Fusion Recalibration Method for Addressing Multiplicative and Additive Effects and Peak Shifts in Analytical Chemistry
Abstract
:1. Introduction
- We have incorporated a new mode in EMSC that specifically eliminates their influence to address the distortion issue caused by the COW and DTW algorithms.
- We propose a fusion recalibration algorithm that combines the EMSC and COW algorithms to address the issues caused by both the multiplicative and additive effects and peak shift phenomenon.
2. Theories and Methods
2.1. Correlation-Optimized Warping
2.2. Extended Multiplicative Signal Correction
2.3. COW Algorithm Challenges with Multiplicative and Additive Effects in Spectral Data
2.4. EMSC Algorithm Challenges with Peak Shifts in Spectral Data
2.5. SNV Algorithm Challenges with Multiplicative and Additive Effects in Spectral Data
2.6. First Derivative Algorithm Challenges with Multiplicative and Additive Effects in Spectral Data
3. Materials and Methods
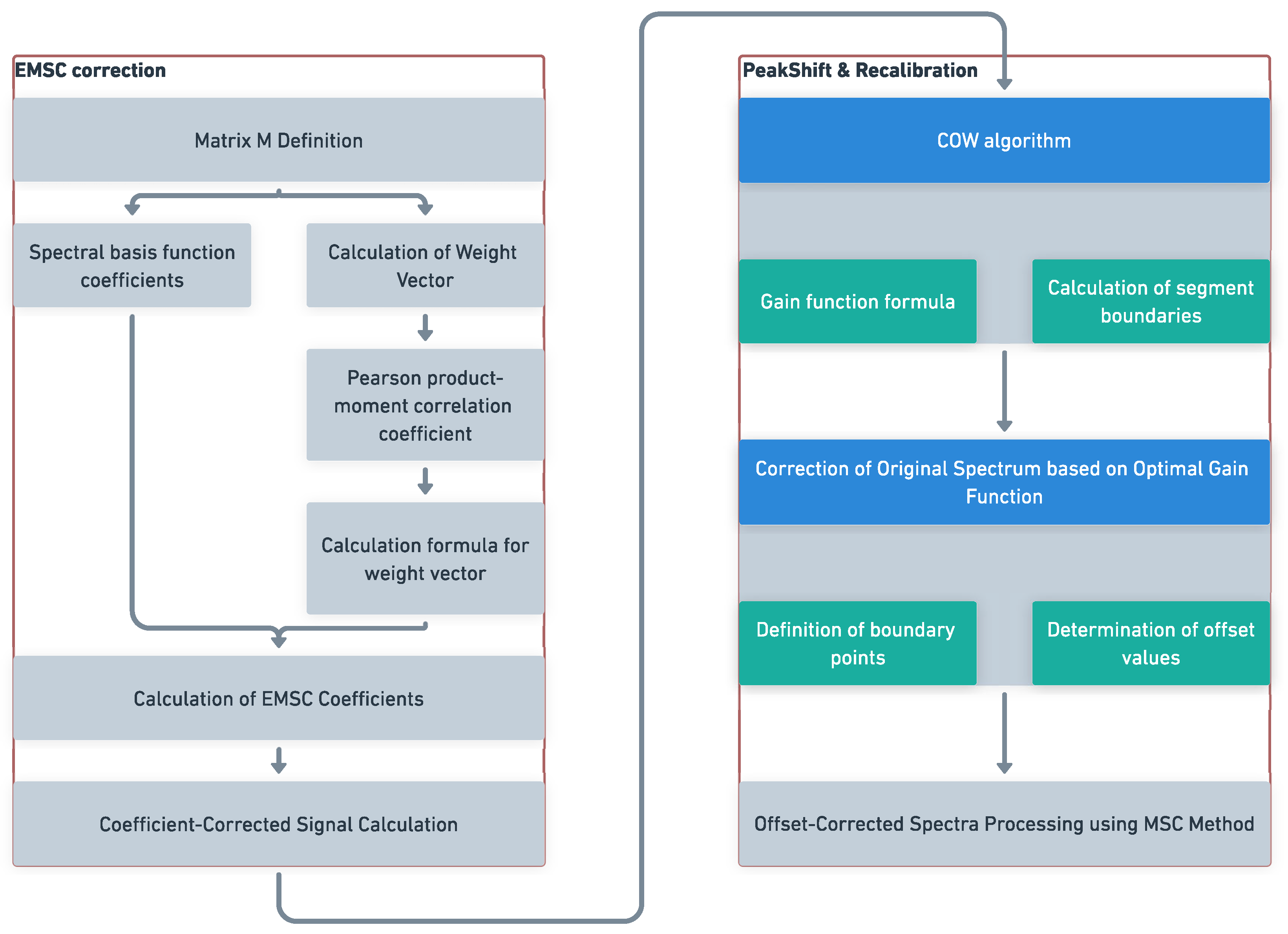
3.1. Spectral Offset Recalibration Method
- The matrix M is defined as:where are the coefficients corresponding to the spectral basis functions. This matrix is used in the calculation of the EMSC coefficients and the subsequent coefficient correction in the EMSC method.
