Abstract
An untargeted Fourier transform infrared (FTIR) metabolomic approach was employed to study metabolic changes and disarrangements, recorded as infrared signatures, in Parkinson’s disease (PD). Herein, the principal aim was to propose an efficient sequential classification strategy based on SELECT-LDA, which enabled optimal stratification of three main categories: PD patients from subjects with Alzheimer’s disease (AD) and healthy controls (HC). Moreover, sub-categories, such as PD at the early stage (PDI) from PD in the advanced stage (PDD), and PDD vs. AD, were stratified. Every classification step with selected wavenumbers achieved 90.11% to 100% correct assignment rates in classification and internal validation. Therefore, selected metabolic signatures from new patients could be used as input features for screening and diagnostic purposes.
1. Introduction
A considerable segment of the ageing population worldwide suffers from Parkinson’s disease, the second most prevalent progressive neurological disorder after Alzheimer’s dementia [1,2,3]. The risk factors for PD are complex and likely interconnected, so the onset of PD is thought to be caused by a combination of genetic predisposition and environmental influences (exposome). Unfortunately, there is still no standard treatment for this disorder. Most of the currently available therapeutic options are focused on treating and mitigating the symptoms, i.e., on palliative care, but curative treatment is not yet a medical reality, with its consequent impact on morbidity and mortality.
All findings towards detecting the disorder’s pathogenesis suggest various metabolites that are modified in PD, from a significant role attributed to α-synuclein [4,5,6] and Lewy’s body [7] to exosome factors that are involved [8,9,10]. Thus, Parkinson’s disease is often associated with defects in lipid metabolism, particularly in the central nervous system [11,12]. It was reported that oxidative stress in the substantia nigra at the time of death in advanced Parkinson’s disease manifests in increased lipid peroxidation [13]. Blood-based biomarkers are also widely studied because they can be more accurate than clinical observations regarding dopamine deficiency effects, such as bradykinesia, rigidity, and tremors. In addition, it was suggested that changes in cholesterol levels and cholesterol derivatives might indirectly be related to the onset of PD.
Meanwhile, higher uric acid (UA) levels were associated with a decreased risk of disorder, and ratio acid uric/creatinine (UA/Cr) was evaluated as a predictive factor for a slower disease progression [14,15]. Elevated blood glucose levels were directly related to a longer duration of PD and a higher score of dysautonomia in moderate to advanced PD patients [16]. Interestingly, increased concentration of histamine in the nervous system and putamen of PD patients were reported from different research groups [17,18]. Despite substantial evidence, no specific biomarker was approved for diagnosis or prognostic purposes of this progressive multisystem condition [19,20,21,22,23,24]. Thus, the conventional diagnosis of Parkinson’s remains essentially clinical, based on the subjective observations of clinicians. Nowadays, none of the available clinical tests have been proven to attain high sensitivity, accuracy, and objectivity for PD detection. In addition, the instruments applied in the clinical environment are expensive and unwieldy.
Nevertheless, the correct diagnosis identifying PD at the early stage is crucial. This is because once the patient appears with clinical symptoms, the damage in the brain is already irreversible. For this reason, the diagnosis must be made as soon as possible to avoid further extensive neuronal loss [25].
In this context, the attention to specific biomarkers shifted to explore the production of perturbations in the metabolome and such variations in studies of Parkinson’s disease [26,27,28,29]. Untargeted metabolomic studies, such as that presented here, aim to extract the metabolic fingerprints of analysed samples to classify them according to biological status or origin based on these unique and individual molecular patterns.
For more than ten years, the vibrational approach, nuclear magnetic resonance (NMR) [30,31] spectroscopy, and a wide range of highly sensitive mass spectrometry (MS)-based methods [32,33] have been proven valuable for the evaluation and classification of normal and pathological samples [34,35,36,37,38,39], especially for Parkinson’s detection [40,41,42]. However, despite the large number of studies found in the literature that support the emerging potential of metabolic fingerprinting in clinics, a still challenging bottleneck of these types of studies is their actual translation to clinical practice. From a technical point of view, breaking down barriers to clinical translation depends on advances in measurement technology. Being these instruments rapid, non-invasive, non-destructive, reliable, and easy to use, they are ideal candidates as high-throughput screening techniques for fingerprinting. In this context, vibrational spectroscopy and NMR spectroscopy remain the prime analytical choices because they fulfil the required characteristics of analytical instruments, making large-scale or routine studies much more feasible than with MS-based applications [43].
Likewise, exhaustive metabolomic fingerprinting research should not be limited to a unique analytical platform but rather should test and combine multiple analytical strategies in order to exploit their respective strengths and overcome their weaknesses. To this end, FTIR and NMR spectroscopies can successfully complement each other.
In previous work from our research group [44], we evaluated the potential of using a non-targeted lipidomic approach to extract NMR-based signatures for the clinical differential diagnosis and stratification of PD. To complete and complement this work, we now propose a metabolomic fingerprint classification strategy also aimed at extracting disease/stage-specific panels of infrared markers, but expanding the search to a broader range of molecular species (not restricted to lipid compounds) thanks to the inherent ability of FTIR to provide biochemical information holistically.
Moreover, IR is particularly suitable for analysing human biofluids that are easily obtainable and reflect several physiological functions of the body, such as blood [29,45,46]. The blood is the primary carrier of metabolites throughout the entire organism, and it is composed of a variety of biological materials, mainly proteins, lipids, and sugars. All these are active in the infrared range, and each biomolecule is determined by its unique structure. The changes in their chemical structure can be investigated simultaneously instead of studying isolated molecules, providing a metabolic signature for PD. Thus, the spectrum recorded by FTIR from a biological sample generates a unique IR spectral signature, reflecting its specificity. Furthermore, the IR spectral modes of plasma may reflect the current status of the organism and could be directly correlated with the presence or absence of the disease. For these reasons, IR is highly exploited to identify possible spectra biomarkers associated with Parkinson’s differentiation [47].
Aim of the Study
Given these perspectives, in this study, the direction of research shifts towards investigating the existence of distinct mid-infrared metabolic fingerprints in PD-related diseases, which would drive PD patient stratification and guide an accurate and early differential diagnosis. Thus, an untargeted metabolomics approach (FTIR application in the fingerprint region coupled with multivariate data analysis) was used to reveal spectroscopic biomarker signatures that define patient subgroups for the clinical diagnosis and classification of PD at different stages of the disease. PD at the initial stage (PDI) should be differentiated and not confused with developed PD-related dementia (PDD). Ideally, an accurate analytical tool should be able to differentiate Parkinson’s disease from other neurological impairments such as Alzheimer’s disease. In most cases, subjects affected with PD share a common profile with Alzheimer’s disease, accumulating abnormally aggregated proteins. The two main chemometric strategies employed in this work are based on the initial selection of discriminant variables and the subsequent development of a linear discriminant analysis (LDA) classification. One of our primary goals was to achieve accurate discrimination between plasma samples of patients affected by PD from subjects affected by AD and from healthy control individuals, confirming that infrared signatures can be associated with metabolomic changes based on different pathological conditions. The second major objective was to obtain accurate discrimination between two PD subgroups, identifying reliable wavenumber predictors for disease stage differentiation. However, we also decided to deepen the problem and apply the developed classification rules to a new problem. The differentiation between patients with Parkinson’s disease dementia and those affected by Alzheimer’s dementia was also studied. With this method, we aimed to reveal that LDA stepwise wavenumber selection may extract significant bio-spectroscopic markers, enable objective diagnosis, and make possible the differentiation of such a multifactorial disease as Parkinson’s.
Considering the potential clinical translation advantages of adopting an IR fingerprint-based classification, the cost-effectiveness and relative ease of access to IR portable devices should be highlighted, which could be ideal for point-of-care testing, primary health care, or wherever required.
