Impedimetric Biosensor Based on a Hechtia argentea Lectin for the Detection of Salmonella spp.
Abstract
1. Introduction
2. Materials and Methods
2.1. Lectin Extraction and Evaluation
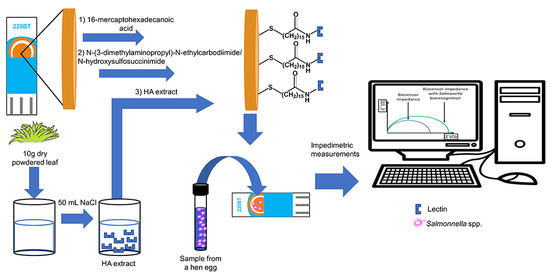
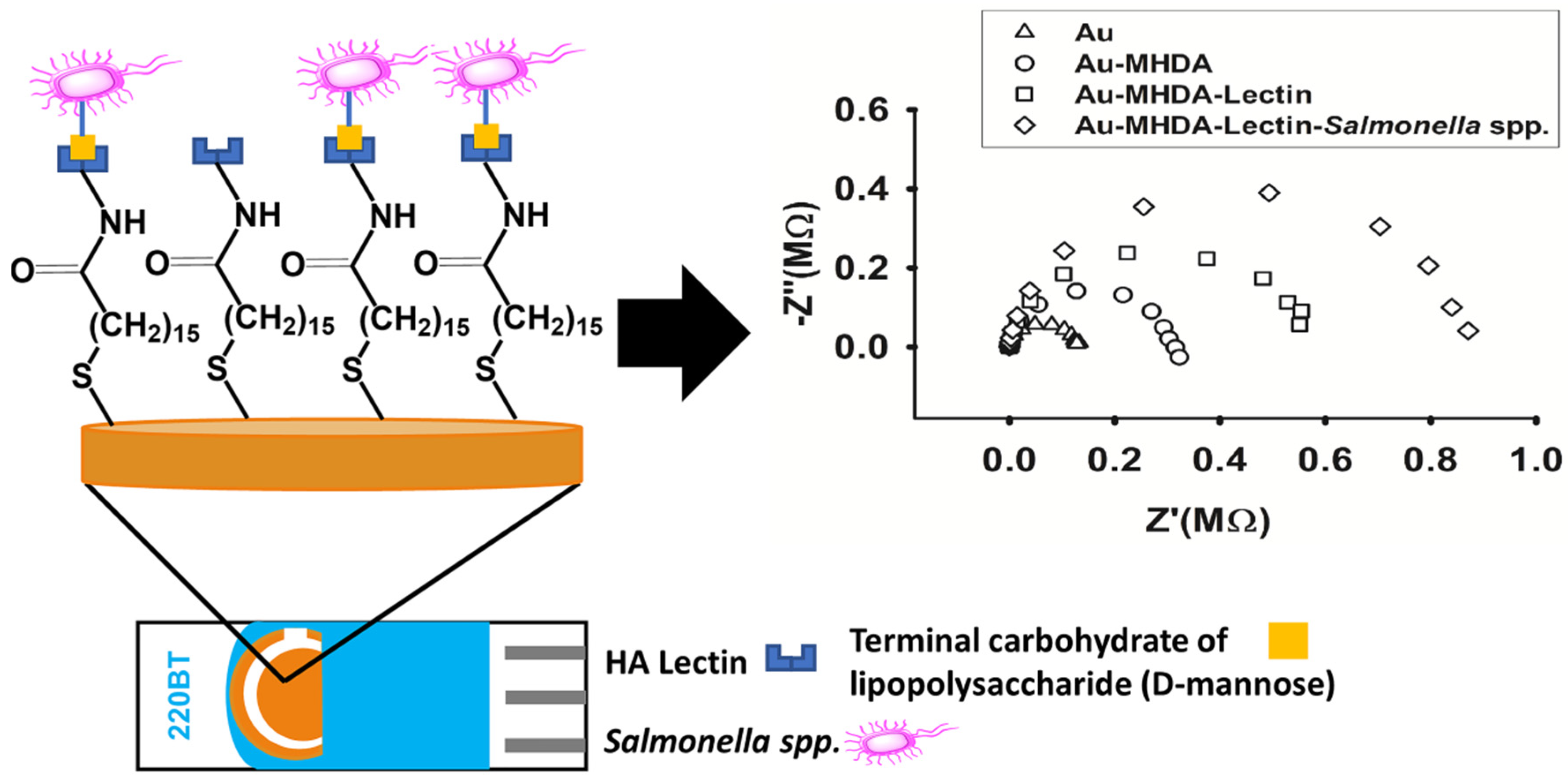
2.2. Biosensor Construction
2.3. Impedimetric Measurements
3. Results and Discussion
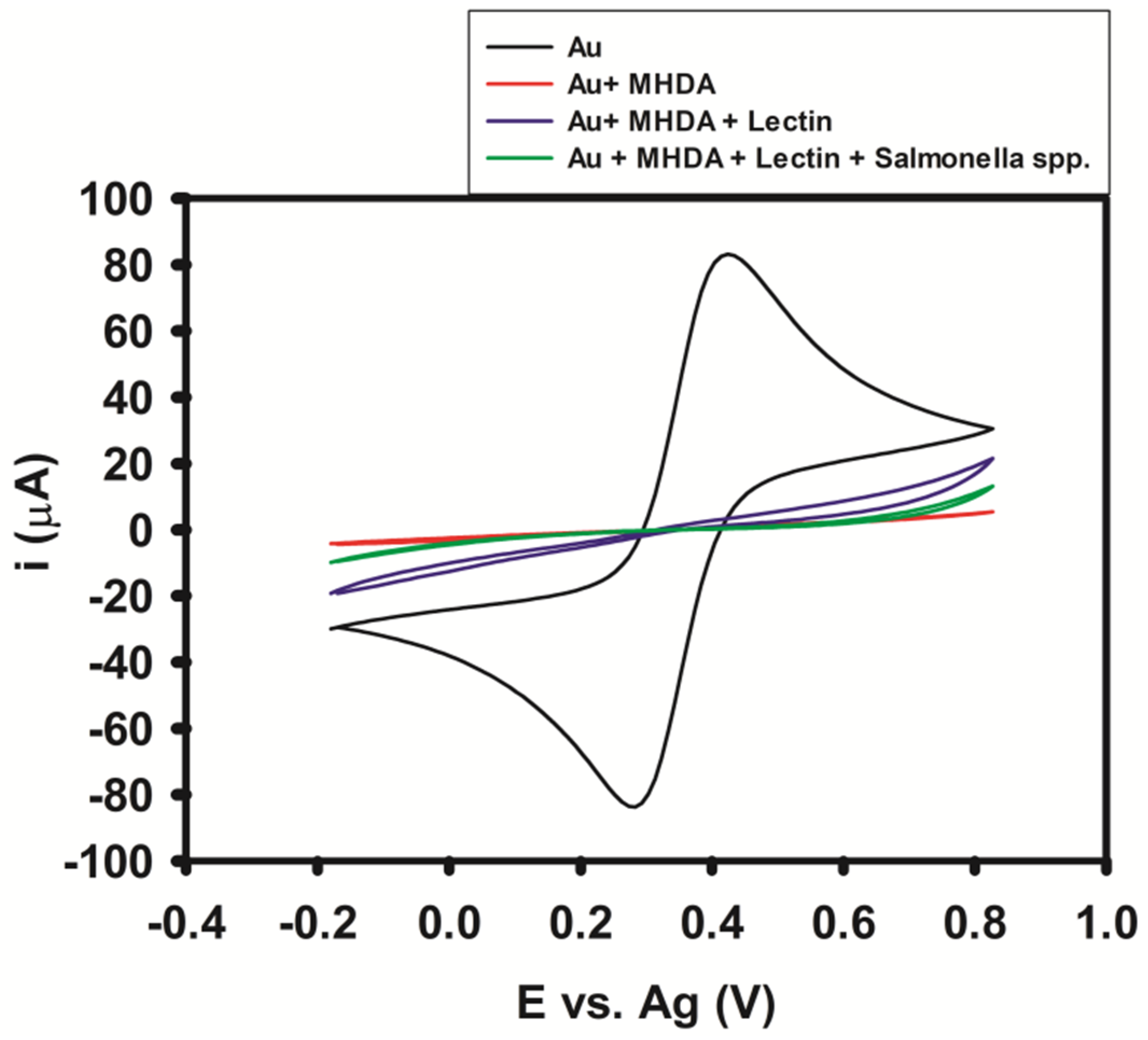
3.1. Lectin Evaluation
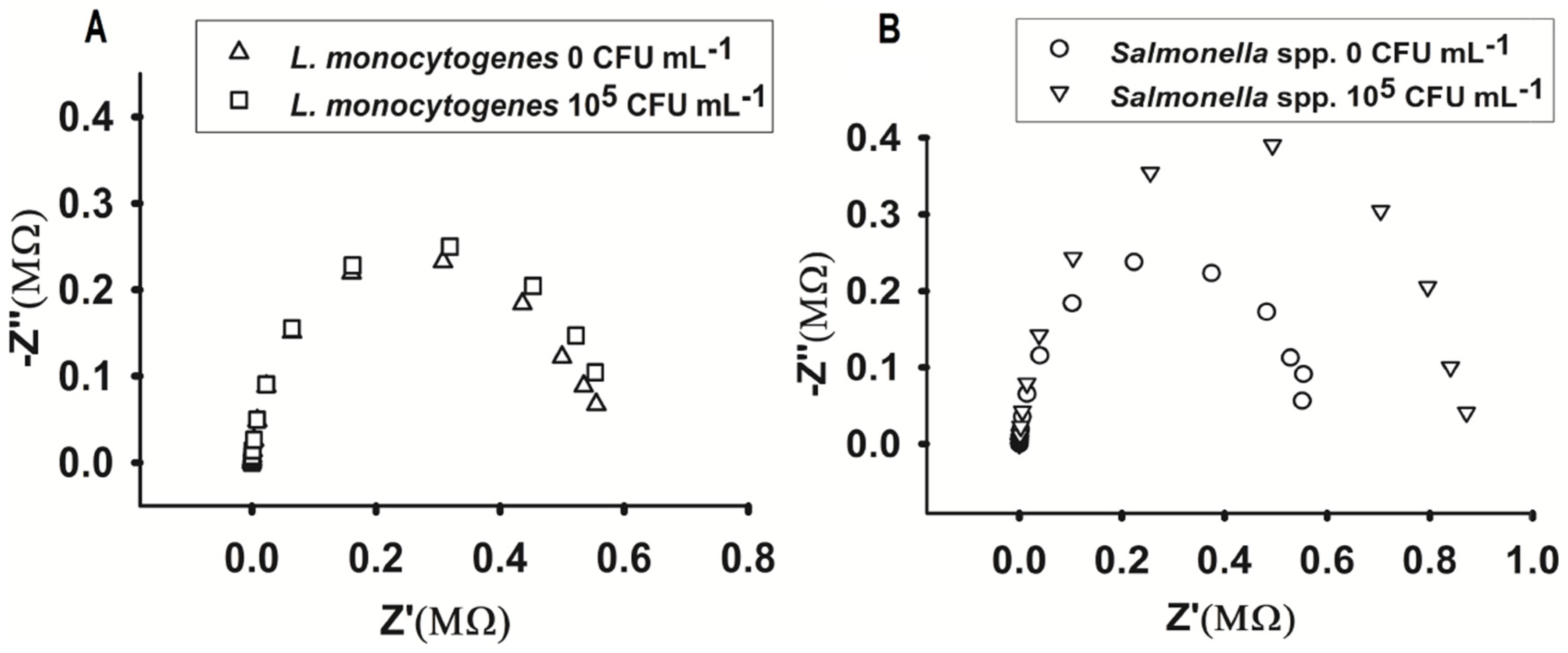