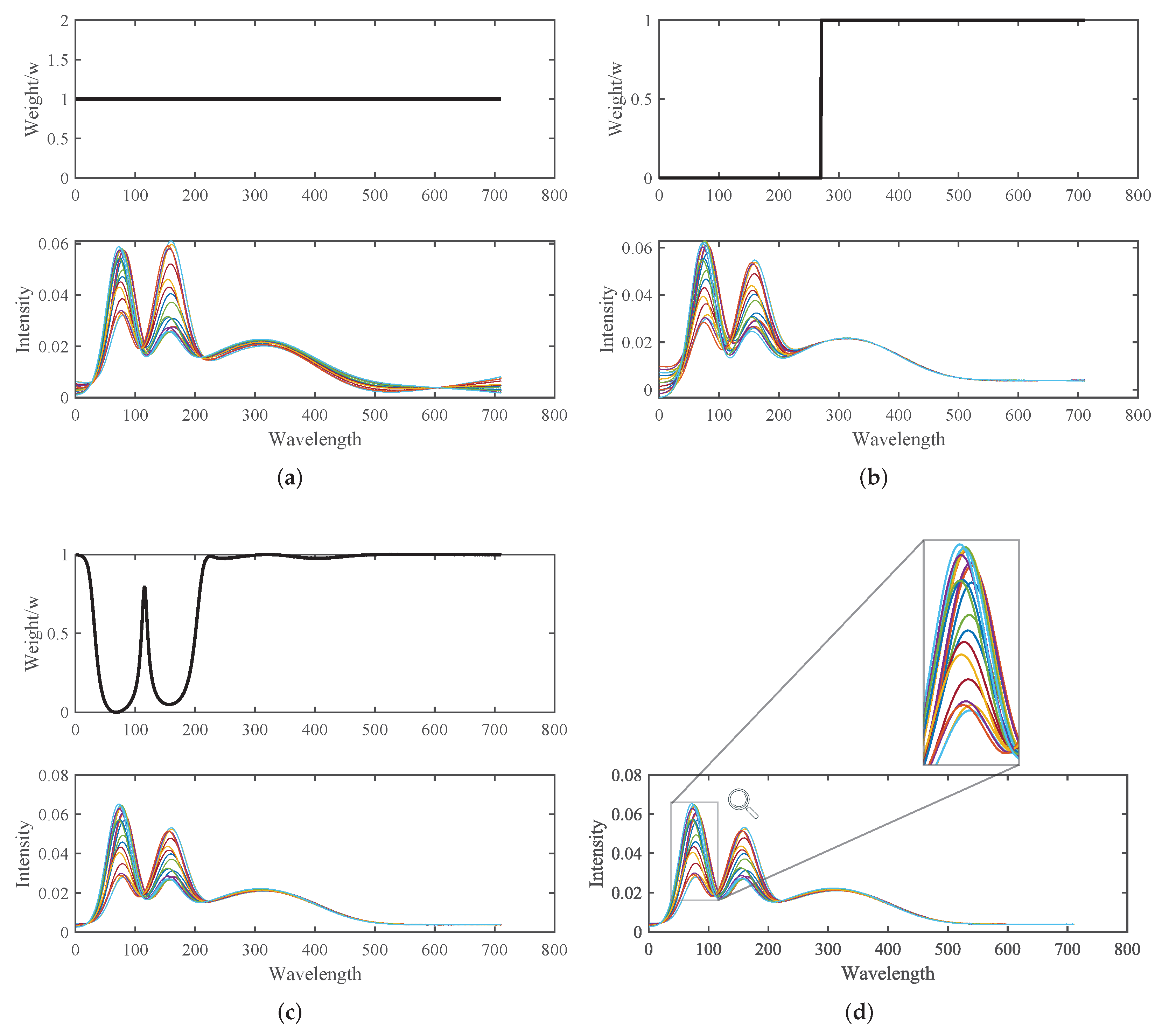
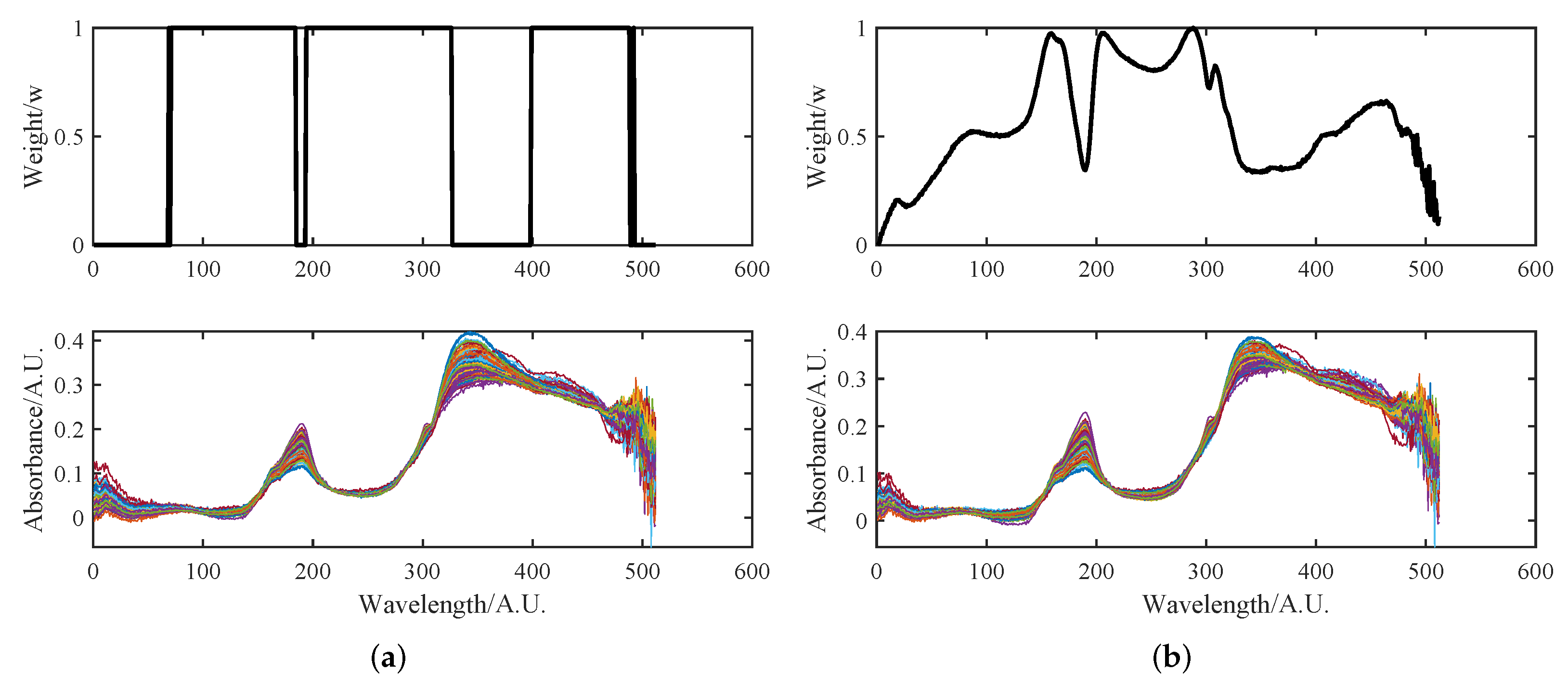
- The Pearson product-moment correlation coefficient is used to calculate the degree of correlation between each spectrum in the original spectrum and the rest of the spectral dataset. These correlation coefficients are then summed and normalized to obtain the weight vector, denoted as W, for the EMSC method. The calculation formula for the weight vector is as follows:
- To calculate the EMSC coefficients, denoted aswhere X is a matrix representing the original spectra, with dimensions , where L is the number of spectral data points; denotes matrix transpose.