2. Materials and Methods
2.1. Study Population
Ethical approval was obtained from the Research Ethics Committee of San Pedro Hospital of La Rioja Province. In the present study, 97 patients were recruited from the Molecular Neurobiology Unit in the Centre for Biomedical Research of La Rioja (CIBIR). Based on the type of neurodegenerative pathology and its advance, a total of 41 patients were classified as PDI, and 9 were considered to have PDD; additionally, 23 AD patients and 24 healthy controls (HC), belonging to the family environment and alike in age to PD patients, enrolled in the study. Written informed consent was achieved from all participants from the Neurology Department of San Pedro Hospital (Logroño, Spain).
2.2. Collection and Handling of Plasma Samples
Approximately 2 mL of blood samples was collected via antecubital venepuncture from each subject in a sitting position. Becton Dickinson (BD) Vacutainer® plastic blood collection tubes with K2EDTA were used for plasma separation. The plasma samples were obtained after centrifugation at 2200× g for 15 min at 4 °C and then stored at −80 °C in Eppendorf tubes as aliquots of 200 μL each for further FTIR processing and analysis. Before FTIR measurement, plasma samples were thawed according to an optimised ultrasound-based protocol for lipidomic analyses recently developed in our research group [48].
2.3. Instrument
For the FTIR analysis, 25 μL of plasma from each patient was manually deposed on a CaF2 windows liquid cell PerkinElmer (Omni Cell, Specac Ltd., Orpington, UK) with a 50 μm Mylar spacer. The absorptions were recorded in the medium infrared range (4000–300 cm−1) under a constant nitrogen purge to remove atmospheric water vapor and CO2 by Spectrum-One ABB Miracle Type MB3000 FTIR Spectrophotometer (Zurich, Switzerland). Measurements were performed in triplicate at 2 cm−1 spectral resolution, and 32 scans were accumulated and averaged. A mean spectrum was subsequently obtained from the replicates recorded for each plasma sample. The sample temperature was maintained at 23.0 °C ± 1.0 °C while recording the signals. The Horizon MBTM program was used for data analysis. Quality control (QC) samples were inserted regularly and processed similarly to the actual samples to monitor reproducibility and repeatability during the analysis.
2.4. Pre-Processing of Spectra
The FTIR dataset was processed and analysed with Parvus [49] and Unscrambler 11 chemometric software package (version 11.0, Camo Software, Oslo, Norway). Pre-processing is imperative in analysing high-dimensionality biological spectral data; it corrects many problems with spectral data acquisition such as random noise, baseline distortions, or light scattering. Spectra were then cut to include the biochemical “fingerprint region” between 1490 and 1155 cm−1; other regions were excluded from further analysis as non-informative zones. Often, spectral wavenumbers have solid correlations and, therefore, are highly amenable [50]. Finally, MIR spectra data was submitted to pre-treatment by Extended Multiple Scatter Correction (EMSC). This method was preferable as a pre-processing step, allowing for the resolution of overlapping peaks, and it showed better results in decreasing scatter effects. The data were always centred before multivariate analysis.
2.5. Data Analysis
2.5.1. PCA
Chemometric methods are increasingly applied to obtain meaningful and reliable information from registered spectra, enabling their characterisation and enhancing process understanding. Principal component analysis (PCA) is one of the most useful preliminary steps for the exploratory analysis of a large number of correlated features such as FTIR spectral data. This unsupervised data analysis step was performed to reduce the dimensionality of our complex spectral dataset into a few Principal Components (PCs), still preserving the majority of information and capturing relevant sources of data variability. However, this step alone was not enough to allow a clear separation of our data. However, it helped to identify outliers. Furthermore, it highlighted the need to resort to a two-step sequential classification strategy based on applying an efficient variable selection technique to find the informative features that would allow successful prediction.
Before setting any classification approach, important parameters should be considered: the study’s objective, the number of categories, and the particular requirements of a sample assigned to a specific class. The proposed classification strategy was seen as a sequence of two consecutive classification problems in this study. The approach used for classification first concerns a generic classification of the disease and then specific discrimination of PD to distinguish the two different stages of the disease. Thus, for a first global classification, three distinct categories have been defined (Parkinson’s disease, Alzheimer’s disease, and control subjects) so that patients with PD at an early stage and those with PD-related dementia were considered as one class (Parkinson’s disease). Preliminary investigations have shown that this approach considerably improves the performance of the classification method. Then, after reaching a clear differentiation between the above three previously designated classes, PD was divided into two classes to further classify these patients according to the severity of the disease. A strategy to differentiate two types of dementia affecting patients with separate neurodegenerative disorders was investigated and applied.
2.5.2. Classification Methods
Linear discriminant analysis (LDA) [51,52], a well-known and extensively applied powerful supervised chemometric classification technique, enabled us to discriminate between prespecified subgroups. Based on LDA classification rules, the objects are always classified in one of the predefined classes [53]. The discrimination performance was optimised by selecting the most significant wavenumbers by a stepwise orthogonalisation of predictors (SELECT) [49]. The SELECT algorithm enabled reliable classification between patients’ classes, providing input features for a further stepwise LDA.
To avoid possible overfitting, one crucial step in LDA-based variable selection should be considered, namely, the number of variables used in classification development. Thus, the number of training objects should be at least three times greater than the number of retained variables by SELECT. In addition, internal cross-validation was applied to access the predictive discrimination of each classification developed. Therefore, essential variables were selected and decorrelated with other variables based on the maximum correlation weight. This step is essential when dealing with big data dimensionality, eliminating the futile features due to noise, and identifying the relevant and important variables to be applied in the following steps. Cross-validation (CV) was used to optimise classifications, and external validation to evaluate prediction ability. The quality of the discriminant rules derived was evaluated according to several parameters:
- -
- Total classification (prediction) rate (TR)
- -
- Category c rate (Rc)where mcc is the number of correct classifications (predictions) for a certain category c, is the total number of correct classifications (predictions), Nc is the number of classifications (predictions) for a certain category c, and N is the total number of classifications (total predictions). Since an object can be classified several times during cross-validation, Nc is not always equal to the number of samples belonging to class c, just as N does not always represent the total number of objects.
3. Results
Using plasma-based vibrational spectroscopy, we achieved results with significant discrimination relevance at multiple levels. FTIR spectroscopy coupled with chemometric strategy has been proven to detect differences between pathological patients and healthy subjects and between different stages of neurodegenerative disease. As contributors to the discrimination between patients, several selected wavenumbers appeared to be of particular interest. Thus, we performed a tentative biochemical assignment of the selected signatures for each classification step. It should be made clear that the tentative band assignment and its speculative nature was performed to attribute a possible chemical reasoning to the reduced fingerprint bands responsible for each stratification. Therefore, the attempt band assignment was not, at any time, the primary objective of this study. However, chemical reasoning could be a starting point in future targeted-based studies.
Thus, numerous selected wavenumbers in the fingerprint region were associated with key molecules, from carbohydrates to nucleic acids. Our results are reassuring and utterly consistent with the formulated theories about PD pathogenesis and metabolic biomarkers. Furthermore, the spectral absorptions implicated in the discrimination between disease principal groups and subgroups were quite similar. Sometimes, the exact wavenumber was associated with a different absorption type but was always in line with the possible contribution found in the literature.
3.1. FTIR Spectral Profiles
FTIR spectroscopy is a promising technique which provides molecular fingerprints of biological samples based on vibrational transitions of chemical bonds in the samples upon interaction with infrared light. [54,55]. The fingerprint region (the segment of the IR spectrum below 1500 cm−1) is a complex area that comprises a characteristic pattern of absorption that is unique to a sample [47], containing a huge amount of valuable information about the metabolic changes that occur during the onset and progression of the disease. Therefore, in this study, the most important spectral zone was limited between 1490 cm−1 and 1155 cm−1. Thus, the changes in band profiles arising from the fundamental vibrations of biomolecules could be related to alteration in the metabolism induced by neurodegenerative diseases.
