Abstract
Microorganism assessment plays a key role in food quality and safety control but conventional techniques are costly and/or time consuming. Alternatively, electronic tongues (E-tongues) can fulfill this critical task. Thus, a potentiometric lab-made E-tongue (40 lipid sensor membranes) was used to differentiate four common food contamination bacteria, including two Gram positive (Enterococcus faecalis, Staphylococcus aureus) and two Gram negative (Escherichia coli, Pseudomonas aeruginosa). Principal component analysis and a linear discriminant analysis-simulated annealing algorithm (LDA-SA) showed that the potentiometric signal profiles acquired during the analysis of aqueous solutions containing known amounts of each studied bacteria allowed a satisfactory differentiation of the four bacterial strains. An E-tongue-LDA-SA model (12 non-redundant sensors) correctly classified 98 ± 5% of the samples (repeated K-fold-CV), the satisfactory performance of which can be attributed to the capability of the lipid membranes to establish electrostatic interactions/hydrogen bonds with hydroxyl, amine and/or carbonyl groups, which are comprised in the bacteria outer membranes. Furthermore, multiple linear regression models, based on selected subsets of E-tongue sensors (12–15 sensors), also allowed quantifying the bacteria contents in aqueous solutions (0.993 ± 0.011 ≤ R2 ≤ 0.998 ± 0.005, for repeated K-fold-CV). In conclusion, the E-tongue could be of great value as a preliminary food quality and safety diagnosis tool.
1. Introduction
Pathogenic microorganisms in foodstuffs form a major source of foodborne diseases in humans. In foods, pathogenic microorganisms are frequently present in low levels and are heterogeneously distributed, which makes their detection a difficult task [1]. Thus, indicator microorganisms are commonly used to assess food quality and safety, whenever there is a positive relationship between the presence of an indicator microorganism and the occurrence of a pathogen [2]. Escherichia coli and Staphylococcus aureus are used worldwide as hygiene and safety indicator microorganisms for several food products [3,4]. Enterococcus faecalis is considered an indicator of fecal contamination, although it also belongs to the natural microbiota of many fermented products (dairy products, meat and vegetables), with E. faecalis as one of the dominant species. Indeed, E. faecalis plays an important role in the development of organoleptic characteristics. In addition, this microorganism contributes to the safety of fermented products since it producers bacteriocins that have antimicrobial activity against foodborne pathogens and spoilage bacteria [5]. Pseudomonas spp. are commonly found in soil, water, plants and foods (e.g., dairy and meat products) as well as on the surfaces of food processing plants, which are all linked to their capacity to form biofilms [6]. Furthermore, these bacteria are psychrotrophic (i.e., they can grow at low temperatures ranging from 0 to 7 °C), and are thus able to become the dominant population in cold stored foods [7]. P. aeruginosa is responsible for serious infections in immunodeficient persons. Most Pseudomonas spp. are resistant to penicillin and related β-lactam antibiotics [8]. In 2013, the U.S. Center for Disease Control and Prevention identified antibiotic-resistant threats in the U.S, and P. aeruginosa was included in the list of serious threats [9]. The recognition of this bacterium as an emerging opportunistic pathogen has been highlighted in the European Union Commission Directive 2015/1787 [10], regarding the water quality for human consumption, which endorsed the identification of P. aeruginosa as a quality indicator microorganism.
Several conventional techniques are commonly applied to detecting microorganisms including conventional culture methods [11,12], immunological techniques (e.g., enzyme-linked immunosorbent assays) [13,14,15], and polymerase chain reactions [16,17]. In spite of their recognition accuracy, most of these techniques are laborious, time-consuming and costly [18,19]. Alternatively, (bio)sensor-based approaches have emerged as cost-effective alternative/complementary identification methods for microorganisms, providing a short analysis time. However, the industrial use of such instruments in detecting food spoilage or the presence of pathogenic microorganisms is still at the early stages and some technical drawbacks still need to be addressed, as reviewed by Ghasemi-Varnamkhasti et al. [19]. As recently pointed out, electrochemical sensor devices, such as electronic noses (E-noses) [20,21] and electronic tongues (E-tongues) [22], are the basis of a wide number of low-cost and fast response analytical strategies for the analysis of gas and liquid matrices, respectively. E-tongues based on different sensing technologies (e.g., potentiometric, voltammetric), comprising sets of multisensors with low selectivity and cross-sensitivity, together with multivariate qualitative and quantitative chemometric tools (e.g., principal component analysis (PCA); soft independent modelling of class analogy (SIMCA); linear discriminant analysis (LDA); partial least-squares regression (PLS); support vector machines (SVMs); artificial neural networks (ANNs)) have been successfully applied to directly or indirectly identify several microorganisms. Voltammetric E-tongues, together with chemometric tools, have been successfully applied to monitoring the growth of molds (e.g., Aspergillus, Penicillium, Mucor and Rhizopus) and to differentiating them, or to estimating the total bacterial count (between 1 and 10 log CFU/mL) in liquid media [23,24,25], the amount of E. coli (from 0.1 to 107 CFU/mL) in drinking water treatment plants [26] or of the number of spoilage bacteria (mesophilic bacteria and Enterobacteriaceae, in the range 4.5 to 9.0 log CFU/g) in fresh cod during storage [27], as well as to differentiating known food microbial contaminants, including yeasts (e.g., Zygosaccharomyces bailii), bacteria (e.g., E. coli, P. aeruginosa, Klebsiella oxytoca, Bacillus subtilis) and/or molds (Aspergillus flavus, A. oryzae, A. ochraceus, A. versicolor, Penicillium commune, Rhizopus stolonifer) [24,26,28,29,30]. However, only a few studies have reported the use of laboratory-made potentiometric E-tongues. For example, lab-made E-tongues, comprising sensors with chalcogenide glass and plasticized polymeric membranes or electrodes containing pastes with different active elements (e.g., platinum, gold, silver, graphite, silver-palladium, copper and ruthenium oxide) have been used to differentiate species of Aspergillus (A. flavus, A. oryzae, A. ochraceus, A. versicolor) and a yeast species (Z. bailii) [29], or to assess the total viable counts (i.e., total number of microorganisms) of sea bream samples [31]. More recently, commercial potentiometric E-tongues (α-Astree device from Alpha MOS Company, Toulouse, France), using sensors based on chemically modified field-effect transistor technology, were capable of accurately assessing the total viable counts in spoiled fish samples [32], detecting and differentiating known bacterial strains (S. aureus, E. coli and P. aeruginosa) [33] and differentiating eight Fusarium isolates and monitoring the fungal growth [34].
