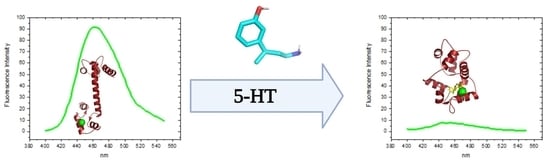
Application of a Fluorescent Biosensor in Determining the Binding of 5-HT to Calmodulin
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Steady-State Fluorescence
2.3. Obtaining and Preparing the Files of the Three-Dimensional Structures
2.4. Docking
2.5. Molecular Dynamics Simulation
2.6. Binding Free Energies Calculated by Molecular Mechanics/Poisson–Boltzmann Surface Area (MM/PBSA)
3. Results and Discussion
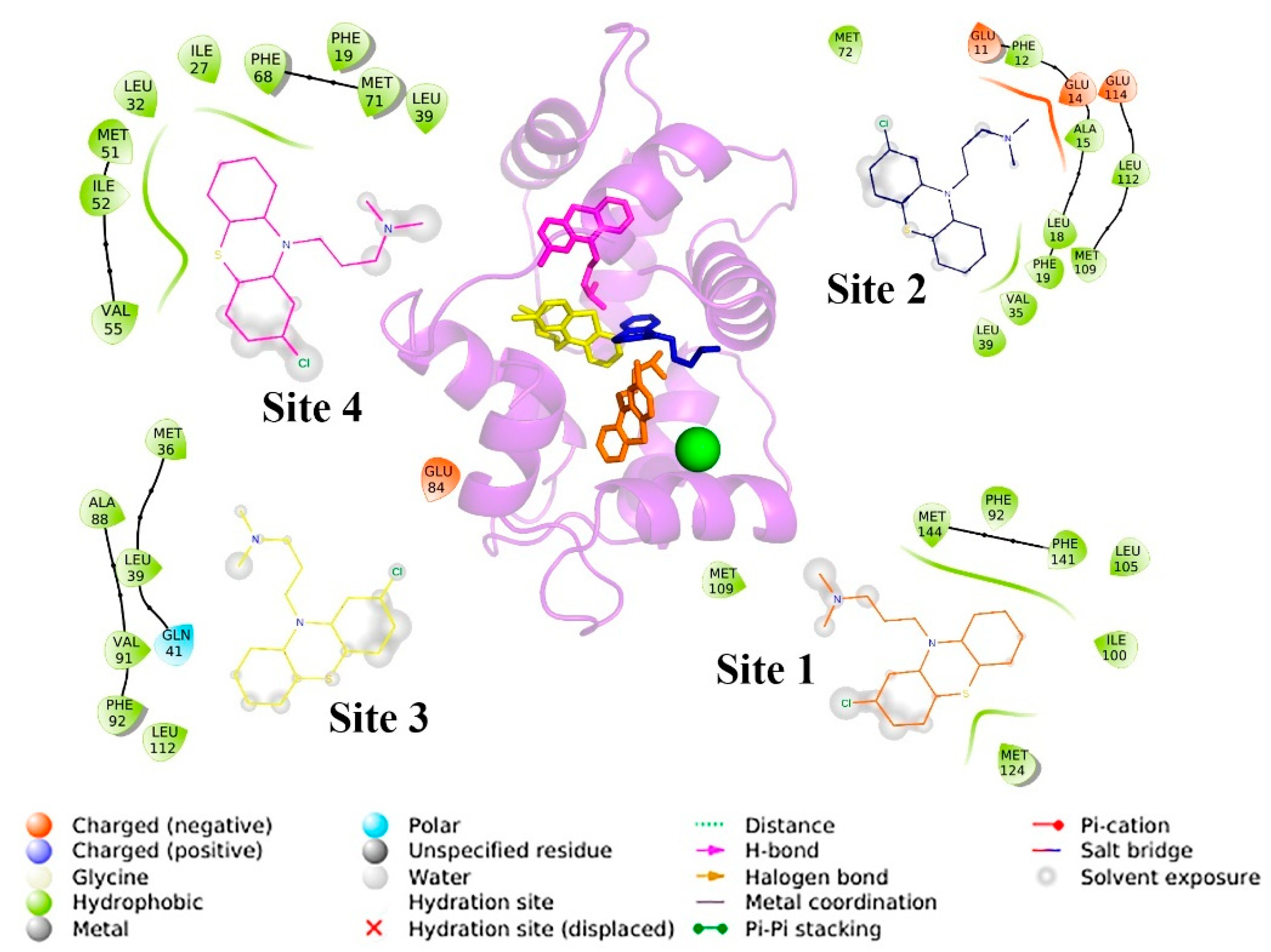
3.1. Analysis of the CaM Complex with Clorpromazine (1:4) for Biosensor Design
3.2. Purification and Chemical Modification with mBBr of the CaM M124C Protein
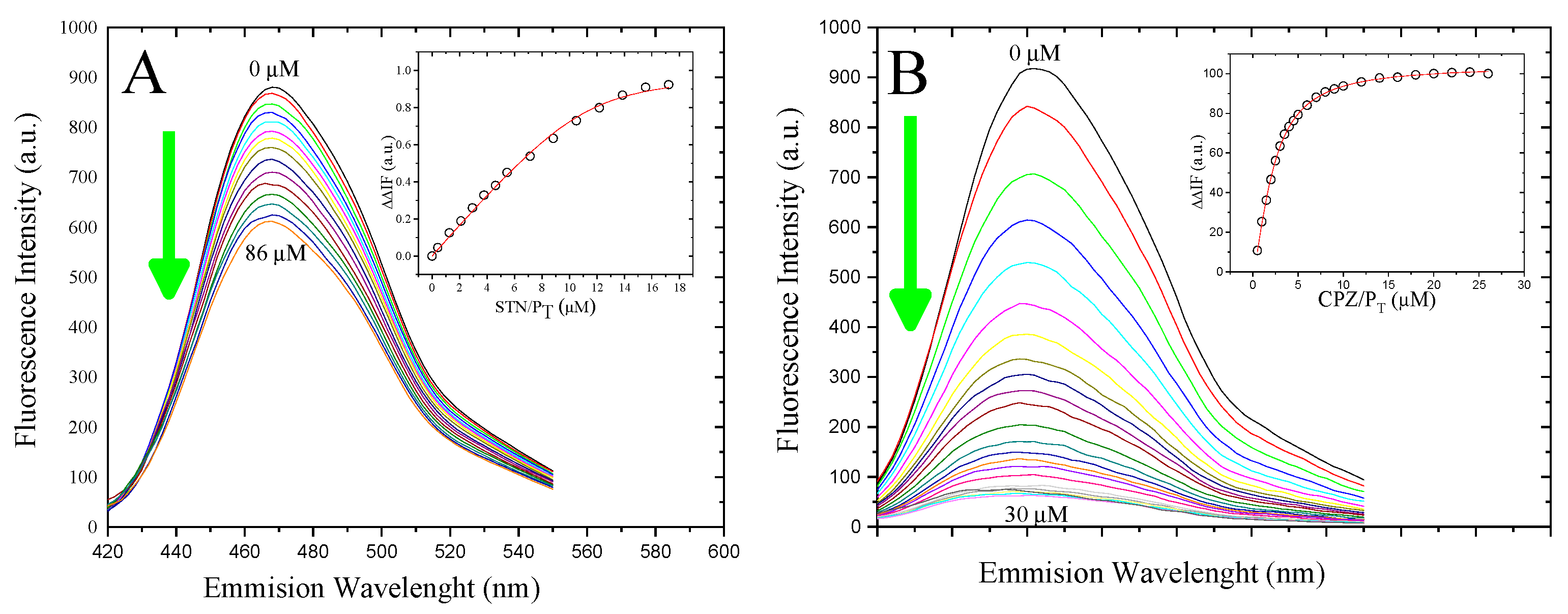
3.3. Determination of the Binding Affinity of 5-HT and CPZ with the Biosensor hCaM M124C-mBBr
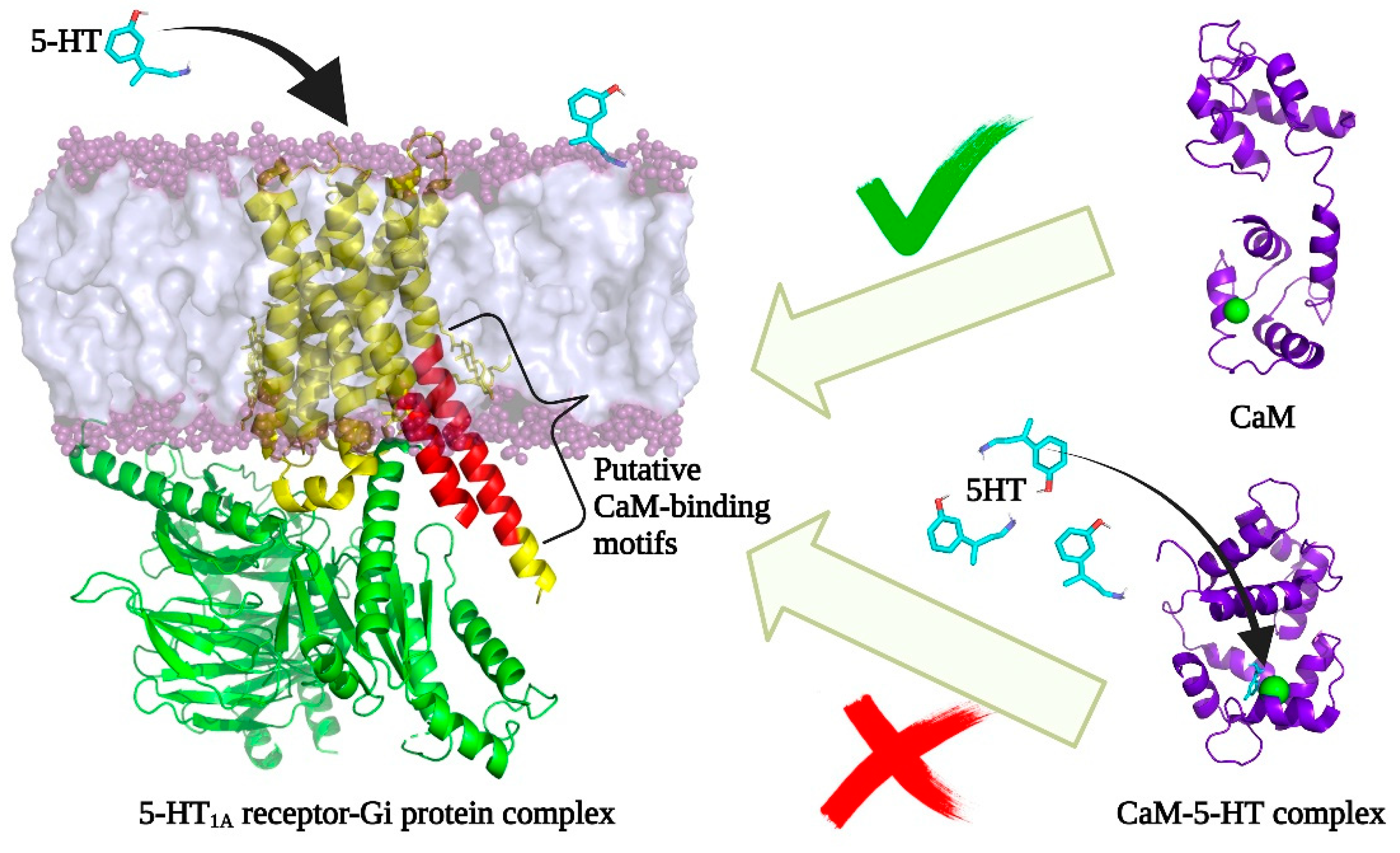
3.4. Relevance of Interaction between CaM–5-HT
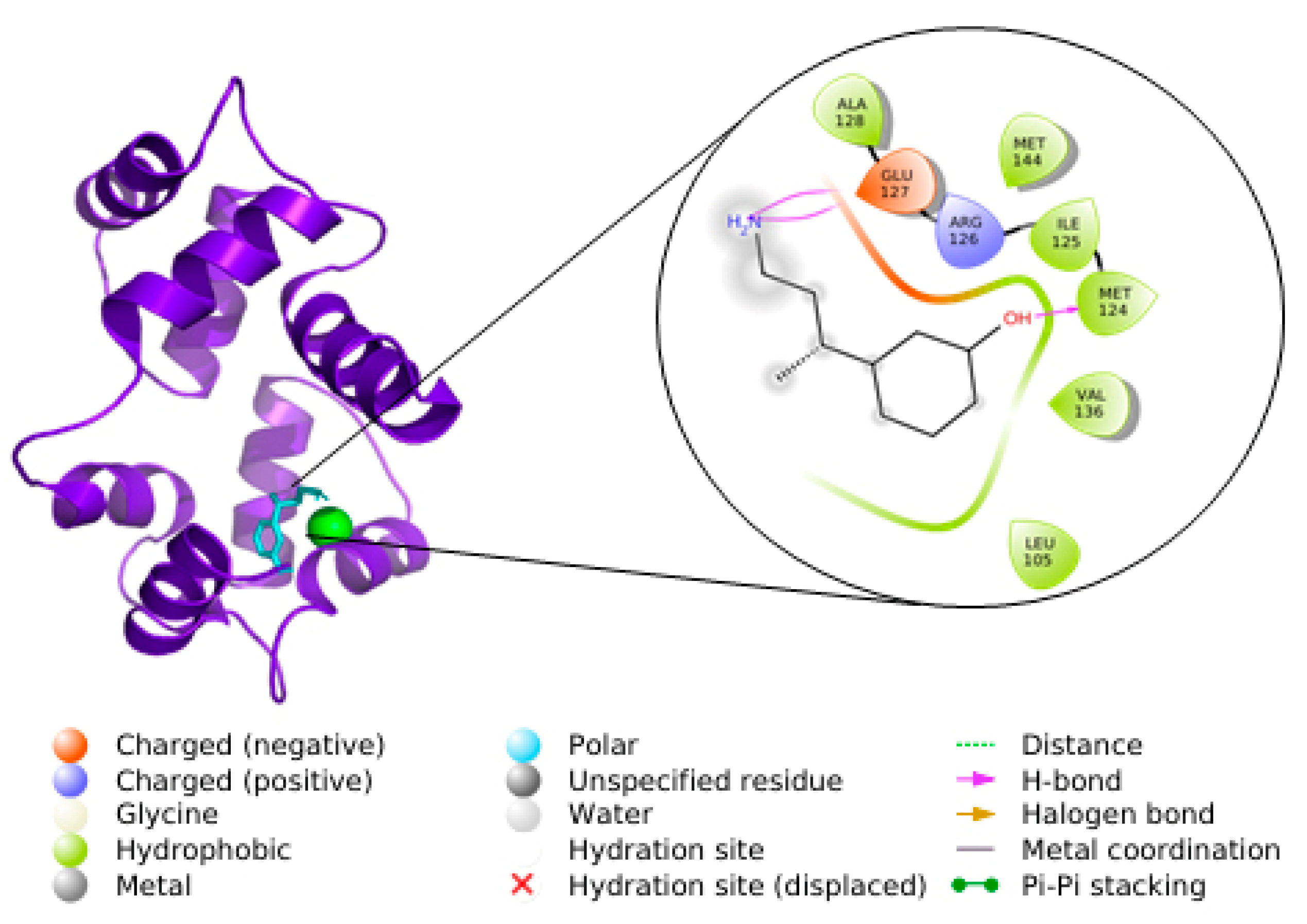
3.5. Docking
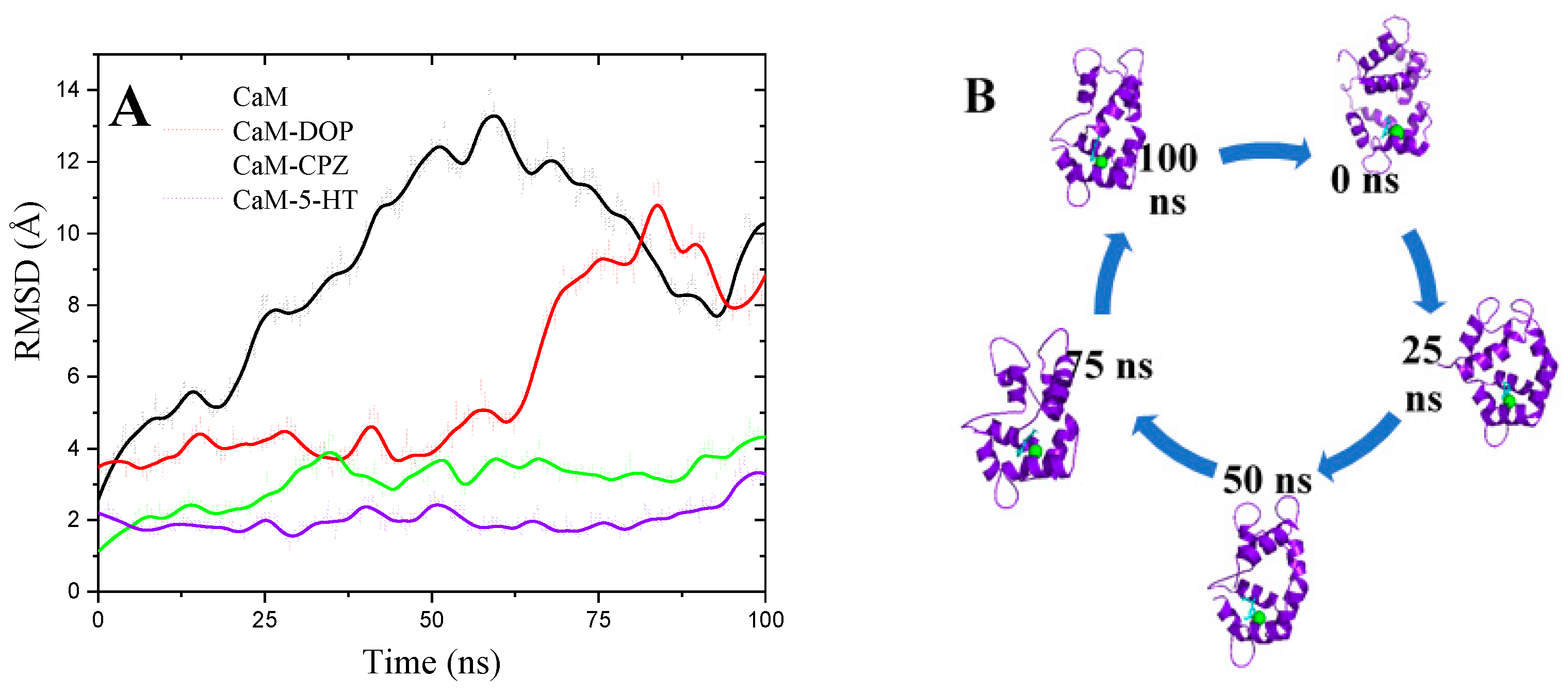
3.6. Molecular Dynamics Simulations
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tainaka, K.; Sakaguchi, R.; Hayashi, H.; Nakano, S.; Liew, F.F.; Morii, T. Design strategies of fluorescent biosensors based on biological macromolecular receptors. Sensors 2010, 10, 1355–1376. [Google Scholar] [CrossRef]
- Keusgen, M. Biosensors: New approaches in drug discovery. Naturwissenschaften 2002, 89, 433–444. [Google Scholar] [CrossRef] [PubMed]
- Orosz, F.; Telegdi, M.; Liliom, K.; Solti, M.; Korbonits, D.; Ovadi, J. Dissimilar mechanisms of action of anticalmodulin drugs: Quantitative analysis. Mol. Pharm. 1990, 38, 910–916. [Google Scholar]
- Martinez-Luis, S.; Perez-Vasquez, A.; Mata, R. Natural products with calmodulin inhibitor properties. Phytochemistry 2007, 68, 1882–1903. [Google Scholar] [CrossRef] [PubMed]
- Horvath, I.; Harmat, V.; Perczel, A.; Palfi, V.; Nyitray, L.; Nagy, A.; Hlavanda, E.; Naray-Szabo, G.; Ovadi, J. The structure of the complex of calmodulin with KAR-2: A novel mode of binding explains the unique pharmacology of the drug. J. Biol. Chem. 2005, 280, 8266–8274. [Google Scholar] [CrossRef] [Green Version]
- Heyduk, T. Measuring protein conformational changes by FRET/LRET. Curr. Opin. Biotechnol. 2002, 13, 292–296. [Google Scholar] [CrossRef]