The EMSC correction effect on spectra is displayed in Figure 1A, underlining how significantly reduced the scatter effect is in all datasets. After EMSC, PCA was applied as a visualisation tool, confirming that the visual discrimination and comparison among categories with different diseases was quite impossible; the overlapping features persisted (Figure 2).
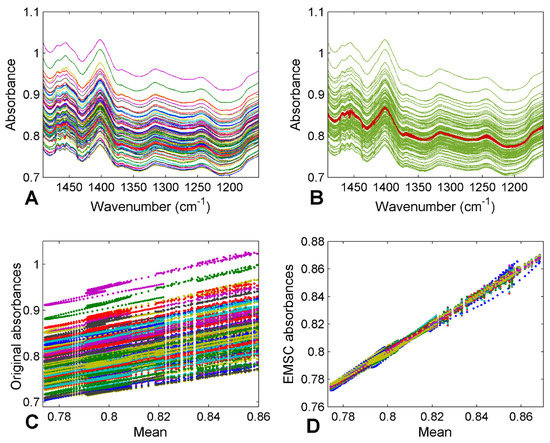
Figure 1.
FTIR spectra of plasma samples before (A) and after (B) EMSC pre-treatment, and the corresponding plots of scattering effects between signals before (C) and after (D) correction.
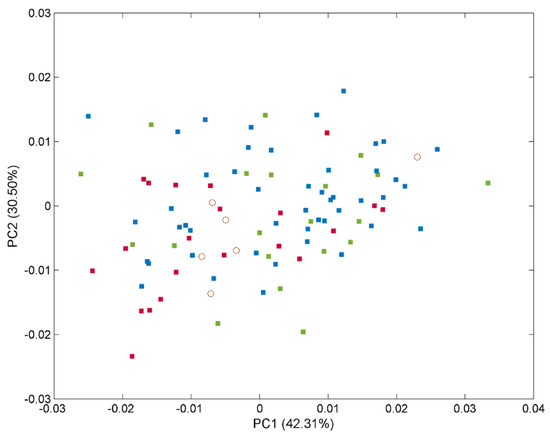
Figure 2.
Scores for the plasma samples from the 97 patients (full data set) on the first 2 principal components explaining the variability in the IR spectral data. The samples are labelled according to their specific pathology: healthy control (■), PD (■), and AD (■) subjects and external test samples (◯).
3.2. Global Classification: 3-Class Approach for Discriminating Patients with PD, AD, and Healthy Controls
Based on the results of the exploratory analyses, considering the classification problem studied here as a unique step approach was rejected, as it would lead to unreliable results. For this aim, a multivariate classification approach based on a multiple-step sequential classification was studied and applied to a patient population.
Firstly, three separate classes were defined; the group of Parkinson’s patients at the early stage was combined with patients accompanied by dementia. Thus, the first global classification approach concerns healthy subjects, PD (PDI + PDD), and AD. The best solution for the global classification problem was obtained when the SELECT-LDA method was applied, so the information responsible for the successful discrimination between PD, AD, and the control group was compressed from 340 to only 30 variables (conforming to a reduced IR signature of disease status), with almost 100% of total correct assignment rates in classification and prediction (Table 1). Excellent discrimination among classes was achieved, providing a 100% level of correctly classified samples for control subjects and patients with Alzheimer’s, and 99.79% correct assignment rates in the case of patients with Parkinson’s disorder. Satisfactory internal prediction performances ranging from 86.36% to 95.24% were achieved for the various categories, but the real strong point was in external prediction. The prediction performance of the SELECT-LDA classification was developed and optimised using ten cancellation groups for CV and a test subset of six samples, distributed randomly in the following way: two for the control group, two for PD, and two for AD. All six test samples provided 100% of correctly classified samples, reinforcing the reliability of the developed classification strategy based on the reduced IR fingerprint extracted wavenumbers (Table 2). Furthermore, a clear interclass separation achieved between the three main categories can also be visually appreciated in the corresponding LDA score plot (Figure 3).

Table 1.
Percentages of correctly classified samples/patients in both classification and internal/external validation corresponding to the SELECT-LDA performed when addressing the global classification approach.

Table 2.
Discriminant wavenumbers (in order of selection) corresponding to the SELECT-LDA classification developed from IR spectra of plasma samples in the global approach.
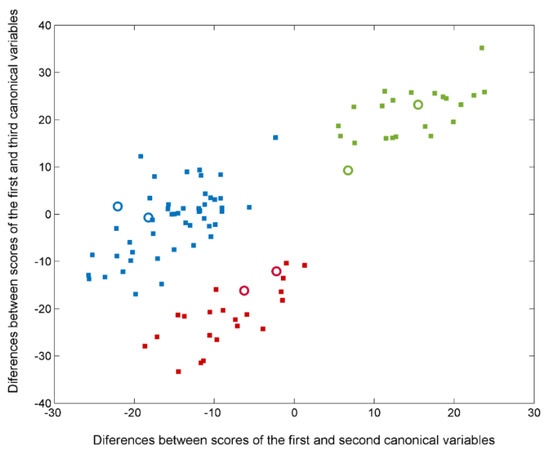
Figure 3.
Plot of the differences between discriminant scores for plasma samples after performing SELECT-LDA in the global classification approach. The 3 categories considered are labelled as healthy control (■), PD (■), and AD (■) subjects. Test samples are displayed as unfilled circles (◯).
The patient profile of three main categories seems to be unequivocally distinguished by the score difference between the first and third canonical variables. Likewise, the maximum difference between healthy subjects and two groups affected by neurodegenerative disorders (PD and AD) is evidenced by the score difference between the first and third canonical variables, suggesting that the two disease-carrying groups do not share a similar profile with the healthy group. In particular, it can be observed that there is a double contribution of both differences between scores; thus, two classes characterised by pathology are separated by an imaginary axis coinciding with the delimiter. Appendix A (Table A1) summarises all relevant absorption bands for the global classification step, including the identification of the bond vibrations involved and the respective biochemical assignments.
3.3. PD Stratification: 2-Class Approach for Discriminating between Patients with Early-Stage PD and PD-Related Dementia
After the differentiation among three classes of patients was obtained, we carried out a sub-analysis, comparing the group of patients affected by Parkinson’s disease depending on the disease’s progress. Therefore, a single undifferentiated group of Parkinson’s was split up into two distinct classes: PD at an early stage and PD-related dementia. The classification strategy followed the same classification rules as described ahead [52]. However, in compliance with rules cited before, the number of discriminant variables changed so that training objects were at least three times greater than the number of final selected wavenumbers. Full CV and a test subset of three samples, selected randomly as two for the PDI group and one for PDD, were performed to optimise and validate classifications, always on autoscaled data. In this case, 100% in classification and 100% in prediction (internal and external) were achieved (Table 3). Herein, the number of features was reduced from 340 to only 15 variables, which makes our approach even more remarkable (Table 4). A discriminative histogram is reported in Figure 4, which shows a clear class separation on the first canonical variable. Thus, the first canonical variable represents the direction with the maximum discrimination power, namely the maximum Fisher ratio (the ratio between the interclass and the intraclass variance). This feature proves an outstanding performance of the method and its reliability and good performance of the classification strategy based on reduced MIR–plasma signatures. All relevant absorption bands for this second classification step are reported in Appendix A (Table A2), including the identification of the bond vibrations involved and the respective biochemical assignments.

Table 3.
Percentages of samples/patients classified correctly in both classification and internal/external validation in the SELECT-LDA performed when addressing the classification approach for PD stratification.

Table 4.
Discriminant wavenumbers (in order of selection) corresponding to the SELECT-LDA classification developed from IR spectra of plasma samples for the differentiation of PD progression stage.
