panel of recombinant inbred mouse strains. Presence or absence of pathologic neurologic traits of one of parental strains. Genetika 2009, 45, 280–283. [Google Scholar]
- Thompson, J.L.; Carl, F.G.; Holmes, G.L. Effects of age on seizure susceptibility in genetically epilepsy-prone rats (GEPR-9s). Epilepsia 1991, 32, 161–167. [Google Scholar] [CrossRef]
- Ribak, C.E.; Roberts, R.C.; Byun, M.Y.; Kim, H.L. Anatomical and behavioral analyses of the inheritance of audiogenic seizures in the progeny of genetically epilepsy-prone and Sprague-Dawley rats. Epilepsy Res. 1988, 2, 345–355. [Google Scholar] [CrossRef]
- Ribak, C.E. An abnormal GABAergic system in the inferior colliculus provides a basis for audiogenic seizures in genetically epilepsy-prone rats. Epilepsy Behav. 2017, 71, 160–164. [Google Scholar] [CrossRef]
- Damasceno, S.; Gómez-Nieto, R.; Garcia-Cairasco, N.; Herrero-Turrión, M.J.; Marín, F.; Lopéz, D.E. Top Common Differentially Expressed Genes in the Epileptogenic Nucleus of Two Strains of Rodents Susceptible to Audiogenic Seizures: WAR and GASH/Sal. Front. Neurol. 2020, 11, 33. [Google Scholar] [CrossRef]
- Damasceno, S.; Menezes, N.B.; Rocha, C.S.; Matos, A.H.B.; Vieira, A.S.; Moraes, M.F.D.; Martins, A.S.; Lopes-Cendes, I.; Godard, A.L.B. Transcriptome of the Wistar audiogenic rat (WAR) strain following audiogenic seizures. Epilepsy Res. 2018, 147, 22–31. [Google Scholar] [CrossRef] [PubMed]
- Díaz-Casado, E.; Gómez-Nieto, R.; de Pereda, J.M.; Muñoz, L.J.; Jara-Acevedo, M.; López, D.E. Analysis of gene variants in the GASH/Sal model of epilepsy. PLoS ONE 2020, 15, e0229953. [Google Scholar] [CrossRef] [PubMed]
- Díaz-Rodríguez, S.M.; López-López, D.; Herrero-Turrión, M.J.; Gómez-Nieto, R.; Canal-Alonso, A.; Lopéz, D.E. Inferior Colliculus Transcriptome After Status Epilepticus in the Genetically Audiogenic Seizure-Prone Hamster GASH/Sal. Front. NeuroSci. 2020, 14, 508. [Google Scholar] [CrossRef] [PubMed]
- López-López, D.; Gómez-Nieto, R.; Herrero-Turrión, M.J.; García-Cairasco, N.; Sánchez-Benito, D.; Ludeña, M.D.; López, D.E. Overexpression of the immediate-early genes Egr1, Egr2, and Egr3 in two strains of rodents susceptible to audiogenic seizures. Epilepsy Behav. 2017, 71, 226–237. [Google Scholar] [CrossRef]
- Roberts, D.S.; Raol, Y.H.; Bandyopadhyay, S.; Lund, I.V.; Budreck, E.C.; Passini, M.A.; Passini, M.J.; Wolfe, J.H.; Brooks-Kayal, A.R.; Russek, S.J. Egr3 stimulation of GABRA4 promoter activity as a mechanism for seizure-induced up-regulation of GABA(A) receptor alpha4 subunit expression. Proc. Natl. Acad. Sci. USA 2005, 102, 11894–11899. [Google Scholar] [CrossRef]
- Brooks-Kayal, A.R.; Shumate, M.D.; Jin, H.; Rikhter, T.Y.; Coulter, D.A. Selective changes in single cell GABA(A) receptor subunit expression and function in temporal lobe epilepsy. Nat. Med. 1998, 4, 1166–1172. [Google Scholar] [CrossRef]
- Ando, Y.; Coelho, T.; Berk, J.L.; Cruz, M.W.; Ericzon, B.G.; Ikeda, S.; Lewis, W.D.; Obici, L.; Planté-Bordeneuve, V.; Rapezzi, C.; et al. Guideline of transthyretin-related hereditary amyloidosis for clinicians. Orphanet J. Rare Dis. 2013, 8, 31. [Google Scholar] [CrossRef]