The satisfactory qualitative and/or quantitative performances reported for both voltammetric and potentiometric E-tongues may be related to their sensing responses towards different primary or secondary metabolites consumed or produced during the growth of microorganisms, which would lead to detectable composition changes at growing media levels [34]. In addition, the differences in the chemical composition of the cell membranes/walls of microorganisms (e.g., different chemical groups, more or less prone to redox reactions and/or to electrostatic and hydrogen bond interactions) could result in numerous and different cell-wall-sensor interactions, leading to specific signal fingerprints of sensors, and thus to the capability of recognizing and distinguishing different microorganisms [35]. Bacteria can be classified into two major groups: Gram positive and Gram negative. The distinction between both groups is based on the Gram staining method, which depends on differences in the structure of the cell walls. The cell walls of Gram-positive bacteria consist of a thick layer of peptidoglycans. These bacteria retain the crystal violet dye during the staining process, leading to a violet color. In the case of Gram-positive bacteria, the major chemical diversity between species is associated with the composition of the peptide crosslinks between glycan strands (a macromolecular network of repeating units of the disaccharide N-acetyl glucosamine-N-acetyl muramic acid) [36,37] and with the high variability in structure and chemical composition of long anionic polymers, which are composed largely of glycerol phosphate, glucosyl phosphate or ribitol phosphate repeats, covalently attached to peptidoglycan, namely to teichoic acids or lipoteichoic acids, anchored to the head groups of the membrane lipids [38]. However, Gram-negative bacteria have a very thin peptidoglycan layer between the inner and outer cell membranes, and thus, do not retain the violet dye during the staining process, resulting in red staining. In the case of Gram-negative bacteria, the layer of peptidoglycan is thinner and is surrounded by an outer membrane and an asymmetric bilayer, with phospholipids in the inner leaflet and lipopolysaccharides in the outer leaflet. The lipopolysaccharide molecules are composed of three moieties; namely, lipid A, a core oligosaccharide and a polysaccharide O-chain, being an O-chain that is highly variable even between strains [39]. The differences in the type and number of chemical groups present on the cell walls of microorganisms may result in changes in the surface potential and the charge density of the membranes of the sensors during the cell-wall-sensor interactions, justifying the capability of the potentiometric E-tongue in detecting different microorganisms. This recognition capability may be enhanced according to the type and concentrations of the lipid additives and plasticizers used for obtaining each lipid sensor membrane. In fact, the sensitivity of the sensor is related to the concentration of the charged lipids inside the membrane being, for example, the sensitivity towards charged compounds promoted by low-density lipids. Meanwhile, the selectivity of a sensor greatly depends on its surface hydrophobicity, which in turn depends on the relative composition and type of lipids and plasticizers used. The use of high lipid concentrations results in a more hydrophilic surface and, conversely, high plasticizer concentration turns the surface more hydrophobic [40].
In this work, a lab-made potentiometric E-tongue, comprising only lipid sensor membranes, was used as a recognition device to discriminate and, for the first time, quantify four typical food contamination microorganisms (E. faecalis, S. aureus, E. coli and P. aeruginosa). Lipid polymeric sensor membranes are not specific to any particular substance [40], but, as previously explained, their sensing characteristics are related to their surface potential and charge density, for which selectivity and sensitivity vary depending on the type and concentrations of the lipid additives and plasticizers used [40]. The versatility of E-tongues containing lipid sensors to respond in the presence of numerous compounds, due to the establishment of electrostatic interactions or hydrogen bonds between the different compounds and the polar and non-polar regions of the lipid membranes, has been widely reported in the literature [41,42,43,44,45,46,47,48].
2. Materials and Methods
2.1. Bacterial Strains and Inoculum Preparation
This work was aimed at detecting and quantifying three food-borne pathogens, including two Gram-positive spherically shaped bacteria S. aureus ATCC653 and E. faecalis ATCC29212, and one Gram-negative rod-shaped bacterium (P. aeruginosa ATCC15442). Additionally, E. coli ATCC29998 was used since it is a broad hygiene/fecal indicator. Bacterial inoculum solutions were made by mixing 300 µL of glycerol (Sigma-Aldrich, Merck, Darmstadt, Germany) and 700 µL of bacterial culture, grown overnight in the nutrient-rich microorganism growing medium, Brain Heart Infusion (BHI; PanReac AppliChem, ITW Reagents, Barcelona, Spain) at 37 °C, under orbital agitation (Orbital incubator S1500, Stuart, Staffordshire, UK) at 90 rpm. The inocula were cryopreserved at −20 °C in a freezer until being used. All reagents were of analytical grade.
2.2. Growth Conditions and Biomass Recovery
Each strain was inoculated aseptically in separate 1-L Erlenmeyer flasks containing 300 mL BHI. Bacterial cultures were grown for 24 h to reach the stationary phase of cells at 37 °C with orbital agitation of 90 rpm. A total of eight independent fermentations was carried out for each microorganism studied. At the end of the individual fermentation, a Gram-staining procedure was performed to determine if there was any culture contamination. Gram staining was performed as described by Gregersen [49]. Heat-fixed smear on slides were flooded with crystal violet solution (Sigma-Aldrich, Merck, Darmstadt, Germany) for 20 s and then washed with tap water. Next, the smear was covered with Gram’s iodine (Sigma-Aldrich, Merck, Darmstadt, Germany), the mordant, for 1 min and decolorized with commercial 96% ethanol for 15 s, followed by washing with tap water and counterstaining with fuchsin (Sigma-Aldrich, Merck, Darmstadt, Germany) for 30 s, and was finally washed with tap water and slides dried at room temperature. Observation was carried out by optical microscope at magnification of 1000×. Biomass yield of each culture growth was assessed by dry weight. For this purpose, each bacterial culture was divided into 4 pre-weighed 50 mL polypropylene centrifuge tubes. Cells were harvested by centrifugation at 4000 rpm (Eppendorf Centrifuge 5810 R, Hamburg, Germany) for 10 min, cells were resuspended in water and centrifuged again. The tubes with biomass were dried at 37 °C (for 48 h) and weighed, after cooling in a desiccator, on an analytical balance with accuracy to 0.1 mg (Pioneer TM, OHAUS, Parsippany, NJ, USA). The biomass concentration was expressed as a mg of cell dry weight per mL of bacterial culture. All reagents used were of analytical grade.