- Torres-Piedra, M.; Figueroa, M.; Hernandez-Abreu, O.; Ibarra-Barajas, M.; Navarrete-Vazquez, G.; Estrada-Soto, S. Vasorelaxant effect of flavonoids through calmodulin inhibition: Ex vivo, in vitro, and in silico approaches. Bioorg. Med. Chem. 2011, 19, 542–546. [Google Scholar] [CrossRef]
- Figueroa, M.; González, M.d.C.; Rodríguez-Sotres, R.; Sosa-Peinado, A.; González-Andrade, M.; Cerda-García-Rojas, C.M.; Mata, R. Calmodulin inhibitors from the fungus Emericella sp. Bioorganic Med. Chem. 2009, 17, 2167–2174. [Google Scholar] [CrossRef] [Green Version]
- Rajendra Prasad, V.V.; Deepak Reddy, G.; Appaji, D.; Peters, G.J.; Mayur, Y.C. Chemosensitizing acridones: In vitro calmodulin dependent cAMP phosphodiesterase inhibition, docking, pharmacophore modeling and 3D QSAR studies. J. Mol. Graph. Model. 2013, 40, 116–124. [Google Scholar] [CrossRef]
- Davis, T.N.; Thorner, J. Vertebrate and yeast calmodulin, despite significant sequence divergence, are functionally interchangeable. Proc. Natl. Acad. Sci. USA 1989, 86, 7909–7913. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoeflich, K.P.; Ikura, M. Calmodulin in action: Diversity in target recognition and activation mechanisms. Cell 2002, 108, 739–742. [Google Scholar] [CrossRef] [Green Version]
- Tripathi, S.; Waxham, M.N.; Cheung, M.S.; Liu, Y. Lessons in Protein Design from Combined Evolution and Conformational Dynamics. Sci. Rep. 2015, 5, 14259. [Google Scholar] [CrossRef] [Green Version]
- Seales, E.C.; Micoli, K.J.; McDonald, J.M. Calmodulin is a critical regulator of osteoclastic differentiation, function, and survival. J. Cell Biochem. 2006, 97, 45–55. [Google Scholar] [CrossRef]
- Junker, J.P.; Ziegler, F.; Rief, M. Ligand-dependent equilibrium fluctuations of single calmodulin molecules. Science 2009, 323, 633–637. [Google Scholar] [CrossRef]
- Weiss, B.; Prozialeck, W.C.; Wallace, T.L. Interaction of drugs with calmodulin. Biochemical, pharmacological and clinical implications. Biochem. Pharmacol. 1982, 31, 2217–2226. [Google Scholar] [CrossRef]
- Mayur, Y.C.; Jagadeesh, S.; Thimmaiah, K.N. Targeting calmodulin in reversing multi drug resistance in cancer cells. Mini Rev. Med. Chem. 2006, 6, 1383–1389. [Google Scholar] [CrossRef] [PubMed]