- Franco, A.; Bentes, C.; de Carvalho, M.; Pereira, P.; Pimentel, J.; Conceição, I. Epileptic seizures as a presentation of central nervous system involvement in TTR Val30Met-FAP. J. Neurol. 2016, 263, 2336–2338. [Google Scholar] [CrossRef]
- Zhou, L.; Tang, X.; Li, X.; Bai, Y.; Buxbaum, J.N.; Chen, G. Identification of transthyretin as a novel interacting partner for the δ subunit of GABAA receptors. PLoS ONE 2019, 14, e0210094. [Google Scholar] [CrossRef]
- Stein, T.D.; Anders, N.J.; DeCarli, C.; Chan, S.L.; Mattson, M.P.; Johnson, J.A. Neutralization of transthyretin reverses the neuroprotective effects of secreted amyloid precursor protein (APP) in APPSW mice resulting in tau phosphorylation and loss of hippocampal neurons: Support for the amyloid hypothesis. J. NeuroSci. 2004, 24, 7707–7717. [Google Scholar] [CrossRef]
- Kajiwara, K.; Sunaga, K.; Tsuda, T.; Sugaya, A.; Sugaya, E.; Kimura, M. Peony root extract upregulates transthyretin and phosphoglycerate mutase in mouse cobalt focus seizure. BioChem. Biophys. Res. Commun. 2008, 371, 375–379. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Buxbaum, J.N. Transthyretin and the brain re-visited: Is neuronal synthesis of transthyretin protective in Alzheimer’s disease? Mol. Neurodegener. 2011, 6, 79. [Google Scholar] [CrossRef] [PubMed]
- Siderovski, D.P.; Heximer, S.P.; Forsdyke, D.R. A human gene encoding a putative basic helix-loop-helix phosphoprotein whose mRNA increases rapidly in cycloheximide-treated blood mononuclear cells. DNA Cell Biol. 1994, 13, 125–147. [Google Scholar] [CrossRef] [PubMed]
- Gold, S.J.; Heifets, B.D.; Pudiak, C.M.; Potts, B.W.; Nestler, E.J. Regulation of regulators of G protein signaling mRNA expression in rat brain by acute and chronic electroconvulsive seizures. J. NeuroChem. 2002, 82, 828–838. [Google Scholar] [CrossRef]
- Milanesi, E.; Cucos, C.A.; Matias-Guiu, J.A.; Piñol-Ripoll, G.; Manda, G.; Dobre, M.; Cuadrado, A. Reduced Blood. Front. Aging NeuroSci. 2021, 13, 738244. [Google Scholar] [CrossRef]
- Noe’, F.; Nissinen, J.; Pitkänen, A.; Gobbi, M.; Sperk, G.; During, M.; Vezzani, A. Gene therapy in epilepsy: The focus on NPY. Peptides 2007, 28, 377–383. [Google Scholar] [CrossRef]
- Ercegovac, M.; Jovic, N.; Sokic, D.; Savic-Radojevic, A.; Coric, V.; Radic, T.; Nikolic, D.; Kecmanovic, M.; Matic, M.; Simic, T.; et al. GSTA1, GSTM1, GSTP1 and GSTT1 polymorphisms in progressive myoclonus epilepsy: A Serbian case-control study. Seizure 2015, 32, 30–36. [Google Scholar] [CrossRef]
- Prabha, T.S.; Kumaraswami, K.; Kutala, V.K. Association of GSTT1 and GSTM1 polymorphisms in South Indian Epilepsy Patients. Indian J. Exp. Biol. 2016, 54, 783–787. [Google Scholar]
- Jiang, X.; Raju, P.K.; D’Avanzo, N.; Lachance, M.; Pepin, J.; Dubeau, F.; Mitchell, W.G.; Bello-Espinosa, L.E.; Pierson, T.M.; Minassian, B.A.; et al. Both gain-of-function and loss-of-function de novo CACNA1A mutations cause severe developmental epileptic encephalopathies in the spectrum of Lennox-Gastaut syndrome. Epilepsia 2019, 60, 1881–1894. [Google Scholar] [CrossRef]