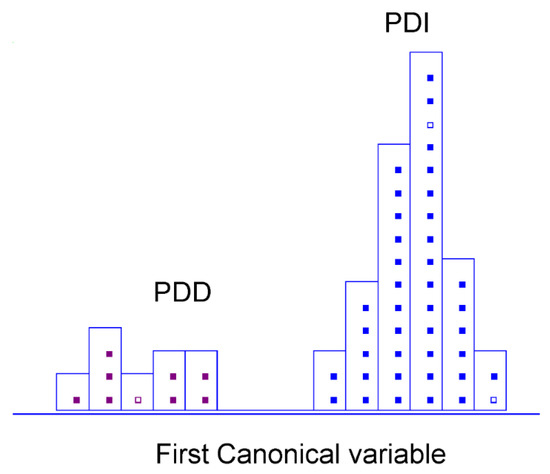
Figure 4.
Histogram of the first canonical variable for the discrimination of PD early stage (■) and PD-related dementia (■) patients after performing SELECT-LDA in the PD stratification approach (y-axis indicates the maximum discrimination power between categories).
3.4. Dementia Type Differentiation: 2-Class Approach for Discriminating between Patients with PD-Related Dementia and Alzheimer’s Dementia
The last sub-classification problem was examined in greater depth to directly discriminate between dementia associated with both pathologies: Parkinson’s disease dementia and Alzheimer’s dementia. The analytical approach was based on the same classification strategy described before, following the rule of extraction of the truly discriminant variables by SELECT. Ten cancellation groups for the CV and a subset of two random samples (each for one category) were established to optimise and validate classifications. The achieved results were auspicious, so the success rate for both categories was 100% in classification, 87.50% and 100% in internal prediction, respectively, and 100% in external prediction (Table 5). Given the results obtained per category, it is essential to highlight that AD patients were effectively discriminated from patients with PD-related dementia (100% correct assignments in both classification and CV). Focusing solely on the numeric value of the obtained percentages made it more challenging to accurately classify PDD subjects into their real category. It should be considered that the slight decrease in internal validation performance observed for this latter class is related to the wrong assignment of a single sample. However, the limited number of available patients for this category indeed contributed to maximising the influence of this deviation. The results can be visually appreciated in Figure 5. Only ten selected variables were needed to provide a remarkable stratification between two different types of dementia (Table 6). Appendix A (Table A3) summarises all relevant absorption bands for this classification step, comprising the identification of the bond vibrations involved and the respective biochemical assignments.

Table 5.
Percentages of samples/patients classified correctly in both classification and internal/external validation in the SELECT-LDA performed when addressing the classification approach for dementia type differentiation.
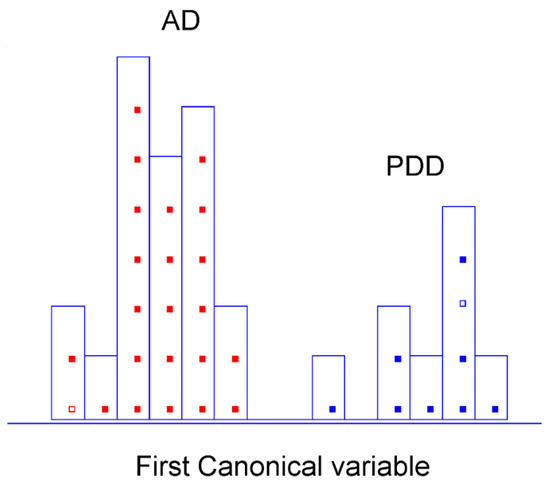
Figure 5.
Histogram of canonical variable for the discrimination of PD-related dementia (■) and Alzheimer’s dementia (■) patients after performing SELECT-LDA in the PD dementia type direct differentiation approach (y-axis indicates the maximum discrimination power between categories).

Table 6.
Percentages of samples/patients classified correctly in both classification and internal/external validation in the SELECT-LDA performed when addressing the classification approach for dementia type differentiation.
4. Discussion
The pathogenesis of Parkinson’s disease remains unclear; it could be seen as a complex convergence of genetic and environmental etiologic factors. For this reason, many hypotheses were formulated to evaluate the meaningful biomarkers that differentiate affected patients from controls. Herein, an attempted assignment of selected bands was performed based on the classification type. It should be precise that this study’s principal aim was unfocused on quantifying the level of the determined compound. Instead, it is focused on determining whether it is possible to differentiate between categories of patients and identify the degree of PD progression. In line with our results, we want to discuss some evidence already found in the literature that reinforces our results after analysis. Numerous selected wavenumbers in the fingerprint region were associated with critical molecules, from carbohydrates to nucleic acids. Our results are reassuring and utterly consistent with the formulated theories about PD pathogenesis and possible biomarkers found in the literature.
In the first classification step, several selected wavenumbers appear to be of particular interest; thus, a tentative biochemical assignment of the selected variables was conducted. Different bands comparing PD, AD, and HC were selected between 1150 and 1000 cm−1, which is usually associated with oxidative stress [39]. Therefore, it could be attributed to different levels of damage caused by free radicals. Many studies have shown that UA plays an essential role as an antioxidant reagent, mainly as urate in the human body. Furthermore, increased levels of this endogenous compound are linked with reduced brain damage caused by reactive oxygen species (ROS) [14,15]. The topic of oxidative stress joins other evidence that we discuss further on.
Close to this region, bands at 1156 and 1159 cm−1 or 1302 cm−1 could be attributed to bending (CH) of Ala. Likewise, selected signatures between 1411 and 1400 cm−1 could correspond to symmetric stretching ( sym) (COO-) and sym. (C = O) of Ala or Glu. Various successful breakthroughs have shown metabolite variations in blood samples of PD. Significant variations in glutamate (Glu) and alanine (Ala) in cerebrospinal fluid were also observed in PD and have been broadly targeted [27].
Discriminant signatures around 1215/1253 cm−1 due to asymmetric stretching ( ass) (P = O) of phospholipids; stretching vibration (COO-) of fatty acids at 1403 cm−1; and bending vibrations (δ) (CH2) at 1455, 1474, 1477, and 1489 cm−1 of lipids could be attributed to controversial theories about the role of cholesterol, lipids, and proteins in Parkinson’s disease [56]. Furthermore, different research findings support the associations with pathological interactions of alpha-synuclein in PD. Thus, the selected protein vibrations also seem to play a key role in patients’ stratification [6]. Multiple bands, such as 1312, 1403, and 1455 cm−1, were assigned to lipids congruent with previous findings.
In this classification strategy, one of the bands was assigned to histidine, the signature at 1215 cm−1, proving some evidence found by Picca et al. [10], where higher concentrations of 3-methyl-histidine, citrulline, and serine were determined in control participants. At the same time, the band at 1182 cm−1 could correspond to bending (C-OH) and stretching (C = O) of serine for the differentiation of three main categories of patients. The role of serine is of great importance, as it takes part in different metabolic pathways, including the generation of phosphatidylserine and phosphoserine, both of which play important roles in the function of the neurodegenerative system [5]. In addition, other studies suggested that histamine could potentially affect neuronal survival and participate in neurotrophic processes aimed to re-establish damaged brain functions [17,18]. From the analysis of this first-step classification, different wavenumbers resulted significantly in the region between 1360 and 1220 cm−1. Therefore, we could speculate that these spectral markers could be attributed to amide III-band stretching and bending and corresponding to histamine.
Many of the selected spectral biomarkers hold the potential to be linked with carbohydrates. For this purpose, it seems particularly important that many influential bands in Parkinson’s classification were also attributed to “sugars”. Thus, from 30 spectral variables, bands at 1156 cm−1 and 1159 cm−1 could be due to stretching (CO-O-C) of carbohydrates. The region 1330–1220 cm−1 seems to be rich in methylene stretching of carbohydrates residues, and therefore, the band at 1424 cm−1 could correspond to polysaccharides. Other studies already evidenced a substantial contribution to the separation between metabolite profiles of unmedicated PD patients and controls of alternative metabolites, such as myoinositol, sorbitol, citrate, acetate, succinate, and pyruvate [27].