- Mayur, Y.C.; Padma, T.; Parimala, B.H.; Chandramouli, K.H.; Jagadeesh, S.; Gowda, N.M.; Thimmaiah, K.N. Sensitization of multidrug resistant (MDR) cancer cells to vinblastine by novel acridones: Correlation between anti-calmodulin activity and anti-MDR activity. Med. Chem. 2006, 2, 63–77. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Andrade, M.; Figueroa, M.; Rodriguez-Sotres, R.; Mata, R.; Sosa-Peinado, A. An alternative assay to discover potential calmodulin inhibitors using a human fluorophore-labeled CaM protein. Anal. Biochem. 2009, 387, 64–70. [Google Scholar] [CrossRef]
- Kim, C.W.; Choe, C.; Kim, E.J.; Lee, J.I.; Yoon, S.Y.; Cho, Y.W.; Han, S.; Tak, H.M.; Han, J.; Kang, D. Dual effects of fluoxetine on mouse early embryonic development. Toxicol. Appl. Pharm. 2012, 265, 61–72. [Google Scholar] [CrossRef]
- Vandonselaar, M.; Hickie, R.A.; Quail, J.W.; Delbaere, L.T. Trifluoperazine-induced conformational change in Ca(2+)-calmodulin. Nat. Struct. Biol. 1994, 1, 795–801. [Google Scholar] [CrossRef]
- Orosz, F.; Comin, B.; Rais, B.; Puigjaner, J.; Kovacs, J.; Tarkanyi, G.; Acs, T.; Keve, T.; Cascante, M.; Ovadi, J. New semisynthetic vinca alkaloids: Chemical, biochemical and cellular studies. Br. J. Cancer 1999, 79, 1356–1365. [Google Scholar] [CrossRef] [Green Version]
- Mata, R.; Figueroa, M.; Gonzalez-Andrade, M.; Rivera-Chavez, J.A.; Madariaga-Mazon, A.; Del Valle, P. Calmodulin inhibitors from natural sources: An update. J. Nat. Prod. 2015, 78, 576–586. [Google Scholar] [CrossRef]
- Lee, C.T.; Huang, Y.W.; Yang, C.H.; Huang, K.S. Drug delivery systems and combination therapy by using vinca alkaloids. Curr Top. Med. Chem. 2015, 15, 1491–1500. [Google Scholar] [CrossRef] [Green Version]
- Hegde, R.; Thimmaiah, P.; Yerigeri, M.C.; Krishnegowda, G.; Thimmaiah, K.N.; Houghton, P.J. Anti-calmodulin acridone derivatives modulate vinblastine resistance in multidrug resistant (MDR) cancer cells. Eur. J. Med. Chem. 2004, 39, 161–177. [Google Scholar] [CrossRef]
- Ouyang, H.; Vogel, H.J. Melatonin and serotonin interactions with calmodulin: NMR, spectroscopic and biochemical studies. Biochim. Biophys. Acta. 1998, 1383, 37–47. [Google Scholar] [CrossRef]