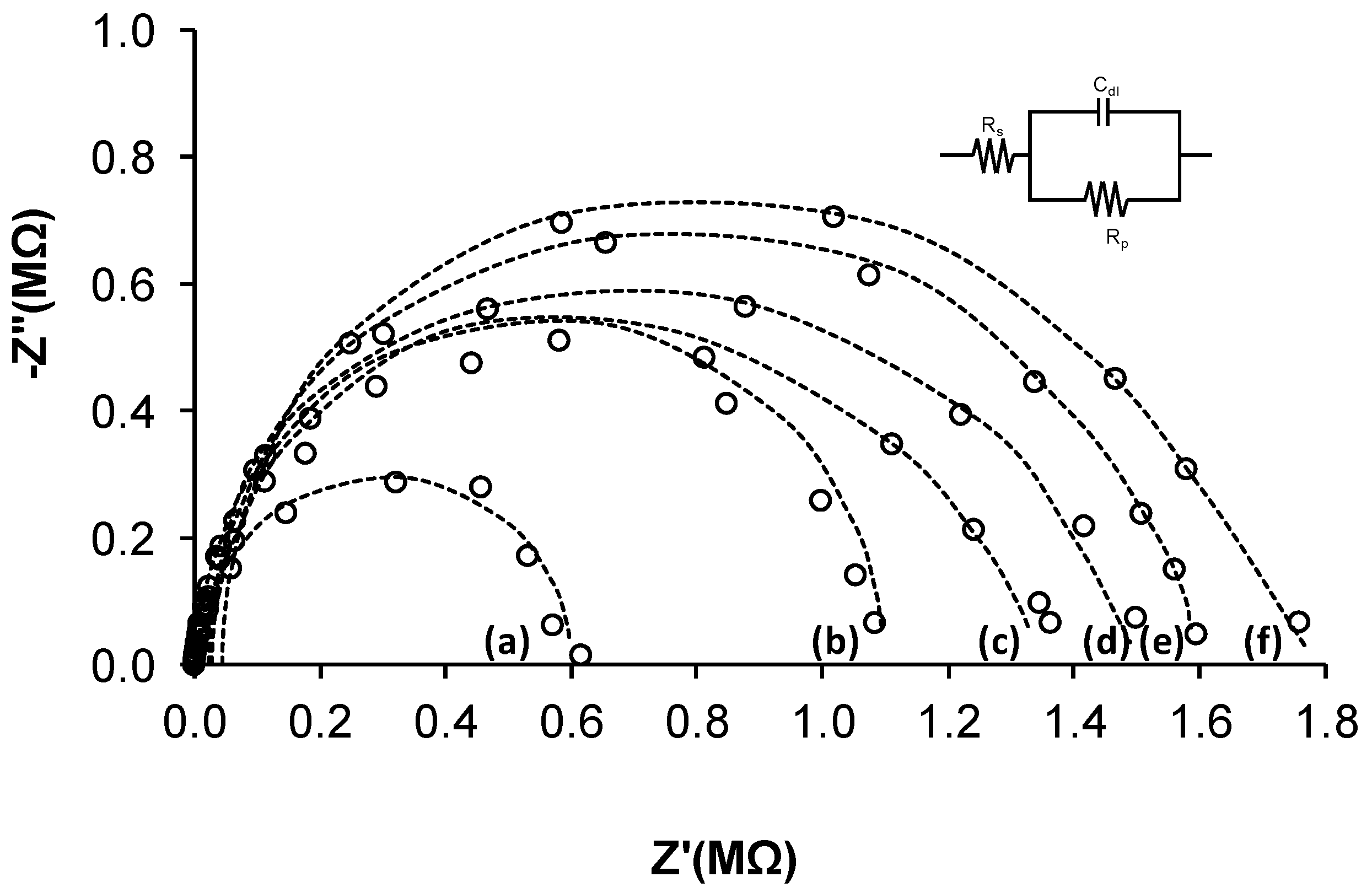
3.2. Biosensor Impedimetric Evaluation
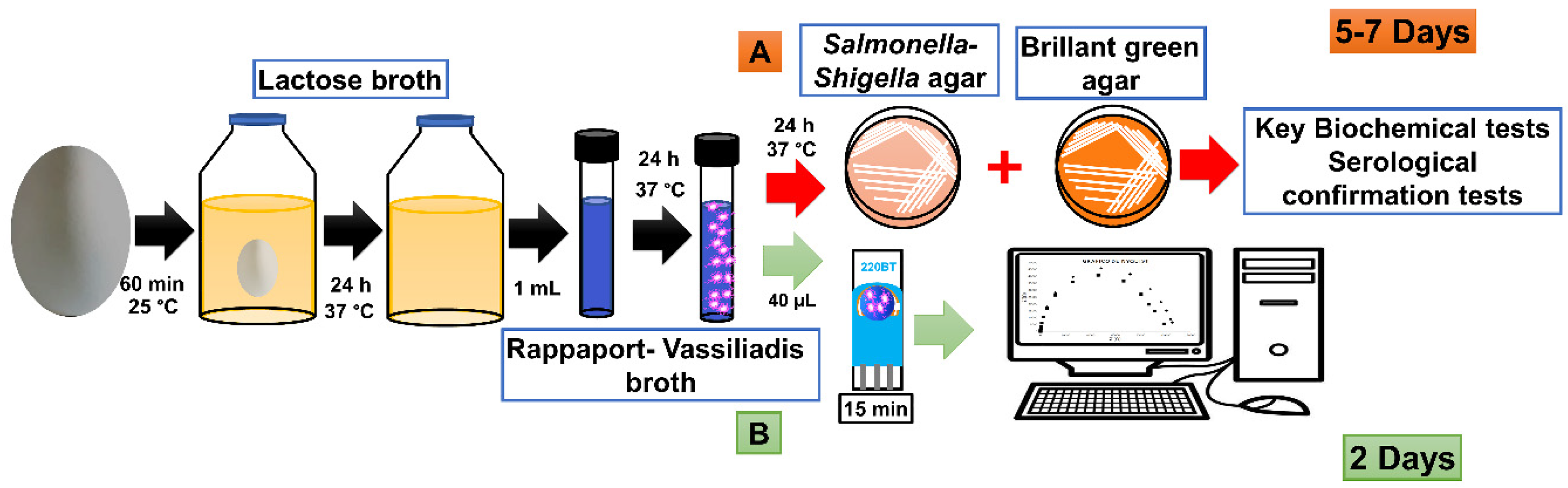
3.3. Sample Analysis
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- World Health Organization. WHO Food Safety. April 2020. Available online: https://www.who.int/news-room/fact-sheets/detail/food-safety (accessed on 8 May 2020).
- Park, S.; Szonyi, B.; Gautam, R.; Nightingale, K.; Anciso, J.; Ivanek, R. Risk factors for microbial contamination in fruits and vegetables at the preharvest level: A systematic review. J. Food Prot. 2012, 75, 2055–2081. [Google Scholar] [CrossRef]
- Pui, C.F.; Wong, W.C.; Chai, L.C.; Nillian, E.; Ghazali, F.M.; Cheah, Y.K.; Radu, S. Simultaneous detection of Salmonella spp., Salmonella Typhi and Salmonella Typhimurium in sliced fruits using multiplex PCR. Food Control 2011, 22, 337–342. [Google Scholar] [CrossRef]
- Allerberger, F.; Liesegang, A.; Grif, K.; Khaschabi, D.; Prager, R.; Danzl, J.; Höck, F.; Ottl, J.; Dierich, M.P.; Berghold, C.; et al. Occurrence of Salmonella enterica serovar Dublin in Austria. Wien. Med. Wochenschr. 2002, 7, 65–70. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. WHO Salmonella (Non-Typhoidal). February 2018. Available online: https://www.who.int/news-room/fact-sheets/detail/salmonella-(non-typhoidal) (accessed on 8 May 2020).