2.3. E-Tongue Apparatus
A lab-made potentiometric E-tongue multisensor device, comprising two cylindrical arrays [50] and a Ag/AgCl reference electrode (Crison, model 5241, Barcelona, Spain), was used to establish the potentiometric fingerprints of each microorganism resuspended in water. The E-tongue device was connected to an Agilent Data Acquisition unit (model 34970A, Agilent Technologies, Loveland, CO, USA), which was controlled by Agilent BenchLink Data Logger software, as shown in Figure 1. Each array comprised 20 lipid polymeric cross-sensitive sensor membranes (40 sensors in total), each corresponding to a different mixture of an additive compound (~3%, methyltrioctylammonium chloride, octadecylamine, oleic acid and oleyl alcohol) and a plasticizer (~32%, bis (1-butylpentyl) adipate, dibutyl sebacate, dioctyl phenylphosphonate, 2-nitrophenyl-octyl ether and tris (2-ethylhexyl) phosphate), plus a high molecular weight polyvinyl chloride (PVC, ~65%). All reagents were of analytical grade. Each sensor was coded with a letter S (for sensor) followed by a number related to the sensor array (1: or 2:) and a number related to the lipid sensor membrane applied (1 to 20), corresponding to different combinations of plasticizer and additive compounds, as described in Table 1. The type of lipid polymeric sensors and the compositions were chosen due to (i) the satisfactory signal stability over time with a lower than 5% relative standard deviation (RSD) over a 5-h period using both standard solutions and real-life samples; (ii) the satisfactory intra- and inter-day repeatability (0.1 ≤ RSD ≤ 15%) towards standard chemical solutions that mimicked the five basic taste sensations (sweet, acid, bitter, salty and umami) or the alcoholic aqueous extracts of olive oils; and (iii) their re-usability during at least a one-year time period without requiring any replacement of the sensor membranes [51].
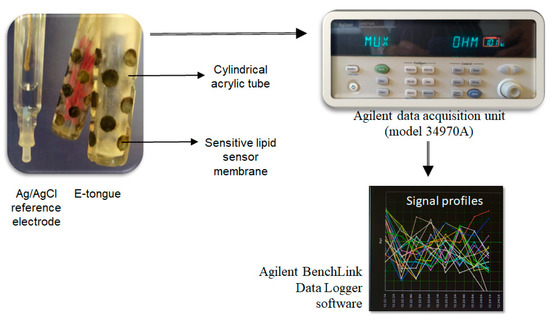
Figure 1.
Lab-made E-tongue device: sensor arrays, data logger (data acquisition unit) and control software installed on a PC.

Table 1.
E-tongue sensor codes and related composition (%) of the respective lipid membranes (type/pair of additive and plasticizer).
2.4. E-Tongue Analysis
For the experimental potentiometric assays, known amounts of each microorganism were resuspended in 100 mL of deionized water and then mixed for 1–2 min in a vortex. Then, the solutions were immediately analyzed with the E-tongue for 5 min to allow time for attaining a pseudo-equilibrium between the E-tongue non-specific lipid polymeric membranes and the microorganism of each solution. In practice, after a 5 min analysis, the potentiometric signal of each lipid polymeric membrane showed an almost constant potential, signaling that equilibrium was established. This facilitated a fast analysis time. After each assay the E-tongue was washed with deionized water and, after 4–5 assays it was immersed in a HCL aqueous solution (0.01 mol/L) to evaluate the signal repeatability, as well as promoting a more efficient cleaning of the sensor arrays. At the end of each day, the E-tongue was stored at room temperature and immersed in an aqueous 0.01 mil/L HCl solution to clean the sensor arrays. This also improved the signal repeatability.
2.5. Statistical Analysis
The unsupervised and supervised classification performance of the E-tongue to identify and differentiate the four microorganisms under study was evaluated using principal component analysis (PCA) and linear discriminant analysis (LDA) coupled with the meta-heuristic simulated annealing (SA) variable selection algorithm [52,53,54]. PCA is a technique that allows reducing the number of variables (in this case 40 E-tongue sensors) into a small number of principal components (PCs) so that a particular pattern in the dataset is easily recognizable when they are plotted in a multidimensional space. LDA is a classification multivariate technique that provides linear models of the classification scores regarding the descriptors under study (i.e., pre-defined groups). These models maximize the ratio of between-class variance and minimize the ratio of within-class variance. LDA assumes a priori knowledge of the group membership of each sample in a training set. The SA meta-heuristic algorithm was used to identify the best sub-set containing the lower number of non-redundant E-tongue sensors [44]. The LDA performance was checked using two internal cross-validation (CV) variants, namely the leave-one-out CV (LOO-CV) and the repeated K-fold-CV. The former is considered an over-optimistic internal validation technique and the latter aims to overcome this possible limitation and to minimize overfitting risks. In the former variant, the dataset minus one observation is used to establish the linear discriminant (LD) functions and then the omitted data are classified, being the procedure repeated for all the observations. For the latter CV variant, data are randomly split more than once, depending on the number of repeats (set equal to 10), each time divided into K folds of approximately equal size (set equal to 4, allowing keeping 25% of the initial data for validation purposes). Each of the folds is left out in turn and the other K-1 folds are used to train the model. The held-out fold is predicted and these predictions are summarized according to a performance measure like the sensitivity (e.g., the percentage of correct classifications). The K estimates are averaged to achieve the overall resampled estimate. Variable scaling and centering procedures were implemented as data normalization procedures. The PCA and LDA model outputs were graphically evaluated using 3D plots of the three most significant PCs or LDs, being the calculated sensitivity values (i.e., the percentage of samples correctly classified into the pre-established groups) for the latter approach. Multiple linear regression (MLR) models, based on selected non-redundant E-tongue sensors identified using the simulated annealing (SA) algorithm, were developed using the potentiometric data acquired during the analysis of the aqueous solutions containing different known amounts of each of the four studied microorganisms. MLR is based on the regression analysis of two or more independent variables. In this sense it is a linear model that describes how a dependent variable (i.e., each microorganism’s amount in aqueous solutions) relates to more than one independent variable (i.e., from 2 to 40 E-tongue sensors). The accuracy of the E-tongue-MLR-SA models was established based on the determination coefficients (R2) and the root-mean-square errors (RMSEs) for training and internal validation (leave-one-out cross-validation, LOO-CV; and repeated K-fold-CV with 4 folds and 10 repeats) procedures. The accuracy of the E-tongue-MLR-SA models was compared against the analytical conventional techniques. The acceptance of the alternative method as a procedure equivalent to a reference procedure (for results) requires that the slope and intercept values of the regression line between the data for both approaches be equal to those of a perfect line (i.e., 1 and 0, respectively) [55,56]. All statistical analyses were performed using the Sub-select [53] and MASS [57] packages of the open-source statistical program R (version 2.15.1), at a 5% significance level.