- Kornhuber, J.; Terfloth, L.; Bleich, S.; Wiltfang, J.; Rupprecht, R. Molecular properties of psychopharmacological drugs determining non-competitive inhibition of 5-HT3A receptors. Eur. J. Med. Chem. 2009, 44, 2667–2672. [Google Scholar] [CrossRef] [PubMed]
- Carro, L.; Torrado, M.; Ravina, E.; Masaguer, C.F.; Lage, S.; Brea, J.; Loza, M.I. Synthesis and biological evaluation of a series of aminoalkyl-tetralones and tetralols as dual dopamine/serotonin ligands. Eur. J. Med. Chem. 2014, 71, 237–249. [Google Scholar] [CrossRef] [PubMed]
- Cortes, A.; Moreno, E.; Rodriguez-Ruiz, M.; Canela, E.I.; Casado, V. Targeting the dopamine D3 receptor: An overview of drug design strategies. Expert Opin. Drug Discov. 2016, 11, 641–664. [Google Scholar] [CrossRef]
- Zajdel, P.; Marciniec, K.; Maslankiewicz, A.; Grychowska, K.; Satala, G.; Duszynska, B.; Lenda, T.; Siwek, A.; Nowak, G.; Partyka, A.; et al. Antidepressant and antipsychotic activity of new quinoline- and isoquinoline-sulfonamide analogs of aripiprazole targeting serotonin 5-HT(1)A/5-HT(2)A/5-HT(7) and dopamine D(2)/D(3) receptors. Eur. J. Med. Chem. 2013, 60, 42–50. [Google Scholar] [CrossRef] [PubMed]
- Adcock, S.A.; McCammon, J.A. Molecular dynamics: Survey of methods for simulating the activity of proteins. Chem. Rev. 2006, 106, 1589–1615. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karplus, M.; McCammon, J.A. Molecular dynamics simulations of biomolecules. Nat. Struct. Biol. 2002, 9, 646–652. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [Green Version]
- Vertessy, B.G.; Harmat, V.; Bocskei, Z.; Naray-Szabo, G.; Orosz, F.; Ovadi, J. Simultaneous binding of drugs with different chemical structures to Ca2+-calmodulin: Crystallographic and spectroscopic studies. Biochemistry 1998, 37, 15300–15310. [Google Scholar] [CrossRef]
- Hanwell, M.D.; Curtis, D.E.; Lonie, D.C.; Vandermeersch, T.; Zurek, E.; Hutchison, G.R. Avogadro: An advanced semantic chemical editor, visualization, and analysis platform. J. Cheminform. 2012, 4, 17. [Google Scholar] [CrossRef] [Green Version]
- Case, D.A.; A, H.M.; Belfon, K.; Ben-Shalom, I.Y.; Brozell, S.R.; Cerutti, D.S.; Cheatham, T.E.; I, I.I.; Cruzeiro, V.W.D.; Darden, T.A.; et al. Amber 2021; University of California: San Francisco, CA, USA, 2021. [Google Scholar]