- Musaj, A.; Llugaxhiu, D.; Hyseni, B. Detection of Salmonella in Eggs. J. Int. Environ. App. Sci. 2018, 13, 1–7. [Google Scholar]
- Odumeru, J.A.; León-Velarde, C.G. Salmonella detection methods for food and food ingredients. In Salmonella—A Dangerous Foodborne Pathogen; Mahmoud, B., Ed.; InTechOpen: Rijeka, Croatia, 2012; pp. 373–392. [Google Scholar] [CrossRef]
- Zourob, M.; Elwary, S.; Turner, A.P. Principles of Bacterial Detection: Biosensors, Recognition Receptors and Microsystems; Springer: New York, NY, USA, 2008. [Google Scholar]
- Grimont, P.A.; Weill, F.X. Antigenic Formulae of the Salmonella Servovars, 9th ed.; WHO Collaborating Centre for Reference and Research on Salmonella: Paris, France, 2007; pp. 1–166. [Google Scholar]
- Shipp, C.R.; Rowe, B. A mechanised microtechnique for Salmonella serotyping. J. Clin. Pathol. 1980, 33, 595–597. [Google Scholar] [CrossRef] [PubMed]
- Waters, S.M.; Murphy, R.A.; Power, R.F. Characterisation of prototype Nurmi cultures using culture-based microbiological techniques and PCR-DGGE. Int. J. Food Microbiol. 2006, 110, 268–277. [Google Scholar] [CrossRef]
- Lynch, M.J.; Leon-Velarde, C.G.; McEwen, S.; Odumeru, J.A. Evaluation of an automated immunomagnetic separation method for the rapid detection of Salmonella species in poultry environmental samples. J. Microbiol. Meth. 2004, 58, 285–288. [Google Scholar] [CrossRef]
- Španová, A.; Rittich, B.; Karpíšková, R.; Čechová, L.; Škapova, D. PCR identification of Salmonella cells in food and stool samples after immunomagnetic separation. Bioseparation 2000, 9, 379–384. [Google Scholar] [CrossRef]
- Rijpens, N.; Herman, L.; Vereecken, F.; Jannes, G.; De Smedt, J.; De Zutter, L. Rapid detection of stressed Salmonella spp. in dairy and egg products using immunomagnetic separation and PCR. Int. J. Food Microbiol. 1999, 46, 37–44. [Google Scholar] [CrossRef]
- Kumar, R.; Surendran, P.K.; Thampuran, N. Evaluation of culture, ELISA and PCR assays for the detection of Salmonella in seafood. Lett. Appl. Microbiol. 2008, 46, 221–226. [Google Scholar] [CrossRef]
- Lee, K.M.; Runyon, M.; Herrman, T.J.; Phillips, R.; Hsieh, J. Review of Salmonella detection and identification methods: Aspects of rapid emergency response and food safety. Food Control 2015, 47, 264–276. [Google Scholar] [CrossRef]
- Mirhosseini, S.A.; Fooladi, A.A.I.; Amani, J.; Sedighian, H. Production of recombinant flagellin to develop ELISA-based detection of Salmonella enteritidis. Braz. J. Microbiol. 2017, 48, 774–781. [Google Scholar] [CrossRef] [PubMed]
- Nicholas, R.A.; Cullen, G.A. Development and application of an ELISA for detecting antibodies to Salmonella enteritidis in chicken flocks. Veter. Rec. 1991, 128, 74. [Google Scholar] [CrossRef] [PubMed]
- Malorny, B.; Paccassoni, E.; Fach, P.; Bunge, C.; Martin, A.; Helmuth, R. Diagnostic real-time PCR for detection of Salmonella in food. App. Environ. Microbiol. 2004, 70, 7046–7052. [Google Scholar] [CrossRef] [PubMed]
- Moosavy, M.H.; Esmaeili, S.; Amiri, F.B.; Mostafavi, E.; Salehi, T.Z. Detection of Salmonella spp. in commercial eggs in Iran. Iran. J. Microbiol. 2015, 7, 50. [Google Scholar] [PubMed]
- Juneja, V.K.; Cherry, J.P.; Tunick, M. Advances in Microbial Food Safety; Symposium Series; American Chemical Society: Washington, DC, USA, 2006; pp. 14–27. [Google Scholar]
- Saleem, M. Biosensors a promising future in measurements. In Proceedings of the 1st International Conference on Sensing for Industry, Control, Communications, & Security Technologies (ICSICCST 2013), Karachi City, Pakistan, 24–26 June 2013; Volume 51. [Google Scholar] [CrossRef]
- Lei, Y.; Chen, W.; Mulchandani, A. Microbial biosensors. Anal. Chim. Acta 2006, 568, 200–210. [Google Scholar] [CrossRef] [PubMed]