3. Results and Discussion
3.1. Biomass Determination by Dry Weight
At the end of each fermentation assay, the culture was checked by evaluating possible contamination with undesirable microorganisms. The microscopic observation of the culture, after Gram staining, allowed verification that at the end of the fermentation, all the cells had the morphology of the microorganism under study and so the presence of the other studied microorganisms was not detected. The biomass determined by dry weight of the culture growth is reported as mean values (standard deviation) for eight fermentation replicas performed for each microorganism: 1.21 (0.24) mg/mL for E. coli; 0.84 (0.13) mg/mL for P. aeruginosa; 0.97 (0.40) mg/mL for E. faecalis; and 1.41 (0.16) mg/mL for S. aureus.
3.2. Microorganism Recognition and Differentiation Based on E-Tongue Potentiometric Profiles
The possibility of recognizing and differentiating the four microorganisms under study, based on the potentiometric fingerprints recorded by the E-tongue during the analysis of solutions containing different amounts of each microorganism, was evaluated. In fact, and as previously suggested, it was expected [40] that the surface potential and charge density of the lipid membranes comprised on the E-tongue would change due to the establishment of electrostatic interactions/hydrogen bonds between the chemical groups of the cell walls of each microorganism and those contained in the lipid sensor membranes. The intensity of the potentiometric signals recorded by each sensor would vary according to type and number of chemical groups on the cell walls of each microorganism under study, as well as the different additive–plasticizer combination (4 additives and 5 plasticizers were used) for each sensor. For each microorganism, eight independent aqueous solutions were prepared by resuspending known amounts of dry weight cells in 100 mL of deionized water (E. coli: 0.309 to 0.666 mg/mL; E. faecalis: 0.279 to 0.538 mg/mL; S. aureus: 0.511 to 0.702 mg/mL; and P. aeruginosa: 0.324 to 0.435 mg/mL), totaling 32 independent aqueous solutions analyzed on the same day to minimize the drifts of signals and to avoid the need for complex signal pre-treatments. Indeed, the intra-day repeatability of the signals of the E-tongue sensors was confirmed by analyzing a fresh aqueous 0.01 mol/L HCl solution (0.5 ≤ RSD ≤ 5.2%) ten times. As shown by a plot of the potentiometric signal for each sensor in Figure 2, the E-tongue signals recorded by the 40 sensor lipid membranes varied within the same potential range (0.2 to 350 mV) for the four bacteria being studied. It could also be inferred that different sensors showed different signal intensities, ranging from 0.2 to 350 mV, highlighting that the different lipid membranes used responded differently in the presence of aqueous solutions containing each of the four microorganisms.
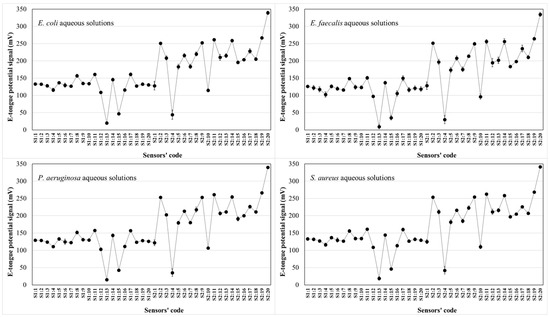
Figure 2.
E-tongue potentiometric signal profiles recorded during the analysis of aqueous 0.1 mol/L HC1 solution containing each of the four microorganisms indicated in the plot.
Next, PCA was applied to the raw E-tongue signals allowing us to obtain a new coordinate system (i.e., the PCs, which are linearly uncorrelated) through an orthogonal linear transformation of the recorded signals. The results obtained are shown in Figure 3A. In this figure, the first three PCs, based on the potentiometric signal profiles acquired by the 40 E-tongue sensors, allowed us to clearly differentiate in the new coordinate space the aqueous solutions containing P. aeruginosa and E. faecalis, although the same was not observed for E. coli and S. aureus, for which a clear overplotting was observed. Thus, the results show that the electronic device could be used to identify aqueous solutions containing P. aeruginosa or E. faecalis, but not the other two studied microorganisms (E. coli and S. aureus). To further assess the E-tongue classification capability, the data were subjected to LDA, for which most discriminant and non-redundant sensors were selected by the SA algorithm. The best E-tongue-LDA-SA model, for which the first three discriminant functions (LDs) explained 100% of the data variability, was established based on 12 E-tongue sensors (1st sensor array: 1st E-tongue array: S1:2, S1:5, S1:7, S1:8 and S1:9; 2nd sensor array: S2:4, S2:9, S2:10, S2:11, S2:13, S2:14 and S2:18). The supervised discriminant model allowed us to correctly classify 100% of the original grouped data (Figure 3B). Moreover, the model showed sensitivities of 100% for the LOO-CV and of 98 ± 5% for the repeated K-fold-CV internal validation procedures. It should be noted that, for the latter CV variant, which ensures that at each run 25% of the dataset is used for validation (i.e., 2 solutions of each microorganism, totaling 8 among 32 independent samples, were kept aside for validation purposes), misclassification was only observed between E. faecalis and P. aeruginosa. The unsupervised and supervised classification performances clearly highlighted the potential use of the E-tongue as an accurate and fast recognition device for the four microorganisms studied, being possible to foresee its use as a preliminary quality/safety control tool, taking into account that these microorganisms are typical water-food contamination indicators.
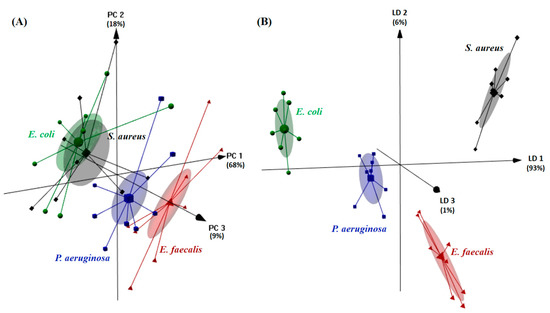
Figure 3.