Thus, among 15 relevant wavenumbers in the second classification step, bands discriminating PDI and PDD at 1214, 1224, and 1226 cm−1 were associated with uric acid ring vibrations, presumably explaining the different levels of brain damage during the disease progression stage.
The selection of different wavenumbers at 1443, 1445, 1475, and 1476 cm−1 due to lipid structures could suggest that oxidative stress in the substantia nigra at the time of death in advanced Parkinson’s disease manifests in terms of increased lipid peroxidation, superoxide dismutase activity, and zinc levels [13]. In addition, previous studies have already performed the analysis with infrared spectroscopy, defining bands due to methylene deformation of lipids or methyl bending of lipids, able to differentiate PD and controls [42]. Thus, our results prove the importance of this region.
Different studies highlight the role of phosphoethanolamine, which is the head group of different lipids, including phosphatidylethanolamine, lysophosphatidylethanolamine, and sphingomyelin. Multiple functions of this molecule in the body and in PD patients were evidenced [8]. Thus, it makes sense that many of the selected spectral markers were assigned to phosphate groups and lipids. Beyond phosphoethanolamine, a circulating amino acid signature encompassing higher amino acids levels was found in older people with PD.
Different research groups reported that impairment in oxidative stress was directly linked with an elevation of plasma sorbitol concentrations in drug-naive patients. As observed, the critical role of oxidative stress in PD is undeniably evident. Several evidence lines have also been found for dysregulation in glucose metabolism in moderate to advanced PD patients [16]. The reduced concentrations of alanine, lactic acid, and glucose were detected and correlated with affected glucose metabolism [24].
Similarly, the selected band at 1297 cm−1 due to bending (CH2) of Ala could have a double significance. Considering that carnosine is a dipeptide of alanine and histidine, which have antioxidant functions in PD, as was already observed before, the decreased levels in alanine could also justify the decrease in the levels of carnosine [56]. Thus, considering the significance of some bands associated with histidine, it is congruent to assume the possible correspondence of these bands to carnosine. Therefore, our findings could be related to the role of a biomarker of carnosine in PD.
To distinguish between spectrochemical profile of patients with PDD and AD, the discriminant band at 1340 cm−1 due to bending (CH2) was selected as the most important. It was assigned to the absorption of collagen [38]. The involvement of determined vibrations attributable to collagen is entirely reasonable. Furthermore, it was shown that the brain’s neurons are the source of the specific type of collagen (collagen VI). Thus, an increased level of this collagen in the brain could have a protective function against AD [57]. Overall, other studies hypothesised that the progressive degradation of nerve cells associated with Alzheimer’s disease duration could potentially undermine their ability to produce collagen [58].
Moreover, in spectroscopic sub-signatures that could further differentiate between PDD and AD, another region at 1402 cm−1 was associated with stretching vibration () (C = C) of UA, which was discussed and justified above. Multiple signatures attributed to stretching vibrations of metile and methylene of lipids and phospholipids at 1379, 1380, 1382, 1402, and 1487 cm−1 and bending (CH2) at 1488 cm−1 have also been selected. Likewise, for the role of the band at 1377 cm−1 in the global classification, in this classification sub-problem, we also speculated that bands at 1379, 1380, and 1382 cm−1 could correspond to amino acids, including proline [59].
Appendix A (Table A1, Table A2 and Table A3) summarises all relevant absorption bands for each classification type, including the identification of the bond vibrations involved and the respective biochemical assignments.
The main highlight of this untargeted FTIR-based metabolomics study focused on discovering if spectroscopic signatures can differentiate PD from other neurodegenerative conditions with shared symptoms, but not on the analysis of the specific contribution of each FTIR-reduced fingerprint component. Nevertheless, the provided biochemical reasoning about the contribution of specific bands in the differentiation of patients seems to be ideally in line with our results, reinforcing the suitability of our classification strategy.
Limit of the Study
Since the samples were retrospectively collected specimens provided by the biobank, any additional data were specified about the patients. One of the most significant limitations of this research is the relatively small number of samples available to perform the analyses. Further research involving a more extensive patient database would be required to confirm the validity of the proposed classification strategy based on reduced IR signatures. However, despite the limited number of samples, the obtained results were validated. The analytical strategy involved both internal cross-validation (to develop optimal classifications) and external validation (to assess the actual predictive performance of the classifications constructed) in preventing overfitting and ensuring the reliability of the results obtained.
5. Conclusions
There is still no standard robust approach for the objective diagnosis of Parkinson’s disease; this field remains underexplored and poorly understood. The reported results highlight the potency of the adopted chemometric strategy, based on a three-step classification approach in the stratification of Parkinson’s patients; the disease was effectively classified and differentiated from the control group and other impairments such as Alzheimer’s dementia. Spectral signatures in human plasma have been successfully identified for differentiation between patient categories by selecting significant wavenumbers closely related to PD pathogenesis and metabolic biomarkers. Moreover, the rapid, high-throughput, and relatively inexpensive method provided optimal discrimination results in both sub-classification problems, succeeding in the stratification of patients with different PD stage progression profiles and those with different dementia type profiles. The reported untargeted metabolomic approach seems to deal significantly with the necessity of developing an alternative screening method to distinguish patient profiles, thus taking vibrational spectroscopy one step forward towards clinical implementation. All the speculations made about the involvement of selected bands in the pathogenesis of PD are immensely reasonable, and their role is perfectly justifiable for patient stratification. A primary limitation of the current work is the relatively small number of available plasma samples, especially for one subgroup (patients with PD-related dementia), preventing more general conclusions. Further investigation is required; a more significant number of enrolled patients could strengthen the validity of the proposed classification strategy as an objective diagnosis of PD.
Author Contributions
The manuscript was written through the contributions of all authors. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by European Union’s H2020 research grant (No. 801586) and Ministry of Science and Innovation: CTQ2011-26603.
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki, and approved by the Ethics Committee of San Pedro Hospital of La Rioja Province (CEICLAR PP-212 01.04.2016). All experiments were performed in accordance with the guidelines: 1. Organic Law 15/199, of 13 December, on the Protection of Personal Data; 2. Law 41/2002, of 14 November, Basic Regulatory of Patient Autonomy and Rights and Obligations in the Field of Information and Clinical Documentation; 3.Order SCO/256/2007, of 5 February, which establishes the principles and detailed guidelines of good clinical practice; and 4. Royal Decree 1716/2011, of 18 November, which establishes the basic requirements for the authorisation and operation of biobanks for the purpose of biomedical research and the treatment of biological samples of human origin for biomedical research.
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
Not applicable.
Acknowledgments
The authors thank Álvarez and Marzo for their kind collaboration.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
Ala: alanine; AD, Alzheimer’s disease; CR, creatinine; CV, cross-validation; Glu, glutamate; FTIR, Fourier transform infrared spectroscopy; HC, healthy controls; LDA, linear discriminant analysis; LOO, leave one out; PCA, principal component analysis; PD, Parkinson’s disease; PDD, Parkinson’s disease dementia; PDI, Parkinson’s initial stage; SELECT, stepwise variable selection method; UA, uric acid.
Appendix A

Table A1.
Biochemical tentative assignments of the most discriminant wavenumbers selected by SELECT-LDA in the global classification approach aimed at differentiating between PD, AD, and healthy patients.
Table A1.