- Sander, T.; Hildmann, T.; Kretz, R.; Fürst, R.; Sailer, U.; Bauer, G.; Schmitz, B.; Beck-Mannagetta, G.; Wienker, T.F.; Janz, D. Allelic association of juvenile absence epilepsy with a GluR5 kainate receptor gene (GRIK1) polymorphism. Am. J. Med. Genet. 1997, 74, 416–421. [Google Scholar] [CrossRef]
- Wu, M.; Katti, P.; Zhao, Y.; Peoples, R.W. Positions in the N-methyl-D-aspartate Receptor GluN2C Subunit M3 and M4 Domains Regulate Alcohol Sensitivity and Receptor Kinetics. Alcohol. Clin. Exp. Res. 2019, 43, 1180–1190. [Google Scholar] [CrossRef] [PubMed]
- Venkateswaran, S.; Myers, K.A.; Smith, A.C.; Beaulieu, C.L.; Schwartzentruber, J.A.; Majewski, J.; Bulman, D.; Boycott, K.M.; Dyment, D.A.; Consortium, F.C. Whole-exome sequencing in an individual with severe global developmental delay and intractable epilepsy identifies a novel, de novo GRIN2A mutation. Epilepsia 2014, 55, e75–e79. [Google Scholar] [CrossRef] [PubMed]
- Gun-Bilgic, D.; Polat, M. Analysis of the Pathogenic Variants of Genes Using a Gene Panel in Turkish Epilepsy Patients. Clin. Lab. 2022, 68. [Google Scholar] [CrossRef] [PubMed]
- Moore, S.W.; Fieggen, K.; Honey, E.; Zaahl, M. Novel Zeb2 gene variation in the Mowat Wilson syndrome (MWS). J. Pediatr. Surg. 2016, 51, 268–271. [Google Scholar] [CrossRef] [PubMed]
- Yamada, Y.; Nomura, N.; Yamada, K.; Matsuo, M.; Suzuki, Y.; Sameshima, K.; Kimura, R.; Yamamoto, Y.; Fukushi, D.; Fukuhara, Y.; et al. The spectrum of ZEB2 mutations causing the Mowat-Wilson syndrome in Japanese populations. Am. J. Med. Genet. A 2014, 164A, 1899–1908. [Google Scholar] [CrossRef] [PubMed]
- Singh, N.A.; Westenskow, P.; Charlier, C.; Pappas, C.; Leslie, J.; Dillon, J.; Anderson, V.E.; Sanguinetti, M.C.; Leppert, M.F.; Consortium, B.P. KCNQ2 and KCNQ3 potassium channel genes in benign familial neonatal convulsions: Expansion of the functional and mutation spectrum. Brain 2003, 126, 2726–2737. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.; Gellert, M.; Yang, W. Mechanism of mismatch recognition revealed by human MutSβ bound to unpaired DNA loops. Nat. Struct. Mol. Biol. 2011, 19, 72–78. [Google Scholar] [CrossRef]
- Francisconi, S.; Codenotti, M.; Ferrari Toninelli, G.; Uberti, D.; Memo, M. Mitochondrial dysfunction and increased sensitivity to excitotoxicity in mice deficient in DNA mismatch repair. J. NeuroChem. 2006, 98, 223–233. [Google Scholar] [CrossRef]
- Rahman, S. Pathophysiology of mitochondrial disease causing epilepsy and status epilepticus. Epilepsy Behav. 2015, 49, 71–75. [Google Scholar] [CrossRef]
- Chernigovskaya, E.V.; Dorofeeva, N.A.; Nasluzova, E.V.; Kulikov, A.A.; Ovsyannikova, V.V.; Glazova, M.V. Apoptosis and proliferation in the inferior colliculus during postnatal development and epileptogenesis in audiogenic Krushinsky-Molodkina rats. Epilepsy Behav. 2018, 88, 227–234. [Google Scholar] [CrossRef]