Differentiation (3D plots) of four common water-food contamination microorganisms based on the E-tongue signal profiles acquired during the analysis of aqueous solutions of E. coli (0.309 to 0.666 mg/mL, symbol ●), E. faecalis (0.279 to 0.538 mg/mL, symbol ▲), S. aureus (0.511 to 0.702 mg/mL, symbol ♦) and P. aeruginosa (0.324 to 0.435 mg/mL, symbol ■). (A) Unsupervised PCA recognition based on the potentiometric signals of 40 E-tongue sensors; and (B) supervised LDA classification based on the potentiometric signals of a set of 12 non-redundant E-tongue sensors, selected by the SA algorithm.
The satisfactory recognition performance of the lab-made potentiometric E-tongue is in line with the results previously reported in the literature for devices based on the same sensing technique. For example, Söderström et al. [29] reported that a potentiometric E-tongue, comprising 27 potentiometric chemical sensors with cross-sensitivity towards inorganic cations and anions as well as to organic substances, could be used to satisfactorily differentiate (PCA) A. versicolor, A. ochraceus and Z. bailii, although it misclassified A. flavus and A. oryzae. A discriminant analysis allowed achieving a correct classification rate of 90% for external validation, it being shown that the potentiometric E-tongue could be used to differentiate all microorganisms in the stationary phase as well as during their growth. More recently, a commercial potentiometric E-tongue (α-Astree device with seven chemically modified solid potential sensors) was used by Al Ramahi et al. [33] to accurately differentiate (PCA) three bacteria (S. aureus, E. coli and P. aeruginosa) at different growing time-periods (from 15 to 24 h, to which corresponded discrimination indices from 83 to 96%). Furthermore, it was shown that a discriminant analysis (based on the information gathered from two sensors) allowed recognizing new E. coli samples, used as an external dataset. Finally, Abu-Khalaf and Rumaila [34] also used an α-Astree device for trying to differentiate (PCA) eight Fusarium isolates, which partially succeeded since the isolates were clustered into two main groups, and to monitor the fungal growth.
3.3. Microorganism Quantification Based on E-Tongue Potentiometric Profiles
The quantitative performance of the lab-made potentiometric E-tongue to quantify the amounts of each of the four studied microorganisms in aqueous solutions was further investigated by applying a MLR approach, based on sets of non-redundant sensors selected using the SA algorithm. Two CV variants were also used to evaluate the performance of the E-tongue-MLR-SA models, i.e., the LOO-CV and repeated K-fold-CV procedures (4 folds × 10 repeats). The parameters of the goodness of fitting of the developed models are shown in Table 2, together with the number and type of sensors included in each MLR model. The very satisfactory R2 and RMSE values (LOO-CV: 0.994 ≤ R2 ≤ 0.998 and 0.028 ≤ RMSE ≤ 0.064 mg/mL; repeated K-fold-CV: 0.993 ± 0.011 ≤ R2 ≤ 0.998 ± 0.002 and 0.032 ± 0.014 ≤ RMSE ≤ 0.076 ± 0.036 mg/mL) provided support for the use of the E-tongue as a quantitative tool for predicting the amounts of the bacteria under study in aqueous solutions. This has opened up the opportunity for the E-tongue to be applied in quality and safety control. The quality of the E-tongue quantitative performance can be further verified in Figure 4, where the results achieved for the repeated K-fold-CV internal validation procedure are plotted. Overall, the referred quantitative performance is in agreement with previous studies, which have shown the possibility of applying a potentiometric E-tongue to quantify the total viable counts of microorganisms in sea bream samples, using a PLS- (R2 = 0.954) based model [31] as well as in spoiled fish samples, using SVM- (R2 = 0.937) and ANN- (R2 = 0.986) based models [32].

Table 2.
Predictive performance of E-tongue-MLR-SA models regarding the quantification of typical water-food contamination microorganisms (contents determined according to cells’ dry weight) resuspended in aqueous solutions.
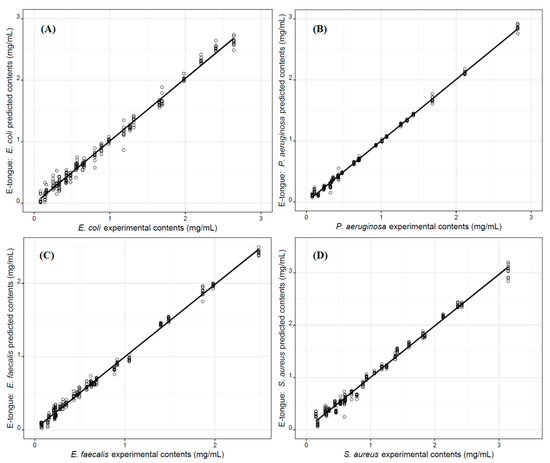
Figure 4.
Predictive performance (repeated K-fold-CV internal validation: 4 folds × 10 repeats) of the potentiometric lab-made E-tongue towards the quantification of the microorganism amounts (mg/mL) resuspended in aqueous solutions: (A) E. coli content assessment using an MLR model based on the signals acquired from 15 E-tongue sensors; (B) P. aeruginosa content assessment using an MLR model based on the signals acquired from 14 E-tongue sensors; (C) E. faecalis content assessment using an MLR model based on the signals acquired from 13 E-tongue sensors; and, (D) S. aureus content assessment using an MLR model based on the signals acquired from 12 E-tongue sensors.
Lastly, the application of E-tongue-MLR-SA models as a possible complementary tool to quantify the amounts of bacteria in aqueous solutions was evaluated following the methodology described by Roig and Thomas [55,56], based on the XPT 90-210 French standard [58] for the comparison of an alternative procedure with a reference method. Thus, single linear regressions were established between the amounts of each bacterium predicted by the E-tongue-MLR-SA model and the experimental known amounts, being investigated if the slope and intercept values were statistically equal to one and zero, respectively (corresponding to a perfect linear fit). Table 3 shows the parameters of the single linear regressions (R2, slope and intercept values and the respective 95% intervals of confidence, IC) for the two cross-validation variants. As can be inferred, at the 5% significance level, the slope and intercept values were statistically equal to the expected theoretical values, except for S. aureus and for the repeated K-fold-CV procedure. Therefore, it could be stated that, in general, the E-tongue coupled with MLR models can be applied as a fast and cost-effective strategy to assess the contents of the four studied bacteria in aqueous solutions.