- Wedemeyer, W.J.; Baker, D. Efficient minimization of angle-dependent potentials for polypeptides in internal coordinates. Proteins 2003, 53, 262–272. [Google Scholar] [CrossRef] [PubMed]
- Morris, G.M.; Goodsell, D.S.; Halliday, R.S.; Huey, R.; Hart, W.E.; Belew, R.K.; Olson, A.J. Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. J. Comput. Chem. 1998, 19, 1639–1662. [Google Scholar] [CrossRef] [Green Version]
- DeLano, W.L. Use of PYMOL as a communications tool for molecular science. Abstr. Pap. Am. Chem. Soc. 2004, 228, U313–U314. [Google Scholar]
- Case, D.A.; Cheatham, T.E., 3rd; Darden, T.; Gohlke, H.; Luo, R.; Merz, K.M., Jr.; Onufriev, A.; Simmerling, C.; Wang, B.; Woods, R.J. The Amber biomolecular simulation programs. J. Comput. Chem. 2005, 26, 1668–1688. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Case, D.A.; Darden, T.A.; Cheatham, T.E.; Simmerling, C.L.I.; Wang, J.; Duke, R.E.; Luo, R.; Crowley, M.; Walker, R.C.; Zhang, W.; et al. Amber 2012; University of California: San Francisco, CA, USA, 2012. [Google Scholar]
- Walker, R.C.; Crowley, M.F.; Case, D.A. The implementation of a fast and accurate QM/MM potential method in Amber. J. Comput. Chem. 2008, 29, 1019–1031. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lindorff-Larsen, K.; Piana, S.; Palmo, K.; Maragakis, P.; Klepeis, J.L.; Dror, R.O.; Shaw, D.E. Improved side-chain torsion potentials for the Amber ff99SB protein force field. Proteins 2010, 78, 1950–1958. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.; Wang, W.; Kollman, P.A.; Case, D.A. Automatic atom type and bond type perception in molecular mechanical calculations. J. Mol. Graph. Model. 2006, 25, 247–260. [Google Scholar] [CrossRef]
- Salomon-Ferrer, R.; Gotz, A.W.; Poole, D.; Le Grand, S.; Walker, R.C. Routine Microsecond Molecular Dynamics Simulations with AMBER on GPUs. Explicit Solvent Particle Mesh Ewald. J. Chem. Theory Comput 2013, 9, 3878–3888. [Google Scholar] [CrossRef]
- Roe, D.R.; Cheatham, T.E., 3rd. PTRAJ and CPPTRAJ: Software for Processing and Analysis of Molecular Dynamics Trajectory Data. J. Chem. Theory Comput. 2013, 9, 3084–3095. [Google Scholar] [CrossRef]