- Chernigovskaya, E.V.; Korotkov, A.A.; Dorofeeva, N.A.; Gorbacheva, E.L.; Kulikov, A.A.; Glazova, M.V. Delayed audiogenic seizure development in a genetic rat model is associated with overactivation of ERK1/2 and disturbances in glutamatergic signaling. Epilepsy Behav. 2019, 99, 106494. [Google Scholar] [CrossRef] [PubMed]
- Fedotova, I.B.; Kostyna, Z.A.; Surina, N.M.; Poletaeva, I.I. Laboratory rat selection for the trait “The absence of audiogenic seizure proneness”. Genetika 2012, 48, 685–691. [Google Scholar] [PubMed]
- Poletaeva, I.I.; Surina, N.M.; Ashapkin, V.V.; Fedotova, I.B.; Merzalov, I.B.; Perepelkina, O.V.; Pavlova, G.V. Maternal methyl-enriched diet in rat reduced the audiogenic seizure proneness in progeny. Pharmacol. BioChem. Behav. 2014, 127, 21–26. [Google Scholar] [CrossRef] [PubMed]
- Parker, M.H.; Smith-Swintosky, V.L.; McComsey, D.F.; Huang, Y.; Brenneman, D.; Klein, B.; Malatynska, E.; White, H.S.; Milewski, M.E.; Herb, M.; et al. Novel, broad-spectrum anticonvulsants containing a sulfamide group: Advancement of N-((benzo[b]thien-3-yl)methyl)sulfamide (JNJ-26990990) into human clinical studies. J Med. Chem. 2009, 52, 7528–7536. [Google Scholar] [CrossRef] [PubMed]
- Nishi, T.; Metcalf, C.S.; Fujimoto, S.; Hasegawa, S.; Miyamoto, M.; Sunahara, E.; Watanabe, S.; Kondo, S.; White, H.S. Anticonvulsive properties of soticlestat, a novel cholesterol 24-hydroxylase inhibitor. Epilepsia 2022, 63, 1580–1590. [Google Scholar] [CrossRef] [PubMed]
- Alachkar, A.; Ojha, S.K.; Sadeq, A.; Adem, A.; Frank, A.; Stark, H.; Sadek, B. Experimental Models for the Discovery of Novel Anticonvulsant Drugs: Focus on Pentylenetetrazole-Ind.duced Seizures and Associated Memory Deficits. Curr. Pharm. Des. 2020, 26, 1693–1711. [Google Scholar] [CrossRef]
- Barker-Haliski, M.; Steve White, H. Validated animal models for antiseizure drug (ASD) discovery: Advantages and potential pitfalls in ASD screening. Neuropharmacology 2020, 167, 107750. [Google Scholar] [CrossRef]
- Singh, T.; Mishra, A.; Goel, R.K. PTZ kindling model for epileptogenesis, refractory epilepsy, and associated comorbidities: Relevance and reliability. Metab. Brain Dis. 2021, 36, 1573–1590. [Google Scholar] [CrossRef]
- Ohno, Y.; Ishihara, S.; Terada, R.; Serikawa, T.; Sasa, M. Antiepileptogenic and anticonvulsive actions of levetiracetam in a pentylenetetrazole kindling model. Epilepsy Res. 2010, 89, 360–364. [Google Scholar] [CrossRef]
- Vinogradova, L.V.; van Rijn, C.M. Anticonvulsive and antiepileptogenic effects of levetiracetam in the audiogenic kindling model. Epilepsia 2008, 49, 1160–1168. [Google Scholar] [CrossRef]
- Kasteleijn-Nolst Trenité, D.G.; Hirsch, E. Levetiracetam: Preliminary efficacy in generalized seizures. Epileptic Disord. 2003, 5 (Suppl. S1), S39–S44. [Google Scholar]
- Löscher, W. Genetic animal models of epilepsy as a unique resource for the evaluation of anticonvulsant drugs. A review. Methods Find. Exp. Clin. Pharmacol. 1984, 6, 531–547. [Google Scholar] [PubMed]