Table 3.
Parameters of the single linear regressions established between the microorganisms’ content in aqueous solutions predicted by the E-tongue-MLR-SA models (LOO-CV and repeated K-fold-CV) and the experimental contents based on the known amounts of dry cells’ weight resuspended in deionized water: coefficient of determination (R2); slopes, intercept values and respective confidence intervals (CI) at 95%.
4. Conclusions
This study showed that a potentiometric E-tongue, comprising lipid polymeric sensor membranes, could be used as a practical, non-invasive and direct tool to discriminate and quantify four typical bacteria (E. coli, E. faecalis, S. aureus and P. aeruginosa) in aqueous solutions, which are recognized as food and/or water contaminant bacteria, related to several worldwide foodborne outbreaks. The satisfactory qualitative and quantitative predictive performances of the sensor device could be tentatively attributed to the known capability of the lipid sensor membranes to establish electrostatic interactions or hydrogen bonds with hydroxyl, carbonyl and/or amine groups that can be found, in different numbers, in the outer membranes of the referred bacteria. Finally, the promising results achieved for aqueous solutions must be further confirmed for other relevant food matrices (e.g., meat and dairy products) in order to ensure that, in the future, the E-tongue may be used as a routine safety/quality control tool in the processed/fermented food production field.
Author Contributions
Conceptualization, T.D. and A.M.P.; methodology, A.C.A.V., T.D. and A.M.P.; formal analysis, A.C.A.V., T.D. and A.M.P.; investigation, H.G. and Í.M.G.M.; resources, A.C.A.V., T.D. and A.M.P.; data curation, H.G. and Í.M.G.M.; writing—original draft preparation, A.C.A.V., T.D. and A.M.P.; writing—review and editing, H.G., Í.M.G.M., A.C.A.V., T.D. and A.M.P.; resources, A.C.A.V., T.D. and A.M.P.; funding acquisition, A.C.A.V., T.D. and A.M.P. All authors have read and agreed to the published version of the manuscript.
Funding
The authors are grateful to the Foundation for Science and Technology (FCT, Portugal) for financial support by national funds FCT/MCTES to CIMO (UIDB/00690/2020) and to CEB (UIDB/04469/2020), as well as to the BioTecNorte operation (NORTE-01-0145-FEDER-000004) funded by the European Regional Development Fund under the scope of Norte2020—Programa Operacional Regional do Norte. Ítala M.G. Marx also acknowledges the Ph.D. research grant (SFRH/BD/137283/2018) provided by FCT.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available on request from the corresponding author.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Pérez-Rodríguez, F.; González-García, P.; Valero, A.; Hernández, M.; Rodríguez-Lázaro, D. Impact of the prevalence of different pathogens on the performance of sampling plans in lettuce products. Int. J. Food Microbiol. 2014, 184, 69–73. [Google Scholar] [CrossRef]
- Pierson, M.; Zink, D.; Smoot, L. Indicator Microorganisms and Microbiological Criteria. In Food Microbiology: Fundamentals and Frontiers, 3rd ed.; Doyle, M., Beuchat, L., Eds.; ASM Press: Washington, DC, USA, 2007; pp. 69–85. [Google Scholar]
- Codex Alimentarius Commission—FAO/WHO. Codex Alimentarius; Food and Agriculture Organization of the United Nations: Rome, Italy; World Health Organization: Geneva, Switzerland, 1992. [Google Scholar]
- European Union. Commission Regulation (EC) 2073/2005 of 15 November 2005 on Microbiological Criteria for Foodstuffs. Off. J. Eur. Union 2015, L338, 1–26. [Google Scholar]
- Hanchi, H.; Mottawea, W.; Sebei, K.; Hammami, R. The Genus Enterococcus: Between Probiotic Potential and Safety Concerns—An Update. Front. Microbiol. 2018, 9, 1791. [Google Scholar] [CrossRef]
- Ahmed, A.B.H.; Abbassi, M.S.; Rojo-Bezares, B.; Ruiz-Roldán, L.; Dhahri, R.; Mehri, I.; Sáenz, Y.; Hassen, A. Characterization of Pseudomonas aeruginosa isolated from various environmental niches: New STs and occurrence of antibiotic susceptible “high-risk clones”. Int. Environ. Health Res. 2020, 30, 643–652. [Google Scholar] [CrossRef]
- Stellato, G.; Utter, D.R.; Voorhis, A.; de Angelis, M.; Eren, A.M.; Ercolini, D. A Few Pseudomonas Oligotypes Dominate in the Meat and Dairy Processing Environment. Front. Microbiol. 2017, 2, 264. [Google Scholar] [CrossRef] [PubMed]
- Croughs, P.D.; Klaassen, C.H.W.; van Rosmalen, J.; Maghdid, D.M.; Boers, S.A.; Hays, J.P.; Goessens, W.H.F. Unexpected mechanisms of resistance in Dutch Pseudomonas aeruginosa isolates collected during 14 years of surveillance. Int. J. Antimicrob. Agents 2018, 52, 407–410. [Google Scholar] [CrossRef] [PubMed]
- Centers for Disease Control and Prevention (CDC). Antibiotic Resistance Threats in the United States. Available online: http://www.cdc.gov/drugresistance/threat-report-2013/pdf/ar-threats-2013-508.pdf (accessed on 26 March 2021).