Biochemical tentative assignments of the most discriminant wavenumbers selected by SELECT-LDA in the global classification approach aimed at differentiating between PD, AD, and healthy patients.
| Spectral Regions in Literature | Peak Position (cm−1) ± 1 | Tentative Band Assignment | Contributions |
|---|---|---|---|
| ~1155 | 1156, 1159 | sym. (CO-O-C) | Carbohydrates |
| ~1185–1120 | 1171, 1182 | (C-C) and (O-P-O); (C-O) ring vibrations | Nucleic acid “sugars” |
| ~1225 | 1215 | asym. (O-P-O) | Nucleic acids, phospholipids |
| ~1250–1220 | 1253 | sym. (P = O) of the PO2 groups | Nucleic acids, phospholipids |
| ~1360–1220 | 1266, 1271, 1284, 1286, 1288, 1289, 1294, 1302, 1312, 1316, 1319, 1335, 1336, 1377 | (C-C) and (C-O) (C-N) and C-(NO2) sym. (PO2), predominantly (C-N) with significant contributions from (CH2) of carbohydrate residues, δ (CH2) | Amide III band, proteins |
| ~1370 | 1377 | sym. def. CH3 and sym. def. CH2 | Proteins, amino acids (cytosine, guanine, proline) lipids, phospholipids |
| ~1400 | 1403 | (C = O) of (COO) group | Fatty acids and amino acids |
| ~1420 | 1424 | sym. (COO), δ asym. (CH2) | Polysaccharides |
| ~1455–1450 | 1455 | δ asym. (CH3) and (CH2) modes | Proteins, lipids |
| ~1490–1470 | 1474, 1477, 1489 | δ (CH2) | Lipids |

Table A2.
Biochemical tentative assignments of the most discriminant wavenumbers selected by SELECT-LDA in the classification approach aimed at stratifying PD patients.
Table A2.
Biochemical tentative assignments of the most discriminant wavenumbers selected by SELECT-LDA in the classification approach aimed at stratifying PD patients.
| Spectral Regions in Literature | Peak Position (cm−1) ± 1 | Tentative Band Assignment | Contributions |
|---|---|---|---|
| ~1185–1120 | 1170, 1171 | (C-C) and (O-P-O); (C-O) ring vibrations | Nucleic acid “sugars” |
|
~1233 ~1225 | 1214, 1224, 1226 | asym. (O-P-O) | Nucleic acids; phospholipids; uric ring vibrations |
| ~1360–1220 | 1292, 1294, 1297, 1342 | ); sym. (PO2) predominantly (C-N) with significant contributions from (CH2) of carbohydrate residues; δ (CH2) | Amide III band; proteins; collagen |
| ~1420 | 1435, 1437 | sym. (COO); δ (CH2) | Polysaccharides |
| ~1455–1450 | 1443, 1445 | δ asym. (CH3) and (CH2) modes | Proteins; lipids |
| ~1490–1470 | 1475, 1476 | δ (CH2) | Lipids |

Table A3.
Biochemical tentative assignments of the most discriminant wavenumbers selected by SELECT-LDA in the classification approach aimed at achieving a direct discrimination between dementia associated with both analysed pathologies (PD-related dementia and Alzheimer’s dementia).
Table A3.
Biochemical tentative assignments of the most discriminant wavenumbers selected by SELECT-LDA in the classification approach aimed at achieving a direct discrimination between dementia associated with both analysed pathologies (PD-related dementia and Alzheimer’s dementia).
| Spectral Regions in Literature | Peak Position (cm−1 | Tentative Band Assignment | Contributions |
|---|---|---|---|
| ~1185–1120 | 1187, 1188 | (C-C) and (O-P-O) (C-O) ring vibrations | Nucleic acid “sugars” |
| ~1360–1220 | 1277 1340 | (C-C) and (C-O) ) sym. (PO2) (C-N) with significant contributions from (CH2) of carbohydrate residues, δ (CH2) | Amide III band; proteins; collagen |
| ~1370 | 1379, 1380, 1382 | sym. def. CH3 and sym. def. CH2 | Proteins; amino acids (cytosine, guanine, proline) Lipids; phospholipids |
| ~1405–1400 | 1402 | (C = O) of (COO) group (C = C) | Fatty acids; amino acids; (aspartate, glutamate) Uric acid |
| ~1490–1470 | 1487, 1488 | δ (CH2) | Lipids |
References
- Mhyre, T.R.; Boyd, J.T.; Hamill, R.W.; Maguire-Zeiss, K.A. Parkinson’s Disease. Subcell. Biochem. 2012, 65, 389–455. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Selkoe, D.J.; Peter, J.; Lansbury, J. Alzheimer’s Disease Is the Most Common Neurodegenerative Disorder. Basic Neurochem. Mol. Cell. Med. Asp. 1999, 6, 101–102. [Google Scholar]
- Rizek, P.; Kumar, N.; Jog, M.S. An Update on the Diagnosis and Treatment of Parkinson Disease. CMAJ 2016, 188, 1157–1165. [Google Scholar] [CrossRef] [Green Version]
- Meade, R.M.; Fairlie, D.P.; Mason, J.M. Alpha-Synuclein Structure and Parkinson’s Disease. Mol. Neurodegener. 2019, 14, 29. [Google Scholar] [CrossRef] [Green Version]
- Rocha, E.M.; De Miranda, B.; Sanders, L.H. Alpha-Synuclein: Pathology, Mitochondrial Dysfunction and Neuroinflammation in Parkinson’s Disease. Neurobiol. Dis. 2018, 109, 249–257. [Google Scholar] [CrossRef]
- Goldman, J.G.; Andrews, H.; Amara, A.; Naito, A.; Alcalay, R.N.; Shaw, L.M.; Taylor, P.; Xie, T.; Tuite, P.; Henchcliffe, C.; et al. Cerebrospinal Fluid, Plasma, and Saliva in the BioFIND Study: Relationships among Biomarkers and Parkinson’s Disease Features. Mov. Disord. 2018, 33, 282–288. [Google Scholar] [CrossRef] [Green Version]
- Gnanalingham, K.K.; Byrne, E.J.; Thornton, A.; Sambrook, M.A.; Bannister, P. Motor and Cognitive Function in Lewy Body Dementia: Comparison with Alzheimer’s and Parkinson’s Diseases. J. Neurol. Neurosurg. Psychiatry 1997, 62, 243–252. [Google Scholar] [CrossRef]
- Tofaris, G.K. A Critical Assessment of Exosomes in the Pathogenesis and Stratification of Parkinson’s Disease. J. Parkinsons Dis. 2017, 7, 569–576. [Google Scholar] [CrossRef] [Green Version]
- D’Andrea, G.; Pizzolato, G.; Gucciardi, A.; Stocchero, M.; Giordano, G.; Baraldi, E.; Leon, A. Different Circulating Trace Amine Profiles in De Novo and Treated Parkinson’s Disease Patients. Sci. Rep. 2019, 9, 6151. [Google Scholar] [CrossRef]
- Picca, A.; Calvani, R.; Landi, G.; Marini, F.; Biancolillo, A.; Gervasoni, J.; Persichilli, S.; Primiano, A.; Urbani, A.; Bossola, M.; et al. Circulating Amino Acid Signature in Older People with Parkinson’s Disease: A Metabolic Complement to the EXosomes in PArkiNson Disease (EXPAND) Study. Exp. Gerontol. 2019, 128, 110766. [Google Scholar] [CrossRef]
- Trošt, M.; Perovnik, M.; Pirtošek, Z. Correlations of Neuropsychological and Metabolic Brain Changes in Parkinson’s Disease and Other α-Synucleinopathies. Front. Neurol. 2019, 10, 1204. [Google Scholar] [CrossRef] [PubMed]