- Klitgaard, H.; Matagne, A.; Lamberty, Y. Use of epileptic animals for adverse effect testing. Epilepsy Res. 2002, 50, 55–65. [Google Scholar] [CrossRef]
- Chung, L.; Bey, A.L.; Towers, A.J.; Cao, X.; Kim, I.H.; Jiang, Y.H. Lovastatin suppresses hyperexcitability and seizure in Angelman syndrome model. Neurobiol. Dis. 2018, 110, 12–19. [Google Scholar] [CrossRef] [PubMed]
- Heulens, I.; D’Hulst, C.; Van Dam, D.; De Deyn, P.P.; Kooy, R.F. Pharmacological treatment of fragile X syndrome with GABAergic drugs in a knockout mouse model. Behav. Brain Res. 2012, 229, 244–249. [Google Scholar] [CrossRef] [PubMed]
- Wildin, J.D.; Pleuvry, B.J. Tolerance to the anticonvulsant effects of clobazam in mice. Neuropharmacology 1992, 31, 129–135. [Google Scholar] [CrossRef]
- Gatta, E.; Cupello, A.; Di Braccio, M.; Grossi, G.; Robello, M.; Scicchitano, F.; Russo, E.; De Sarro, G. Anticonvulsive Activity in Audiogenic DBA/2 Mice of 1,4-Benzodiazepines and 1,5-Benzodiazepines with Different Activities at Cerebellar Granule Cell GABA. J. Mol. NeuroSci. 2016, 60, 539–547. [Google Scholar] [CrossRef]
- Muzzi, M.; Coppi, E.; Pugliese, A.M.; Chiarugi, A. Anticonvulsant effect of AMP by direct activation of adenosine A1 receptor. Exp. Neurol. 2013, 250, 189–193. [Google Scholar] [CrossRef]
- De Sarro, G.; De Sarro, A.; Di Paola, E.D.; Bertorelli, R. Effects of adenosine receptor agonists and antagonists on audiogenic seizure-sensible DBA/2 mice. Eur. J. Pharmacol. 1999, 371, 137–145. [Google Scholar] [CrossRef]
- Sparks, D.L.; Buckholtz, N.S. Effects of 6-methoxy-1,2,3,4-tetrahydro-beta-carboline (6-MeO-THbetaC) on audiogenic seizures in DBA/2J mice. Pharmacol. BioChem. Behav. 1980, 12, 119–124. [Google Scholar] [CrossRef]
- Brown, J.W.; Moeller, A.; Schmidt, M.; Turner, S.C.; Nimmrich, V.; Ma, J.; Rueter, L.E.; van der Kam, E.; Zhang, M. Anticonvulsant effects of structurally diverse GABA(B) positive allosteric modulators in the DBA/2J audiogenic seizure test: Comparison to baclofen and utility as a pharmacodynamic screening model. Neuropharmacology 2016, 101, 358–369. [Google Scholar] [CrossRef]
- Gareri, P.; Condorelli, D.; Belluardo, N.; Gratteri, S.; Ferreri, G.; Donato Di Paola, E.; De Sarro, A.; De Sarro, G. Influence of carbenoxolone on the anticonvulsant efficacy of conventional antiepileptic drugs against audiogenic seizures in DBA/2 mice. Eur. J. Pharmacol. 2004, 484, 49–56. [Google Scholar] [CrossRef]
- Russo, E.; Donato di Paola, E.; Gareri, P.; Siniscalchi, A.; Labate, A.; Gallelli, L.; Citraro, R.; De Sarro, G. Pharmacodynamic potentiation of antiepileptic drugs’ effects by some HMG-CoA reductase inhibitors against audiogenic seizures in DBA/2 mice. Pharmacol. Res. 2013, 70, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Madsen, K.K.; Clausen, R.P.; Larsson, O.M.; Krogsgaard-Larsen, P.; Schousboe, A.; White, H.S. Synaptic and extrasynaptic GABA transporters as targets for anti-epileptic drugs. J. NeuroChem. 2009, 109 (Suppl. S1), 139–144. [Google Scholar] [CrossRef] [PubMed]