- Hou, T.; Wang, J.; Li, Y.; Wang, W. Assessing the performance of the MM/PBSA and MM/GBSA methods. The accuracy of binding free energy calculations based on molecular dynamics simulations. J. Chem. Inf. Model. 2011, 51, 69–82. [Google Scholar] [CrossRef] [PubMed]
- Harmat, V.; Bocskei, Z.; Naray-Szabo, G.; Bata, I.; Csutor, A.S.; Hermecz, I.; Aranyi, P.; Szabo, B.; Liliom, K.; Vertessy, B.G.; et al. A new potent calmodulin antagonist with arylalkylamine structure: Crystallographic, spectroscopic and functional studies. J. Mol. Biol. 2000, 297, 747–755. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Andrade, M.; Rodriguez-Sotres, R.; Madariaga-Mazon, A.; Rivera-Chavez, J.; Mata, R.; Sosa-Peinado, A.; Del Pozo-Yauner, L.; Arias, O., II. Insights into molecular interactions between CaM and its inhibitors from molecular dynamics simulations and experimental data. J. Biomol. Struct. Dyn. 2016, 34, 78–91. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Andrade, M.; Mata, R.; Madariaga-Mazon, A.; Rodriguez-Sotres, R.; Del Pozo-Yauner, L.; Sosa-Peinado, A. Importance of the interaction protein-protein of the CaM-PDE1A and CaM-MLCK complexes in the development of new anti-CaM drugs. J. Mol. Recognit. 2013, 26, 165–174. [Google Scholar] [CrossRef]
- Figueroa, M.; Gonzalez-Andrade, M.; Sosa-Peinado, A.; Madariaga-Mazon, A.; Del Rio-Portilla, F.; Gonzalez Mdel, C.; Mata, R. Fluorescence, circular dichroism, NMR, and docking studies of the interaction of the alkaloid malbrancheamide with calmodulin. J. Enzym. Inhib. Med. Chem. 2011, 26, 378–385. [Google Scholar] [CrossRef] [Green Version]
- Abdelmonem, M.; Zarrin, B.; Asif, N. High Throughput Screening and Molecular Docking of Calmodulin with Antagonists of Trifluoperazine and Phenothiazine Chemical Class. Lett. Drug Des. Discov. 2018, 15, 136–142. [Google Scholar]
- Tidow, H.; Nissen, P. Structural diversity of calmodulin binding to its target sites. FEBS J. 2013, 280, 5551–5565. [Google Scholar] [CrossRef]
- Sosa-Peinado, A.; Leon-Cruz, E.; Velazquez-Lopez, I.; Matuz-Mares, D.; Cano-Sanchez, P.; Gonzalez-Andrade, M. Theoretical-experimental studies of calmodulin-peptide interactions at different calcium equivalents. J. Biomol. Struct. Dyn. 2020, 1–12. [Google Scholar] [CrossRef]