- Galvagnion, C. The Role of Lipids Interacting with α-Synuclein in the Pathogenesis of Parkinson’s Disease. J. Parkinsons Dis. 2017, 7, 433–450. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jenner, P.; Dexter, D.T.; Sian, J.; Schapira, A.H.V.; Marsden, C.D. Oxidative Stress as a Cause of Nigral Cell Death in Parkinson’s Disease and Incidental Lewy Body Disease. Ann. Neurol. 1992, 32, S82–S87. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.; Zhang, S.; Wang, D.; Fan, M.; Gao, F.; Sun, W.; Li, Z.; Li, S. The significance of uric acid in the diagnosis and treatment of Parkinson disease: An updated systemic review. Medicine 2017, 96, e8502. [Google Scholar] [CrossRef] [PubMed]
- Zhong, L.-L.; Song, Y.-Q.; Tian, X.-Y.; Cao, H.; Ju, K.-J. Level of Uric Acid and Uric Acid/Creatinine Ratios in Correlation with Stage of Parkinson Disease. Medicine 2018, 97, e10967. [Google Scholar] [CrossRef] [PubMed]
- Marques, A.; Dutheil, F.; Durand, E.; Rieu, I.; Mulliez, A.; Fantini, M.L.; Boirie, Y.; Durif, F. Glucose Dysregulation in Parkinson’s Disease: Too Much Glucose or Not Enough Insulin? Parkinsonism Relat. Disord. 2018, 55, 122–127. [Google Scholar] [CrossRef] [PubMed]
- Anichtchik, O.V.; Peitsaro, N.; Anichtchik, O.V.; Rinne, J.O.; Kalimo, H.; Kalimo, H.; Panula, P. Distribution and Modulation of Histamine H3 Receptors in Basal Ganglia and Frontal Cortex of Healthy Controls and Patients with Parkinson’s Disease. Neurobiol. Dis. 2001, 8, 707–716. [Google Scholar] [CrossRef] [Green Version]
- Shan, L.; Swaab, D.F.; Bao, A.M. Neuronal Histaminergic System in Aging and Age-Related Neurodegenerative Disorders. Exp. Gerontol. 2013, 48, 603–607. [Google Scholar] [CrossRef]
- Hertel, J.; Harms, A.C.; Heinken, A.; Baldini, F.; Thinnes, C.C.; Glaab, E.; Vasco, D.A.; Pietzner, M.; Stewart, I.D.; Wareham, N.J.; et al. Integrated Analyses of Microbiome and Longitudinal Metabolome Data Reveal Microbial-Host Interactions on Sulfur Metabolism in Parkinson’s Disease. Cell Rep. 2019, 29, 1767–1777.e8. [Google Scholar] [CrossRef] [Green Version]
- Kataoka, H.; Sugie, K. Serum Adiponectin Levels between Patients with Parkinson’s Disease and Those with PSP. Neurol. Sci. 2020, 41, 1125–1131. [Google Scholar] [CrossRef]
- Maass, F.; Michalke, B.; Willkommen, D.; Leha, A.; Schulte, C.; Tönges, L.; Mollenhauer, B.; Trenkwalder, C.; Rückamp, D.; Börger, M.; et al. Elemental Fingerprint: Reassessment of a Cerebrospinal Fluid Biomarker for Parkinson’s Disease. Neurobiol. Dis. 2020, 134, 104677. [Google Scholar] [CrossRef] [PubMed]
- Espay, A.J.; Kalia, L.V.; Gan-Or, Z.; Williams-Gray, C.H.; Bedard, P.L.; Rowe, S.M.; Morgante, F.; Fasano, A.; Stecher, B.; Kauffman, M.A.; et al. Disease Modification and Biomarker Development in Parkinson Disease: Revision or Reconstruction? Neurology 2020, 94, 481–494. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Santaella, A.; Kuiperij, H.B.; Van Rumund, A.; Esselink, R.A.J.; Van Gool, A.J.; Bloem, B.R.; Verbeek, M.M. Inflammation Biomarker Discovery in Parkinson’s Disease and Atypical Parkinsonisms. BMC Neurol. 2020, 20, 26. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Öhman, A.; Forsgren, L. NMR Metabonomics of Cerebrospinal Fluid Distinguishes between Parkinson’s Disease and Controls. Neurosci. Lett. 2015, 594, 36–39. [Google Scholar] [CrossRef] [PubMed]
- Obeso, J.A.; Stamelou, M.; Goetz, C.G.; Poewe, W.; Lang, A.E.; Weintraub, D.; Burn, D.; Halliday, G.M.; Bezard, E.; Przedborski, S.; et al. Past, Present, and Future of Parkinson’s Disease: A Special Essay on the 200th Anniversary of the Shaking Palsy. Mov. Disord. 2017, 32, 1264–1310. [Google Scholar] [CrossRef]
- Ohmichi, T.; Mitsuhashi, M.; Tatebe, H.; Kasai, T.; Ali El-Agnaf, O.M.; Tokuda, T. Quantification of Brain-Derived Extracellular Vesicles in Plasma as a Biomarker to Diagnose Parkinson’s and Related Diseases. Parkinsonism Relat. Disord. 2019, 61, 82–87. [Google Scholar] [CrossRef]
- Ahmed, S.S.; Santosh, W.; Kumar, S.; Christlet, H.T.T. Metabolic Profiling of Parkinson’s Disease: Evidence of Biomarker from Gene Expression Analysis and Rapid Neural Network Detection. J. Biomed. Sci. 2009, 16, 63. [Google Scholar] [CrossRef] [Green Version]
- Shao, Y.; Le, W. Recent Advances and Perspectives of Metabolomics-Based Investigations in Parkinson’s Disease. Mol. Neurodegener. 2019, 14, 3. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Figura, M.; Kuśmierska, K.; Bucior, E.; Szlufik, S.; Koziorowski, D.; Jamrozik, Z.; Janik, P. Serum Amino Acid Profile in Patients with Parkinson’s Disease. PLoS ONE 2018, 13, e191670. [Google Scholar] [CrossRef]
- Dashti, H.; Westler, W.M.; Tonelli, M.; Wedell, J.R.; Markley, J.L.; Eghbalnia, H.R. Spin System Modeling of Nuclear Magnetic Resonance Spectra for Applications in Metabolomics and Small Molecule Screening. Anal. Chem. 2017, 89, 12201–12208. [Google Scholar] [CrossRef]
- Antcliffe, D.; Jiménez, B.; Veselkov, K.; Holmes, E.; Gordon, A.C. Metabolic Profiling in Patients with Pneumonia on Intensive Care. EBioMedicine 2017, 18, 244–253. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bereman, M.S.; Kirkwood, K.I.; Sabaretnam, T.; Furlong, S.; Rowe, D.B.; Guillemin, G.J.; Mellinger, A.L.; Muddiman, D.C. Metabolite Profiling Reveals Predictive Biomarkers and the Absence of β-Methyl Amino-l-Alanine in Plasma from Individuals Diagnosed with Amyotrophic Lateral Sclerosis. J. Proteome Res. 2020, 19, 3276–3285. [Google Scholar] [CrossRef] [PubMed]
- Ros-Mazurczyk, M.; Jelonek, K.; Marczyk, M.; Binczyk, F.; Pietrowska, M.; Polanska, J.; Dziadziuszko, R.; Jassem, J.; Rzyman, W.; Widlak, P. Serum Lipid Profile Discriminates Patients with Early Lung Cancer from Healthy Controls. Lung Cancer 2017, 112, 69–74. [Google Scholar] [CrossRef] [PubMed]
- Yao, L.; Lyu, N.; Chen, J.; Pan, T.; Yu, J. Joint Analyses Model for Total Cholesterol and Triglyceride in Human Serum with Near-Infrared Spectroscopy. Spectrochim. Acta-Part A Mol. Biomol. Spectrosc. 2016, 159, 53–59. [Google Scholar] [CrossRef]
- Khalil, S.K.H.; Azooz, M.A.; Division, P. Application of Vibrational Spectroscopy in Identification of the Composition of the Urinary Stones. J. Appl. Sci. Res. 2007, 3, 387–391. [Google Scholar]