- Thévenot, D.R.; Toth, K.; Durst, R.A.; Wilson, G.S. Electrochemical biosensors: Recommended definitions and classification. Biosens. Bioelectron. 2001, 16, 121–131. [Google Scholar] [CrossRef]
- Silva, N.F.; Magalhães, J.M.; Freire, C.; Delerue-Matos, C. Electrochemical biosensors for Salmonella: State of the art and challenges in food safety assessment. Biosens. Bioelectron. 2018, 99, 667–682. [Google Scholar] [CrossRef]
- Zeng, X.; Andrade, C.A.; Oliveira, M.D.; Sun, X.L. Carbohydrate–protein interactions and their biosensing applications. Anal. Bioanal. Chem. 2012, 402, 3161–3176. [Google Scholar] [CrossRef]
- Preston, A.; Mandrell, R.E.; Gibson, B.W.; Apicella, M.A. The lipooligosaccharides of pathogenic gram-negative bacteria. Crit. Rev. Microbiol. 1996, 22, 139–180. [Google Scholar] [CrossRef]
- Helander, I.M.; Mamat, U.; Rietschel, E.T. Lipopolysaccharides. eLS 2001. [Google Scholar] [CrossRef]
- Lam, J.S.; Graham, L.L.; Lightfoot, J.; Dasgupta, T.; Beveridge, T.J. Ultrastructural examination of the lipopolysaccharides of Pseudomonas aeruginosa strains and their isogenic rough mutants by freeze-substitution. J. Bacteriol. 1992, 174, 7159–7167. [Google Scholar] [CrossRef] [PubMed]
- Devyatyarova-Johnson, M.; Rees, I.H.; Robertson, B.D.; Turner, M.W.; Klein, N.J.; Jack, D.L. The Lipopolysaccharide Structures of Salmonella enterica Serovar Typhimurium and Neisseria gonorrhoeae Determine the Attachment of Human Mannose-Binding Lectin to Intact Organisms. Infect. Immun. 2000, 68, 3894–3899. [Google Scholar] [CrossRef] [PubMed]
- Van Damme, E.J.M.; Peumans, W.J.; Pusztai, A.; Bardocz, S. Handbook of Plant lectins: Properties and Biomedical Applications; John Wiley & Sons: Chichester, UK, 1998. [Google Scholar]
- Yagi, F.; Hidaka, M.; Minami, Y.; Tadera, K. A lectin from leaves of Neoregelia flandria recognizes D-glucose, D-mannose and N-acetyl D-glucosamine, differing from the mannose-specific lectins of other monocotyledonous plants. Plant Cell Physiol. 1996, 37, 1007–1012. [Google Scholar] [CrossRef][Green Version]
- Ramírez-Morillo, I.M.; Hornung-Leoni, C.T.; González Ledesma, M.; Romero-Soler, K.J. “The Old White Lady of Kew Gardens”, Hechtia argentea (Bromeliaceae: Hechtioideae), found her homeland in Mexico. Taxon 2020. [Google Scholar] [CrossRef]
- Santana, G.M.S.; Albuquerque, L.P.; Simões, D.A.; Coelho, L.C.B.B.; Paiva, P.M.G.; Gusmão, N.B. Isolation of lectin from Opuntia ficus-indica cladodes. Acta Hortic. 2009, 811, 281–286. [Google Scholar] [CrossRef]
- Kruger, N.J. The Bradford method for protein quantitation. In The Protein Protocols Handbook, 3rd ed.; Walker, J.M., Ed.; Humana Press: Totowa, NJ, USA, 2009; pp. 17–24. [Google Scholar]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef]
- Moreira, R.D.A.; Perrone, J.C. Purification and partial characterization of a lectin from Phaseolus vulgaris. Plant Physiol. 1977, 59, 783–787. [Google Scholar] [CrossRef]
- Silva, M.L.S.; Gutiérrez, E.; Rodríguez, J.A.; Gomes, C.; David, L. Construction and validation of a Sambucus nigra biosensor for cancer-associated STn antigen. Biosens. Bioelectron. 2014, 57, 254–261. [Google Scholar] [CrossRef]
- Secretaria de Salud. NORMA Oficial Mexicana NOM-210-SSA1-2014, Products and Services. Microbiological Test Methods. Determination of Indicator Microorganisms. Determination of Pathogenic Microorganisms for Official Mexican Standard; Diario Oficial de la Federacion: Distrito Federal, Mexico, 2015; pp. 1–96. [Google Scholar]
- ISO. ISO 6579-1:2017. Microbiology of the Food Chain—Horizontal Method for the Detection, Enumeration and Serotyping of Salmonella—Part 1: Detection of Salmonella spp., 1st ed.; International Organization for Standardization: Geneva, Switzerland, 2017. [Google Scholar]