- Dikici, E.; Deo, S.K.; Daunert, S. Drug detection based on the conformational changes of calmodulin and the fluorescence of its enhanced green fluorescent protein fusion partner. Anal. Chim. Acta 2003, 500, 237–245. [Google Scholar] [CrossRef]
- Douglass, P.M.; Salins, L.L.E.; Dikici, E.; Daunert, S. Class-selective drug detection: Fluorescently-labeled calmodulin as the biorecognition element for phenothiazines and tricyclic antidepressants. Bioconjugate Chem. 2002, 13, 1186–1192. [Google Scholar] [CrossRef]
- Li, H.; Villalobo, A. Evidence for the direct interaction between calmodulin and the human epidermal growth factor receptor. Biochem. J. 2002, 362, 499–505. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andrews, R.K.; Suzuki-Inoue, K.; Shen, Y.; Tulasne, D.; Watson, S.P.; Berndt, M.C. Interaction of calmodulin with the cytoplasmic domain of platelet glycoprotein VI. Blood 2002, 99, 4219–4221. [Google Scholar] [CrossRef] [Green Version]
- Bofill-Cardona, E.; Kudlacek, O.; Yang, Q.; Ahorn, H.; Freissmuth, M.; Nanoff, C. Binding of Calmodulin to the D2-Dopamine Receptor Reduces Receptor Signaling by Arresting the G Protein Activation Switch*. J. Biol. Chem. 2000, 275, 32672–32680. [Google Scholar] [CrossRef] [Green Version]
- Wang, D.; Sadee, W.; Quillan, J.M. Calmodulin binding to G protein-coupling domain of opioid receptors. J. Biol. Chem. 1999, 274, 22081–22088. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nickols, H.H.; Shah, V.N.; Chazin, W.J.; Limbird, L.E. Calmodulin interacts with the V2 vasopressin receptor: Elimination of binding to the C terminus also eliminates arginine vasopressin-stimulated elevation of intracellular calcium. J. Biol. Chem. 2004, 279, 46969–46980. [Google Scholar] [CrossRef] [Green Version]
- Turner, J.H.; Raymond, J.R. Interaction of calmodulin with the serotonin 5-hydroxytryptamine2A receptor. A putative regulator of G protein coupling and receptor phosphorylation by protein kinase C. J. Biol. Chem. 2005, 280, 30741–30750. [Google Scholar] [CrossRef] [Green Version]