- Quansah, H.; N’Gouemo, P. Amiloride and SN-6 suppress audiogenic seizure susceptibility in genetically epilepsy-prone rats. CNS NeuroSci. Ther. 2014, 20, 860–866. [Google Scholar] [CrossRef] [PubMed]
- De Sarro, C.; Tallarico, M.; Pisano, M.; Gallelli, L.; Citraro, R.; De Sarro, G.; Leo, A. Liraglutide chronic treatment prevents development of tolerance to antiseizure effects of diazepam in genetically epilepsy prone rats. Eur. J. Pharmacol. 2022, 928, 175098. [Google Scholar] [CrossRef]
- Vinogradova, L.V.; Kuznetsova, G.D.; Shatskova, A.B.; van Rijn, C.M. Vigabatrin in low doses selectively suppresses the clonic component of audiogenically kindled seizures in rats. Epilepsia 2005, 46, 800–810. [Google Scholar] [CrossRef]
- François, J.; Boehrer, A.; Nehlig, A. Effects of carisbamate (RWJ-333369) in two models of genetically determined generalized epilepsy, the GAERS and the audiogenic Wistar AS. Epilepsia 2008, 49, 393–399. [Google Scholar] [CrossRef]
- Damasceno, D.D.; Ferreira, A.J.; Doretto, M.C.; Almeida, A.P. Anticonvulsant and antiarrhythmic effects of nifedipine in rats prone to audiogenic seizures. Braz J. Med. Biol. Res. 2012, 45, 1060–1065. [Google Scholar] [CrossRef][Green Version]
- Lazarini-Lopes, W.; Do Val-da Silva, R.A.; da Silva-Júnior, R.M.P.; Silva-Cardoso, G.K.; Leite-Panissi, C.R.A.; Leite, J.P.; Garcia-Cairasco, N. Chronic cannabidiol (CBD) administration induces anticonvulsant and antiepileptogenic effects in a genetic model of epilepsy. Epilepsy Behav. 2021, 119, 107962. [Google Scholar] [CrossRef]
- Barrera-Bailón, B.; Oliveira, J.A.C.; López, D.E.; Muñoz, L.J.; Garcia-Cairasco, N.; Sancho, C. Pharmacological and neuroethological study of the acute and chronic effects of lamotrigine in the genetic audiogenic seizure hamster (GASH:Sal). Epilepsy Behav. 2017, 71, 207–217. [Google Scholar] [CrossRef] [PubMed]
- Barrera-Bailón, B.; Oliveira, J.A.; López, D.E.; Muñoz, L.J.; Garcia-Cairasco, N.; Sancho, C. Pharmacological and neuroethological studies of three antiepileptic drugs in the Genetic Audiogenic Seizure Hamster (GASH:Sal). Epilepsy Behav. 2013, 28, 413–425. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Turner, T.J.; Zourray, C.; Schorge, S.; Lignani, G. Recent advances in gene therapy for neurodevelopmental disorders with epilepsy. J. NeuroChem. 2021, 157, 229–262. [Google Scholar] [CrossRef] [PubMed]
- Nadadhur, A.G.; Alsaqati, M.; Gasparotto, L.; Cornelissen-Steijger, P.; van Hugte, E.; Dooves, S.; Harwood, A.J.; Heine, V.M. Neuron-Glia Interactions Increase Neuronal Phenotypes in Tuberous Sclerosis Complex Patient iPSC-Derived Models. Stem Cell Rep. 2019, 12, 42–56. [Google Scholar] [CrossRef]
- Schauwecker, P.E. The relevance of individual genetic background and its role in animal models of epilepsy. Epilepsy Res. 2011, 97, 1–11. [Google Scholar] [CrossRef]
- Schauwecker, P.E. Complications associated with genetic background effects in models of experimental epilepsy. Prog Brain Res. 2002, 135, 139–148. [Google Scholar] [CrossRef]