- Kennedy, J.F.; Palva, P.M.G.; Corella, M.T.S.; Cavalcanti, M.S.M.; Coelho, L.C.B.B. Lectins, versatile proteins of recognition: A review. Carbohydr. Polym. 1995, 26, 219–230. [Google Scholar] [CrossRef]
- Sharon, N.; Lis, H. History of lectins: From hemagglutinins to biological recognition molecules. Glycobiology 2004, 14, 53R–62R. [Google Scholar] [CrossRef] [PubMed]
- Van Damme, E.J.; Lannoo, N.; Peumans, W.J. Plant lectins. Adv. Bot. Res. 2008, 48, 107–209. [Google Scholar] [CrossRef]
- Boyd, W.C.; Shapleigh, E. Antigenic relations of blood group antigens as suggested by tests with lectins. J. Immun. 1954, 73, 226–231. [Google Scholar] [PubMed]
- Goldstein, I.J.; Hayes, C.E. The lectins: Carbohydrate-binding proteins of plants and animals. Adv. Carbohydr. Chem. Biochem. 1978, 35, 127–340. [Google Scholar] [CrossRef] [PubMed]
- Nilsson, C. Lectins: Analytical Technologies, 1st ed.; Elsevier: Amsterdam, The Netherlands, 2008. [Google Scholar] [CrossRef]
- Correia, M.T.; Coelho, L. C Purification of a glucose/mannose specific lectin, isoform 1, from seeds of Cratylia mollis Mart. (Camaratu bean). Appl. Biochem. Biotech. 1995, 55, 261–273. [Google Scholar] [CrossRef]
- Oliveira, M.D.; Andrade, C.A.; Correia, M.T.; Coelho, L.C.; Singh, P.R.; Zeng, X. Impedimetric biosensor based on self-assembled hybrid cystein-gold nanoparticles and CramoLL lectin for bacterial lipopolysaccharide recognition. J. Colloid Interface Sci. 2011, 362, 194–201. [Google Scholar] [CrossRef] [PubMed]
- Payne, M.J.; Campbell, S.; Kroll, R.G. Lectin-magnetic separation can enhance methods for the detection of Staphylococcus aureus, Salmonella enteritidis and Listeria monocytogenes. Food Microbiol. 1993, 10, 75–83. [Google Scholar] [CrossRef]
- Karunakaran, C.; Pandiaraj, M.; Santharaman, P. Immunosensors. In Biosensors and Bioelectronics, 1st ed.; Karunakaran, C., Bhargava, K., Benjamin, R., Eds.; Elsevier: Amsterdam, The Netherlands, 2015; pp. 205–245. [Google Scholar] [CrossRef]
- Yang, L.; Bashir, R. Electrical/electrochemical impedance for rapid detection of foodborne pathogenic bacteria. Biotech. Adv. 2008, 26, 135–150. [Google Scholar] [CrossRef]
- Putzbach, W.; Ronkainen, N.J. Immobilization techniques in the fabrication of nanomaterial-based electrochemical biosensors: A review. Sensors 2013, 13, 4811–4840. [Google Scholar] [CrossRef]
- Wang, C.; Yan, Q.; Liu, H.B.; Zhou, X.H.; Xiao, S.J. Different EDC/NHS activation mechanisms between PAA and PMAA brushes and the following amidation reactions. Langmuir 2011, 27, 12058–12068. [Google Scholar] [CrossRef]
- Serra, B.; Gamella, M.; Reviejo, A.J.; Pingarron, J.M. Lectin-modified piezoelectric biosensors for bacteria recognition and quantification. Anal. Bioanal. Chem. 2008, 391, 1853–1860. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Ye, Z.; Si, C.; Ying, Y. Monitoring of Escherichia coli O157: H7 in food samples using lectin based surface plasmon resonance biosensor. Food Chem. 2013, 136, 1303–1308. [Google Scholar] [CrossRef] [PubMed]
- Doyle, R.J.; Dziarski, R. The bacterial cell: Peptidoglycan. In Molecular Medical Microbiology; Sussman, M., Ed.; Academic Press: Cambridge, MA, USA, 2002; pp. 137–154. [Google Scholar] [CrossRef]
- Su, L.; Jia, W.; Hou, C.; Lei, Y. Microbial biosensors: A review. Biosens. Bioelectron. 2011, 26, 1788–1799. [Google Scholar] [CrossRef] [PubMed]
- Reina, J.J.; Díaz, I.; Nieto, P.M.; Campillo, N.E.; Paez, J.A.; Tabarani, G.; Fieschi, F.; Rojo, J. Docking, synthesis, and NMR studies of mannosyl trisaccharide ligands for DC-SIGN lectin. Org. Biomol. Chem. 2008, 6, 2743–2754. [Google Scholar] [CrossRef] [PubMed]