- Selvaraju, R.; Raja, A.; Thiruppathi, G. FT-Raman Spectral Analysis of Human Urinary Stones. Spectrochim. Acta-Part A Mol. Biomol. Spectrosc. 2012, 99, 205–210. [Google Scholar] [CrossRef]
- Roy, S.; Perez-Guaita, D.; Bowden, S.; Heraud, P.; Wood, B.R. Spectroscopy Goes Viral: Diagnosis of Hepatitis B and C Virus Infection from Human Sera Using ATR-FTIR Spectroscopy. Clin. Spectrosc. 2019, 1, 100001. [Google Scholar] [CrossRef]
- Lilo, T.; Morais, C.L.M.; Ashton, K.M.; Pardilho, A.; Davis, C.; Dawson, T.P.; Gurusinghe, N.; Martin, F.L. Spectrochemical Differentiation of Meningioma Tumours Based on Attenuated Total Reflection Fourier-Transform Infrared (ATR-FTIR) Spectroscopy. Anal. Bioanal. Chem. 2020, 412, 1077–1086. [Google Scholar] [CrossRef] [Green Version]
- Carmona, P.; Molina, M.; Calero, M.; Bermejo-Pareja, F.; Martínez-Martín, P.; Toledano, A. Discrimination Analysis of Blood Plasma Associated with Alzheimer’s Disease Using Vibrational Spectroscopy. J. Alzheimer’s Dis. 2013, 34, 911–920. [Google Scholar] [CrossRef] [Green Version]
- Ahmed, S.S.S.J.; Santosh, W.; Kumar, S.; Thanka Christlet, T.H. Neural Network Algorithm for the Early Detection of Parkinson’s Disease from Blood Plasma by FTIR Micro-Spectroscopy. Vib. Spectrosc. 2010, 53, 181–188. [Google Scholar] [CrossRef]
- Li, S.; Liu, J.; Li, G.; Zhang, X.; Xu, F.; Fu, Z.; Teng, L.; Li, Y.; Sun, F. Near-Infrared Light-Responsive, Pramipexole-Loaded Biodegradable PLGA Microspheres for Therapeutic Use in Parkinson’s Disease. Eur. J. Pharm. Biopharm. 2019, 141, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Wu, Q.; Li, C.; Zhou, Y.; Xu, F.; Zong, L.; Ge, S. A Study of Parkinson’s Disease Patients’ Serum Using FTIR Spectroscopy. Infrared Phys. Technol. 2020, 106, 103279. [Google Scholar] [CrossRef]
- Perez-Guaita, D.; Garrigues, S.; de la, M.; Guardia. Infrared-Based Quantification of Clinical Parameters. TrAC-Trends Anal. Chem. 2014, 62, 93–105. [Google Scholar] [CrossRef]
- Pizarro, C.; Esteban-Díez, I.; Espinosa, M.; Rodríguez-Royo, F.; González-Sáiz, J.M. An NMR-Based Lipidomic Approach to Identify Parkinson’s Disease-Stage Specific Lipoprotein-Lipid Signatures in Plasma. Analyst 2019, 144, 1334–1344. [Google Scholar] [CrossRef] [PubMed]
- Lawton, M.; Baig, F.; Toulson, G.; Morovat, A.; Evetts, S.G.; Ben-Shlomo, Y.; Hu, M.T. Blood Biomarkers with Parkinson’s Disease Clusters and Prognosis: The Oxford Discovery Cohort. Mov. Disord. 2020, 35, 279–287. [Google Scholar] [CrossRef] [Green Version]
- Zhao, H.W.; Lin, J.; Wang, X.B.; Cheng, X.; Wang, J.Y.; Hu, B.L.; Zhang, Y.; Zhang, X.; Zhu, J.H. Assessing Plasma Levels of Selenium, Copper, Iron and Zinc in Patients of Parkinson’s Disease. PLoS ONE 2013, 8, e83060. [Google Scholar] [CrossRef]
- Barth, A. Infrared Spectroscopy of Proteins. Biochim. Biophys. Acta-Bioenerg. 2007, 1767, 1073–1101. [Google Scholar] [CrossRef] [Green Version]
- Pizarro, C.; Arenzana-Rámila, I.; Pérez-del-Notario, N.; Pérez-Matute, P.; González-Sáiz, J.M. Thawing as a Critical Pre-Analytical Step in the Lipidomic Profiling of Plasma Samples: New Standardized Protocol. Anal. Chim. Acta 2016, 912, 1–9. [Google Scholar] [CrossRef]
- Forina, M.; Lanteri, S.; Armanino, C.; Oliveos, M.C.C.; Casolino, M.C.; Casale, M. V-Parvus 2011, an Extendable Package of Programs for Explorative Data Analysis, Classification and Regression Analysis, Dip. Chimica e Tecnologie Farmaceutiche ed Alimentari, University of Genova, Genova (Italy). 2011. Available online: https://iris.unige.it/handle/11567/202703 (accessed on 18 March 2022).
- Tabora, J.E.; Domagalski, N. Multivariate Analysis and Statistics in Pharmaceutical Process Research and Development. Annual Review of Chemical and Biomolecular Engineering. Annu. Rev. Chem. Biomol. Eng. 2017, 8, 403–426. [Google Scholar] [CrossRef]
- Forina, M.; Oliveri, P.; Casale, M. Complete Validation for Classification and Class Modeling Procedures with Selection of Variables and/or with Additional Computed Variables. Chemom. Intell. Lab. Syst. 2010, 102, 110–122. [Google Scholar] [CrossRef]
- Casale, M.; Sáiz Abajo, M.J.; González Sáiz, J.M.; Pizarro, C.; Forina, M. Study of the Aging and Oxidation Processes of Vinegar Samples from Different Origins during Storage by Near-Infrared Spectroscopy. Anal. Chim. Acta 2006, 557, 360–366. [Google Scholar] [CrossRef]
- Worsfold, P.J. Chemometrics: A Textbook (Data Handling in Science and Technology, Vol. 2). Anal. Chim. Acta 1989, 225, 457–458. [Google Scholar] [CrossRef]
- Bury, D.; Morais, C.L.M.; Paraskevaidi, M.; Ashton, K.M.; Dawson, T.P.; Martin, F.L. Spectral Classification for Diagnosis Involving Numerous Pathologies in a Complex Clinical Setting: A Neuro-Oncology Example. Spectrochim. Acta-Part A Mol. Biomol. Spectrosc. 2019, 206, 89–96. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, A.L.; Gajjar, K.B.; Theophilou, G.; Martin, F.L.; Martin-Hirsch, P.L. Vibrational Spectroscopy of Biofluids for Disease Screening or Diagnosis: Translation from the Laboratory to a Clinical Setting. J. Biophotonics 2014, 7, 153–165. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Shi, L.; Zhang, L.R. Neuroprotective Effect of Carnosine against Salsolinol-Induced Parkinson’s Disease. Exp. Ther. Med. 2017, 14, 664–670. [Google Scholar] [CrossRef] [Green Version]
- Cheng, J.S.; Dubal, D.B.; Kim, D.H.; Legleiter, J.; Cheng, I.H.; Yu, G.-Q.; Tesseur, I.; Wyss-Coray, T.; Bonaldo, P.; Mucke, L. Collagen VI Protects Neurons against Aβ Toxicity. Nat. Neurosci. 2009, 12, 119–121. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paraskevaidi, M.; Morais, C.L.M.; Lima, K.M.G.; Snowden, J.S.; Saxon, J.A.; Richardson, A.M.T.; Jones, M.; Mann, D.M.A.; Allsop, D.; Martin-Hirsch, P.L.; et al. Differential Diagnosis of Alzheimer’s Disease Using Spectrochemical Analysis of Blood. Proc. Natl. Acad. Sci. USA 2017, 114, E7929–E7938. [Google Scholar] [CrossRef] [Green Version]
- Jim Enez-Jim Enez, F.J.; Alonso-Navarro, H.; Garc Ia-Mart, E.; Ag Undez, J.A.G. Cerebrospinal and Blood Levels of Amino Acids as Potential Biomarkers for Parkinson’s Disease: Review and Meta-Analysis. Eur. J. Neurol. 2020, 2020, 2336–2347. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).