- Combi, R.; Dalprà, L.; Ferini-Strambi, L.; Tenchini, M.L. Frontal lobe epilepsy and mutations of the corticotropin-releasing hormone gene. Ann. Neurol. 2005, 58, 899–904. [Google Scholar] [CrossRef]
- Alakurtti, K.; Virtaneva, K.; Joensuu, T.; Palvimo, J.J.; Lehesjoki, A.E. Characterization of the cystatin B gene promoter harboring the dodecamer repeat expanded in progressive myoclonus epilepsy, EPM1. Gene 2000, 242, 65–73. [Google Scholar] [CrossRef]
- Pathak, S.; Miller, J.; Morris, E.C.; Stewart, W.C.L.; Greenberg, D.A. DNA methylation of the BRD2 promoter is associated with juvenile myoclonic epilepsy in Caucasians. Epilepsia 2018, 59, 1011–1019. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Pitkänen, A.; Löscher, W.; Vezzani, A.; Becker, A.J.; Simonato, M.; Lukasiuk, K.; Gröhn, O.; Bankstahl, J.P.; Friedman, A.; Aronica, E.; et al. Advances in the development of biomarkers for epilepsy. Lancet Neurol. 2016, 15, 843–856. [Google Scholar] [CrossRef]
- Li, M.M.; Li, X.M.; Zheng, X.P.; Yu, J.T.; Tan, L. MicroRNAs dysregulation in epilepsy. Brain Res. 2014, 1584, 94–104. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y. The Challenge of microRNA as a Biomarker of Epilepsy. Curr. NeuroPharmacol. 2018, 16, 37–42. [Google Scholar] [CrossRef] [PubMed]
- Henshall, D.C.; Hamer, H.M.; Pasterkamp, R.J.; Goldstein, D.B.; Kjems, J.; Prehn, J.H.M.; Schorge, S.; Lamottke, K.; Rosenow, F. MicroRNAs in epilepsy: Pathophysiology and clinical utility. Lancet Neurol. 2016, 15, 1368–1376. [Google Scholar] [CrossRef]
- Karnati, H.K.; Panigrahi, M.K.; Gutti, R.K.; Greig, N.H.; Tamargo, I.A. miRNAs: Key Players in Neurodegenerative Disorders and Epilepsy. J. Alzheimers Dis. 2015, 48, 563–580. [Google Scholar] [CrossRef]
- Liu, S.; Fan, M.; Ma, M.; Ge, J.; Chen, F. Long noncoding RNAs: Potential therapeutic targets for epilepsy. Front. NeuroSci. 2022, 16, 986874. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Garbuz, D.G.; Davletshin, A.A.; Litvinova, S.A.; Fedotova, I.B.; Surina, N.M.; Poletaeva, I.I. Rodent Models of Audiogenic Epilepsy: Genetic Aspects, Advantages, Current Problems and Perspectives. Biomedicines 2022, 10, 2934. https://doi.org/10.3390/biomedicines10112934
Garbuz DG, Davletshin AA, Litvinova SA, Fedotova IB, Surina NM, Poletaeva II. Rodent Models of Audiogenic Epilepsy: Genetic Aspects, Advantages, Current Problems and Perspectives. Biomedicines. 2022; 10(11):2934. https://doi.org/10.3390/biomedicines10112934
Chicago/Turabian StyleGarbuz, David G., Artem A. Davletshin, Svetlana A. Litvinova, Irina B. Fedotova, Natalya M. Surina, and Inga I. Poletaeva. 2022. "Rodent Models of Audiogenic Epilepsy: Genetic Aspects, Advantages, Current Problems and Perspectives" Biomedicines 10, no. 11: 2934. https://doi.org/10.3390/biomedicines10112934
APA StyleGarbuz, D. G., Davletshin, A. A., Litvinova, S. A., Fedotova, I. B., Surina, N. M., & Poletaeva, I. I. (2022). Rodent Models of Audiogenic Epilepsy: Genetic Aspects, Advantages, Current Problems and Perspectives. Biomedicines, 10(11), 2934. https://doi.org/10.3390/biomedicines10112934