- European Union. Commission Directive (EU) 2015/1787 of 6 October 2015 on the quality of water intended for human consumption. Off. J. Eur. Union 2015, L260, 6–17. [Google Scholar]
- Gracias, K.S.; McKillip, J.L. A review of conventional detection and enumeration methods for pathogenic bacteria in food. Can. J. Microbiol. 2004, 50, 883–890. [Google Scholar] [CrossRef]
- Hameed, S.; Xie, L.; Ying, Y. Conventional and emerging detection techniques for pathogenic bacteria in food science: A review. Trends Food Sci. Technol. 2018, 81, 61–73. [Google Scholar] [CrossRef]
- Ali, Y.M.; Sim, R.B.; Schwaeble, W.; Shaaban, M.I. Enterococcus faecalis Escapes Complement-Mediated Killing via Recruitment of Complement Factor H. J. Infect. Dis. 2019, 220, 1061–1070. [Google Scholar] [CrossRef] [PubMed]
- Hashemi, E.; Forouzandeh, M. Designing a new biosensor “DNA ELISA” to detect Escherichia coli using genomic DNA and comparison of this method to PCR-ELISA. J. Enzyme Inhib. Med. Chem. 2018, 33, 722–725. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; He, J.; Cao, X.; Huang, K.; Luo, Y.; Xu, W. Development of a double-antibody sandwich ELISA for rapid detection of Bacillus Cereus in food. Sci. Rep. 2016, 6, 16092. [Google Scholar] [CrossRef] [PubMed]
- Jordana-Lluch, E.; Giménez, M.; Quesada, M.D.; Ausina, V.; Martró, E. Improving the diagnosis of bloodstream infections: PCR coupled with mass spectrometry. BioMed Res. Int. 2014, 2014, 501214. [Google Scholar] [CrossRef] [PubMed]
- Ma, K.; Deng, Y.; Bai, Y.; Xu, D.; Chen, E.; Wu, H.; Li, B.; Gao, L. Rapid and simultaneous detection of Salmonella, Shigella, and Staphylococcus aureus in fresh pork using a multiplex real-time PCR assay based on immunomagnetic separation. Food Cont. 2014, 42, 87–93. [Google Scholar] [CrossRef]
- Liu, X.; Hu, Y.; Zheng, S.; Liu, Y.; He, Z.; Luo, F. Surface plasmon resonance immunosensor for fast, highly sensitive, and in situ detection of the magnetic nanoparticles-enriched Salmonella enteritidis. Sens. Actuators B Chem. 2016, 230, 191–198. [Google Scholar] [CrossRef]
- Ghasemi-Varnamkhasti, M.; Apetrei, C.; Lozano, J.; Anyogu, A. Potential use of electronic noses, electronic tongues and biosensors as multisensor systems for spoilage examination in foods. Trends Food Sci. Technol. 2018, 80, 71–92. [Google Scholar] [CrossRef]
- Liao, Y.-H.; Yu, Y.-N.; Abbod, M.F.; Shih, C.-H.; Shieh, J.-S. Using artificial neural network to predict a variety of pathogenic microorganisms. Sens. Mater. 2020, 32, 2375–2385. [Google Scholar]
- Capuano, R.; Paba, E.; Mansi, A.; Marcelloni, A.M.; Chiominto, A.; Proietto, A.R.; Zampetti, E.; Macagnano, A.; Lvova, L.; Catini, A.; et al. Aspergillus species discrimination using a gas sensor array. Sensors 2020, 20, 4004. [Google Scholar] [CrossRef]
- Martínez-Bisbal, M.C.; Carbó Mestre, N.; Martínez-Máñez, R.; Bauzá, J.; Alcañiz Fillol, M. Microalgae degradation follow up by voltammetric electronic tongue, impedance spectroscopy and NMR spectroscopy. Sens. Actuators B Chem. 2019, 281, 44–52. [Google Scholar] [CrossRef]
- Söderström, C.; Borén, H.; Winquist, F.; Krantz-Rülcker, C. Use of an electronic tongue to analyze mold growth in liquid media. Int. J. Food Microbiol. 2003, 83, 253–261. [Google Scholar] [CrossRef]
- Zhao, G.; Lin, X.; Dou, W.; Tian, S.; Deng, S.; Shi, J. Use of the smart tongue to monitor mold growth and discriminate between four mold species grown in liquid media. Anal. Chim. Acta 2011, 690, 240–247. [Google Scholar] [CrossRef]
- Wei, Z.; Wang, J.; Zhang, X. Monitoring of quality and storage time of unsealed pasteurized milk by voltammetric electronic tongue. Electrochim. Acta 2013, 88, 231–239. [Google Scholar] [CrossRef]
- Gomez, J.K.C.; Acevedo, C.M.D.; Garcia, R.O. Application of an E-Tongue and E-nose for a rapid E. coli detection in a drinking water treatment plant. In Proceedings of the ISOEN 2019—18th International Symposium on Olfaction and Electronic Nose, Fukuoka, Japan, 26–29 May 2019. [Google Scholar]
- Ruiz-Rico, M.; Fuentes, A.; Masot, R.; Alcañiz, M.; Fernández-Segovia, I.; Barat, J.M. Use of the voltammetric tongue in fresh cod (Gadus morhua) quality assessment. Innov. Food Sci. Emerg. Technol. 2013, 18, 256–263. [Google Scholar] [CrossRef]
- Söderström, C.; Winquist, F.; Krantz-Rülcker, C. Recognition of six microbial species with an electronic tongue. Sens. Actuators B Chem. 2003, 89, 248–255. [Google Scholar] [CrossRef]
- Söderström, C.; Rudnitskaya, A.; Legin, A.; Krantz-Rülcker, C. Differentiation of four Aspergillus species and one Zygosaccharomyces with two electronic tongues based on different measurement techniques. J. Biotechnol. 2005, 119, 300–308. [Google Scholar] [CrossRef] [PubMed]
- Hong, M.; Jianping, S.; Jing, Z.; Yuming, G.; Zhiming, X. The compare of microbial electronic tongue data based on direct and two-stage processing. Appl. Mech. Mater. 2010, 20, 331–336. [Google Scholar]
- Gil, L.; Barat, J.M.; Escriche, I.; Garcia-Breijo, E.; Martínez-Máñez, R.; Soto, J. An electronic tongue for fish freshness analysis using a thick-film array of electrodes. Microchim. Acta 2008, 163, 121–129. [Google Scholar] [CrossRef]
- Han, F.; Huang, X.; Teye, E.; Haiyang, G. Quantitative Analysis of Fish Microbiological Quality Using Electronic Tongue Coupled with Nonlinear Pattern Recognition Algorithms. J. Food Saf. 2015, 35, 336–344. [Google Scholar] [CrossRef]