- El Far, O.; Bofill-Cardona, E.; Airas, J.M.; O’Connor, V.; Boehm, S.; Freissmuth, M.; Nanoff, C.; Betz, H. Mapping of calmodulin and Gbetagamma binding domains within the C-terminal region of the metabotropic glutamate receptor 7A. J. Biol. Chem. 2001, 276, 30662–30669. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yesylevskyy, S.O.; Ramseyer, C.; Savenko, M.; Mura, S.; Couvreur, P. Low-Density Lipoproteins and Human Serum Albumin as Carriers of Squalenoylated Drugs: Insights from Molecular Simulations. Mol. Pharm. 2018, 15, 585–591. [Google Scholar] [CrossRef] [PubMed]


| Experimental Studies | Docking Studies | MD Studies | |||||
|---|---|---|---|---|---|---|---|
| Complex | ΔGexp (kcal mol−1) | Kd1 (M) | EFEB 2 (kcal mol−1) | Κd3 (M) | ΔH (kcal mol−1) | ΤΔS (kcal mol−1) | ΔGcal (kcal mol−1) |
| CaM–CPZ | −8.19 | 0.97 × 10−6 | −8.24 | 0.90 × 10−6 | −22.27 ± 2.5 | −17.75 ± 7.7 | −4.52 ± 1.8 |
| CaM–5-HT | −8.38 | 0.71 × 10−6 | −7.56 | 2.84 × 10−6 | −21.38 ± 2.7 | −16.52 ± 3.4 | −4.85 ± 0.4 |
| CaM–DOP | - | - | −7.21 | 2.96 × 10−6 | −11.85 ± 3.5 | −13.60 ± 5.24 | +1.74 ± 0.8 |
| CaM–AMA | - | - | −7.74 | 2.13 × 10−6 | +133.30 ± 13.8 | +15.60 ± 5.16 | +117.69 ± 21.2 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vásquez-Bochm, L.X.; Velázquez-López, I.; Mata, R.; Sosa-Peinado, A.; Cano-Sánchez, P.; González-Andrade, M. Application of a Fluorescent Biosensor in Determining the Binding of 5-HT to Calmodulin. Chemosensors 2021, 9, 250. https://doi.org/10.3390/chemosensors9090250
Vásquez-Bochm LX, Velázquez-López I, Mata R, Sosa-Peinado A, Cano-Sánchez P, González-Andrade M. Application of a Fluorescent Biosensor in Determining the Binding of 5-HT to Calmodulin. Chemosensors. 2021; 9(9):250. https://doi.org/10.3390/chemosensors9090250
Chicago/Turabian StyleVásquez-Bochm, L. X., Isabel Velázquez-López, Rachel Mata, Alejandro Sosa-Peinado, Patricia Cano-Sánchez, and Martin González-Andrade. 2021. "Application of a Fluorescent Biosensor in Determining the Binding of 5-HT to Calmodulin" Chemosensors 9, no. 9: 250. https://doi.org/10.3390/chemosensors9090250
APA StyleVásquez-Bochm, L. X., Velázquez-López, I., Mata, R., Sosa-Peinado, A., Cano-Sánchez, P., & González-Andrade, M. (2021). Application of a Fluorescent Biosensor in Determining the Binding of 5-HT to Calmodulin. Chemosensors, 9(9), 250. https://doi.org/10.3390/chemosensors9090250