- Cesewski, E.; Johnson, B.N. Electrochemical biosensors for pathogen detection. Biosens. Bioelectron. 2020, 159, 112214. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Zhou, H.; Hao, H.; Gong, Q.; Nie, K. Detection of Escherichia coli with a label-free impedimetric biosensor based on lectin functionalized mixed self-assembled monolayer. Sens. Actuators B Chem. 2016, 229, 297–304. [Google Scholar] [CrossRef]
- Ma, F.; Rehman, A.; Liu, H.; Zhang, J.; Zhu, S.; Zeng, X. Glycosylation of quinone-fused polythiophene for reagentless and label-free detection of E. coli. Anal. Chem. 2015, 87, 1560–1568. [Google Scholar] [CrossRef]
- Xi, F.; Gao, J.; Wang, J.; Wang, Z. Discrimination and detection of bacteria with a label-free impedimetric biosensor based on self-assembled lectin monolayer. J. Electroanal. Chem. 2011, 656, 252–257. [Google Scholar] [CrossRef]
- Jantra, J.; Kanatharana, P.; Asawatreratanakul, P.; Hedstrom, M.; Mattiasson, B.; Thavarungkul, P. Real-time label-free affinity biosensors for enumeration of total bacteria based on immobilized concanavalin A. J. Environ. Sci. Health 2011, 46, 1450–1460. [Google Scholar] [CrossRef]
- Wan, Y.; Zhang, D.; Hou, B. Monitoring microbial populations of sulfate-reducing bacteria using an impedimetric immunosensor based on agglutination assay. Talanta 2009, 80, 218–223. [Google Scholar] [CrossRef]
- Yang, G.J.; Huang, J.L.; Meng, W.J.; Shen, M.; Jiao, X.A. A reusable capacitive immunosensor for detection of Salmonella spp. based on grafted ethylene diamine and self-assembled gold nanoparticle monolayers. Anal. Chim. Acta 2009, 647, 159–166. [Google Scholar] [CrossRef] [PubMed]


| Lectin Source | Recognition Carbohydrate | Bacteria | Detection Technique | LOD | Ref |
|---|---|---|---|---|---|
| Arachis hypogaea | N-acetyl-D-galactoseamine and D-galactose | L. monocytogenes | Surface plasmon resonance | - | [55] |
| Canavalia ensiformis | D-glucose and D-mannose | E. coli DH5α | EIS | 75 cell/mL | [60] |
| E. coli W1485 | Square wave voltammetry | 25 cell/mL | [61] | ||
| E. coli | Surface plasmon resonance | 12 CFU/mL | [62] | ||
| Desulforibrio caledoiensis | EIS | 1.8 CFU/mL | [63] | ||
| Cratylia mollis | D-glucose | Serratia. marcescens, E. coli, S. enterica and K. pneumoniae | EIS | - | [48] |
| Ricinus communis | N-acetyl-D-galactosamine and D-galactose | E. coli DH5α Enterobacter cloacae, and Bacillus subtilis | EIS | - | [64] |
| Triticum vulgaris | N-acetylneuraminic acid | E. coli O157: H7 | Surface plasmon resonance | 3 × 103 CFU/mL | [55] |
| Hechtia argentea | N-acetyl-D-glucosamine, N-acetylneuraminic acid and D-mannose | Salmonella spp. | EIS | 5 CFU/mL | This work |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lopez-Tellez, J.; Sanchez-Ortega, I.; Hornung-Leoni, C.T.; Santos, E.M.; Miranda, J.M.; Rodriguez, J.A. Impedimetric Biosensor Based on a Hechtia argentea Lectin for the Detection of Salmonella spp. Chemosensors 2020, 8, 115. https://doi.org/10.3390/chemosensors8040115
Lopez-Tellez J, Sanchez-Ortega I, Hornung-Leoni CT, Santos EM, Miranda JM, Rodriguez JA. Impedimetric Biosensor Based on a Hechtia argentea Lectin for the Detection of Salmonella spp. Chemosensors. 2020; 8(4):115. https://doi.org/10.3390/chemosensors8040115
Chicago/Turabian StyleLopez-Tellez, Jorge, Irais Sanchez-Ortega, Claudia Teresa Hornung-Leoni, Eva Maria Santos, Jose Manuel Miranda, and Jose Antonio Rodriguez. 2020. "Impedimetric Biosensor Based on a Hechtia argentea Lectin for the Detection of Salmonella spp." Chemosensors 8, no. 4: 115. https://doi.org/10.3390/chemosensors8040115
APA StyleLopez-Tellez, J., Sanchez-Ortega, I., Hornung-Leoni, C. T., Santos, E. M., Miranda, J. M., & Rodriguez, J. A. (2020). Impedimetric Biosensor Based on a Hechtia argentea Lectin for the Detection of Salmonella spp. Chemosensors, 8(4), 115. https://doi.org/10.3390/chemosensors8040115