- Al Ramahi, R.; Zaid, A.N.; Abu-Khalaf, N. Evaluating the potential use of electronic tongue in early identification and diagnosis of bacterial infections. Infect. Drug Resist. 2019, 12, 2445–2451. [Google Scholar] [CrossRef]
- Abu-Khalaf, N.; Rumaila, B.A. Electronic tongue and box-PCR for categorization of different Fusarium strains. Plant Cell Biotechnol. Mol. Biol. 2020, 21, 121–128. [Google Scholar]
- Saylan, Y.; Erdem, O.; Cihangir, N.; Denizli, A. Detecting fingerprints of waterborne bacteria on a sensor. Chemosensors 2019, 7, 33. [Google Scholar] [CrossRef]
- Yang, H.; Singh, M.; Kim, S.J.; Schaefer, J. Characterization of the tertiary structure of the peptidoglycan of Enterococcus faecalis. Biochim. Biophys. Acta Biomembr. 2017, 1859, 2171–2180. [Google Scholar] [CrossRef] [PubMed]
- Desmarais, S.M.; De Pedro, M.A.; Cava, F.; Huang, K.C. HPLC analyses of bacterial cell wall composition. Mol. Microbiol. 2013, 89, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Dalen, V.R.; Peschel, A.; Sorge, N.M.V. Wall Teichoic Acid in Staphylococcus aureus Host Interaction. Trends Microbiol. 2020, 28, 985–996. [Google Scholar] [CrossRef]
- Liu, B.; Furevi, A.; Perepelov, A.V.; Guo, X.; Cao, H.; Wang, Q.; Reeves, P.R.; Knirel, Y.A.; Wang, L.; Widmalm, G. Structure and genetics of Escherichia coli O antigens. FEMS Microbiol. Rev. 2020, 44, 655–683. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Tahara, Y.; Yatabe, R.; Toko, K. Taste sensor: Electronic tongue with lipid membranes. Anal. Sci. 2020, 36, 147–159. [Google Scholar] [CrossRef] [PubMed]
- Toko, K.; Hara, D.; Tahara, Y.; Yasuura, M.; Ikezaki, H. Relationship between the amount of bitter substances adsorbed onto lipid/polymer membrane and the electric response of taste sensors. Sensors 2014, 14, 16274–16286. [Google Scholar] [CrossRef]
- Yasuura, M.; Shen, Q.; Tahara, Y.; Yatabe, R.; Toko, K. Development and investigation of a sweetness sensor for sugars—Effect of lipids. Sens. Mater. 2015, 27, 351–358. [Google Scholar]
- Sharma, G.; Kumar, S.; Kumar, A.; Sharma, A.; Kumar, R.; Kaur, R.; Bhondekar, A.P. Development of Lipid Membrane Based Taste Sensors for Electronic Tongue. Procedia Comput. Sci. 2015, 70, 146–152. [Google Scholar] [CrossRef]
- Veloso, A.C.A.; Dias, L.G.; Rodrigues, N.; Pereira, J.A.; Peres, A.M. Sensory intensity assessment of olive oils using an electronic tongue. Talanta 2016, 146, 585–593. [Google Scholar] [CrossRef]
- Slim, S.; Rodrigues, N.; Dias, L.G.; Veloso, A.C.A.; Pereira, J.A.; Oueslati, S.; Peres, A.M. Application of an electronic tongue for Tunisian olive oils’ classification according to olive cultivar or physicochemical parameters. Eur. Food Res. Technol. 2017, 243, 1459–1470. [Google Scholar] [CrossRef]
- Arca, V.C.; Peres, A.M.; Machado, A.A.; Bona, E.; Dias, L.G. Sugars’ quantifications using a potentiometric electronic tongue with cross-selective sensors: Influence of an ionic background. Chemosensors 2019, 7, 43. [Google Scholar] [CrossRef]
- Jeong, H.-M.; Kwon, H.-C.; Xu, B.; Jung, D.; Han, M.; Kwon, D.-H.; Kang, S.-W. Taste sensor based on the floating gate structure of a lateral double-diffused metal-oxide semiconductor. Sens. Actuators B Chem. 2020, 308, 127661. [Google Scholar] [CrossRef]
- Wu, X.; Miyake, K.; Tahara, Y.; Fujimoto, H.; Iwai, K.; Narita, Y.; Hanzawa, T.; Kobayashi, T.; Kakiuchi, M.; Ariki, S.; et al. Quantification of bitterness of coffee in the presence of high-potency sweeteners using taste sensors. Sens. Actuators B Chem. 2020, 309, 127784. [Google Scholar] [CrossRef]
- Gregersen, T. Rapid method for distinction of gram-negative from gram-positive bacteria. Eur. J. Appl. Microbiol. Biotechnol. 1978, 5, 123–127. [Google Scholar] [CrossRef]
- Rodrigues, N.; Marx, I.M.G.; Casal, S.; Dias, L.G.; Veloso, A.C.A.; Pereira, J.A.; Peres, A.M. Application of an electronic tongue as a single-run tool for olive oils’ physicochemical and sensory simultaneous assessment. Talanta 2019, 197, 363–373. [Google Scholar] [CrossRef] [PubMed]
- Dias, L.A.; Peres, A.M.; Veloso, A.C.A.; Reis, F.S.; Vilas-Boas, M.; Machado, A.A.S.C. An electronic tongue taste evaluation: Identification of goat milk adulteration with bovine milk. Sens. Actuators B Chem. 2009, 136, 209–217. [Google Scholar] [CrossRef]
- Bertsimas, D.; Tsitsiklis, J. Simulated annealing. Stat. Sci. 1993, 8, 10–15. [Google Scholar] [CrossRef]
- Cadima, J.; Cerdeira, J.O.; Minhoto, M. Computational aspects of algorithms for variable selection in the context of principal components. Comput. Stat. Data Anal. 2004, 47, 225–236. [Google Scholar] [CrossRef]
- Kirkpatrick, S.; Gelatt, C.D.; Vecchi, M.P. Optimization by simulated annealing. Science 1983, 220, 671–680. [Google Scholar] [CrossRef]
- Roig, B.; Thomas, O. Rapid estimation of global sugars by UV photodegradation and UV spectrophotometry. Anal. Chim. Acta 2003, 477, 325–329. [Google Scholar] [CrossRef]
- Roig, B.; Thomas, O. UV monitoring of sugars during wine making. Carbohydr. Res. 2003, 338, 79–83. [Google Scholar] [CrossRef]
- Venables, W.N.; Ripley, R.B. Modern Applied Statistics with S—Statistics and Computing, 4th ed.; Springer: New York, NY, USA, 2002. [Google Scholar]
- Association Française de Normalisation (AFNOR). Recueil des Normes Françaises, Qualité de l’Eau, 2nd ed.; Association Française de Normalisation (AFNOR): Paris, France, 1997; Volume 1, p. 293. [Google Scholar]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).