- The coefficient-corrected signal can be obtained using the formula:where:
- x represents the original spectrum.
- a, b, c, and d are the EMSC coefficients obtained from the previous step.
- The matrix represents the matrix of spectral basis functions, with p indicating the number of basis functions used.
- Let us define the alignment quality, also known as the gain function, as the correlation coefficient between the corrected spectrum z and the spectrum average. The gain function is denoted as and is measured as , where I represents the segment number. The gain function is defined by the formula:Here, the i-th segment of the spectrum is defined by the boundary points and . Given and , where is a constant and t is the tolerance, we have .
- To correct the original spectrum x based on the optimal gain function, we can follow these steps:
- Define the boundary points based on the desired segmentation of the spectrum: , , , …, , .
- Determine the values of within the range for each segment, where represents the offset or shift to be applied to the corresponding segment. Here, is a constant and t is the tolerance.
- Calculate the corrected spectrum by applying the following equation for each segment i: , where denotes the section defined by border points and .
- Repeat the above step for all segments of the spectrum.
By applying this correction process based on the optimal gain function, the original spectrum x can be effectively aligned and corrected. - To further process the offset-corrected near-infrared spectra using the EMSC method, the influence of multiplicative scatter effects are mitigated and further processed spectra for analysis or comparison are obtained.
3.2. Processing Chart Summary
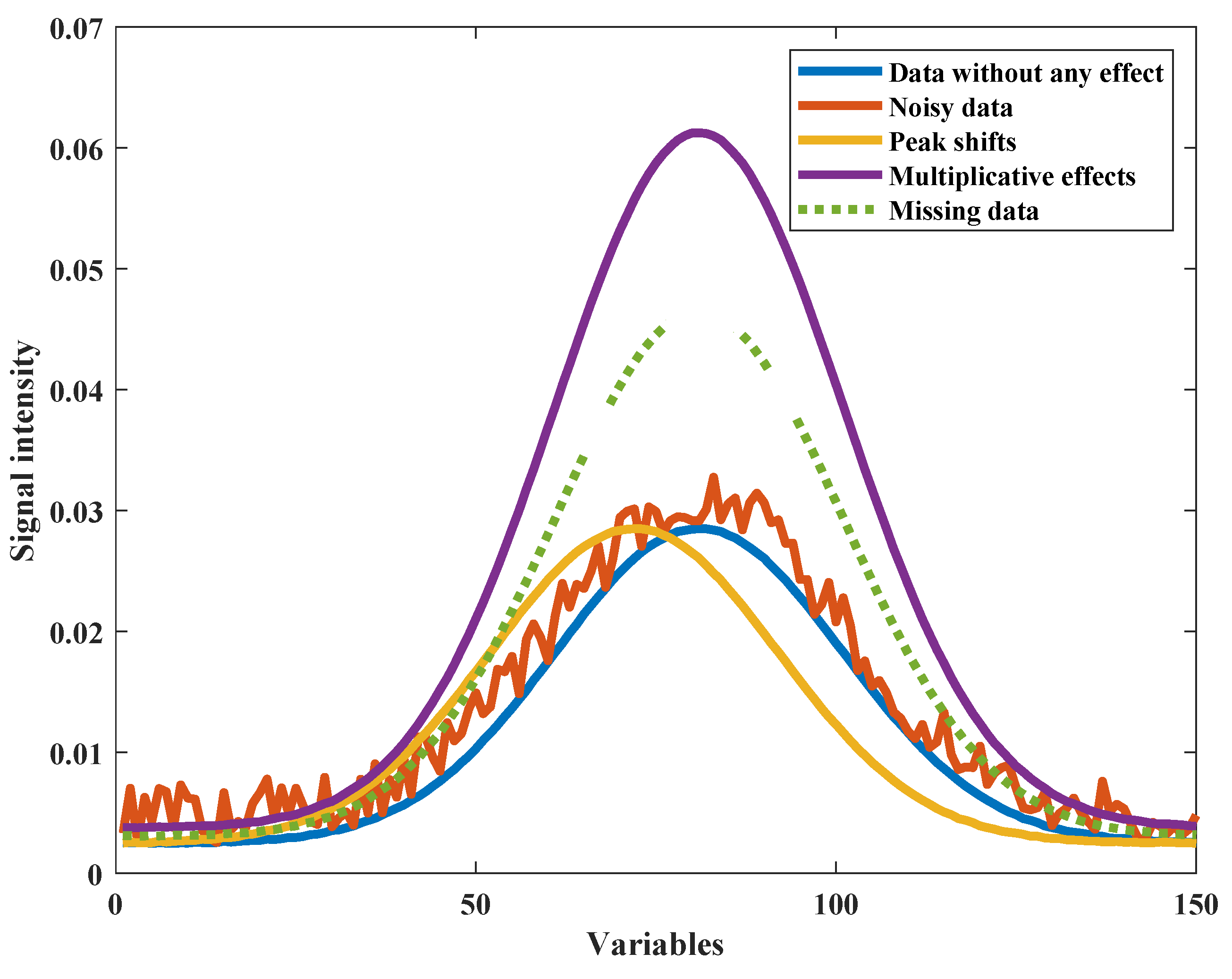
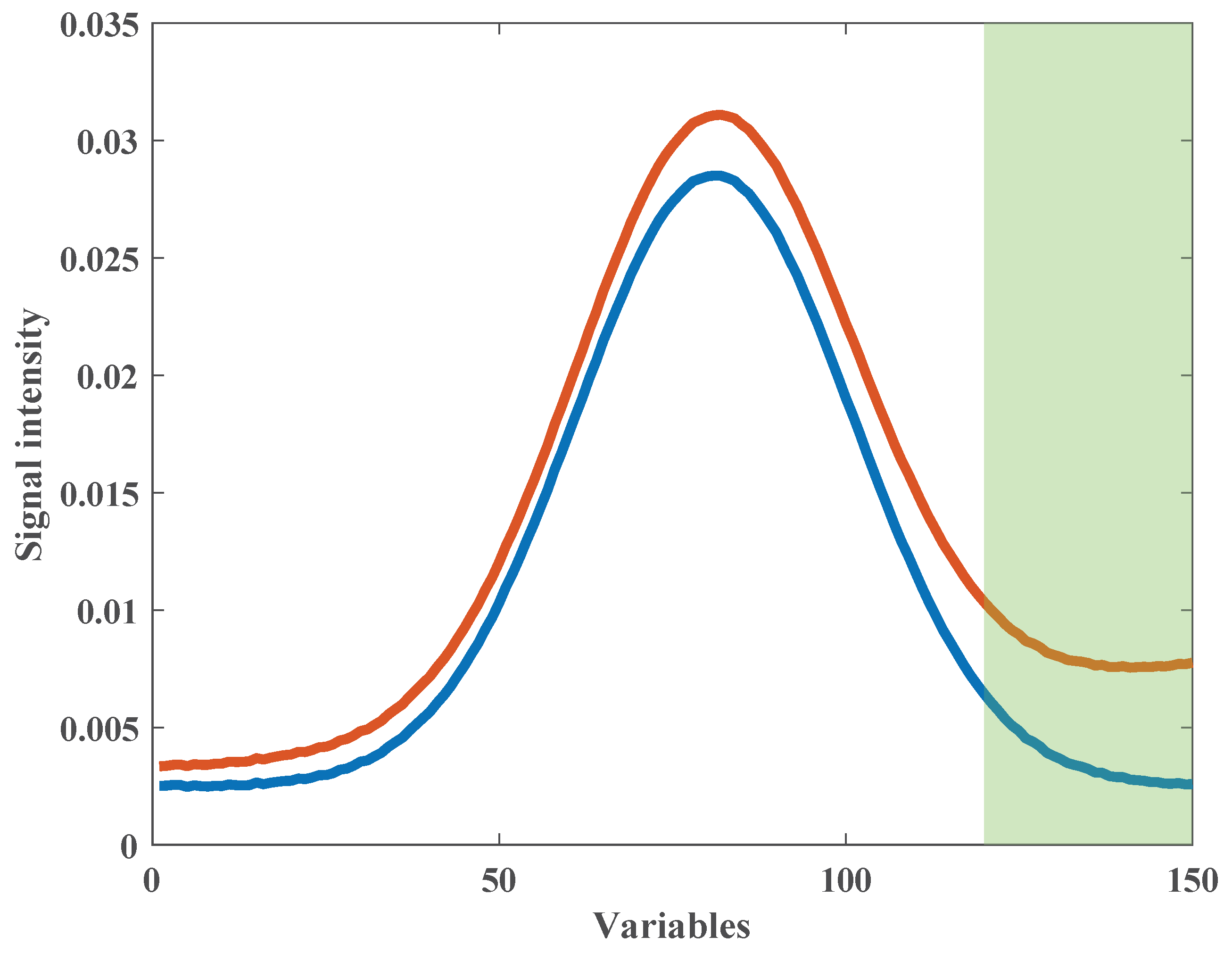
3.3. Simulated Spectra
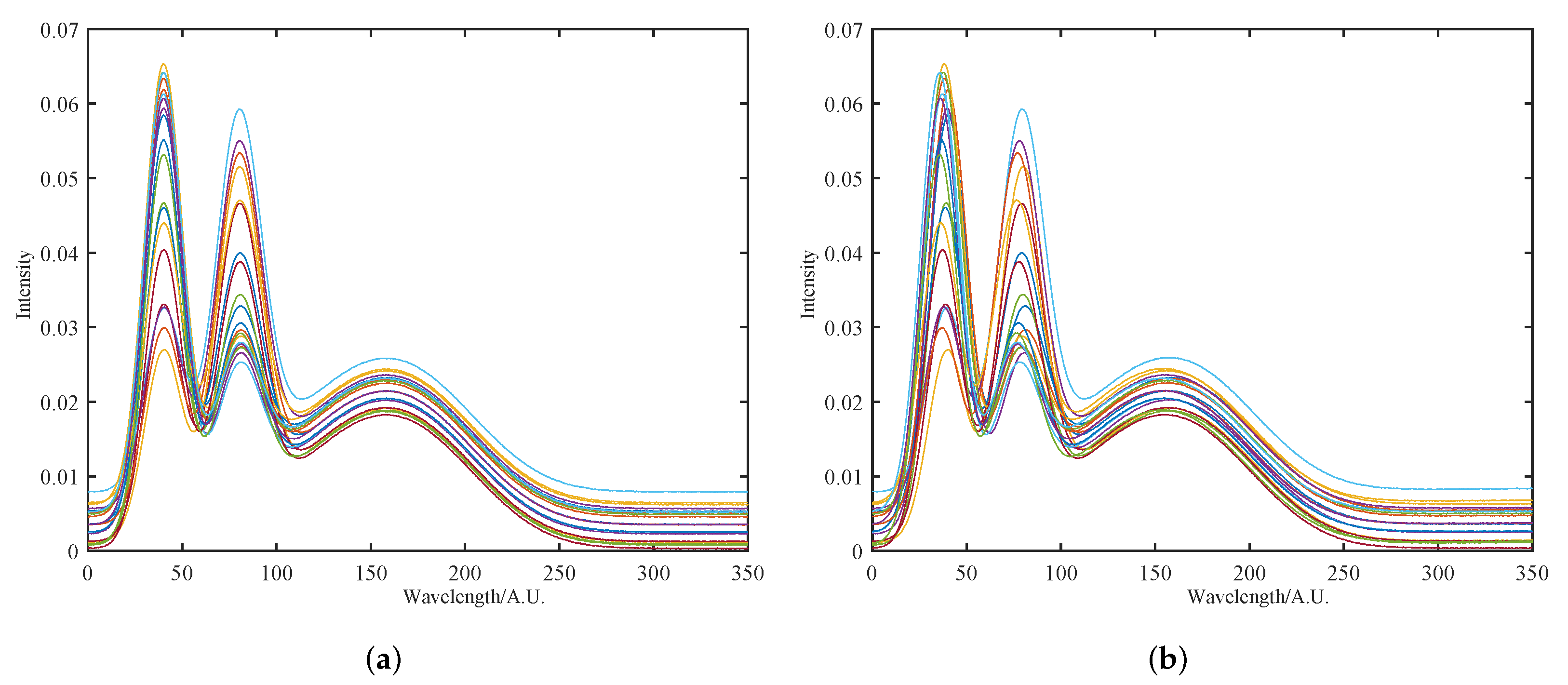
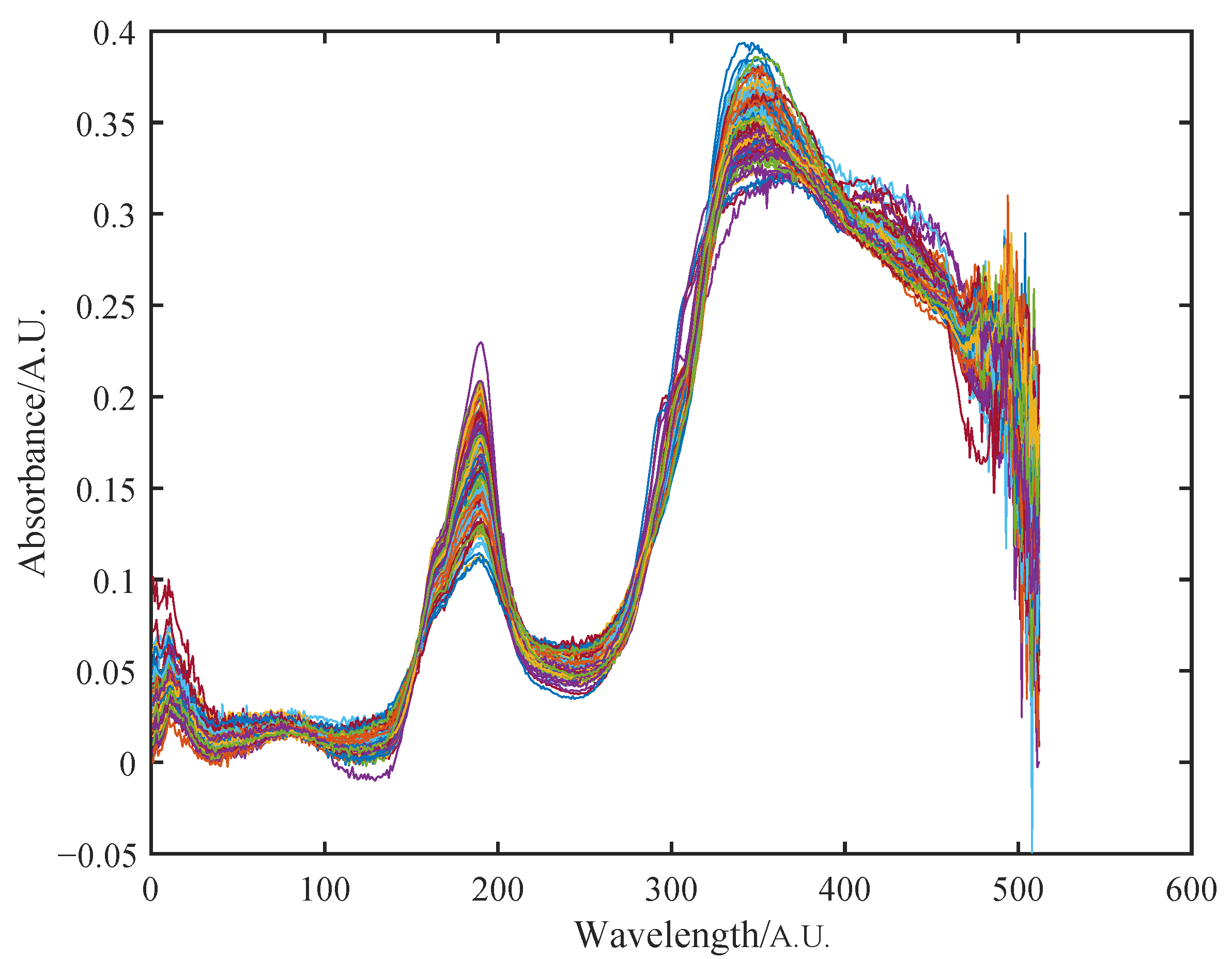
3.4. NIR Spectra of Wood
4. Result and Discussion
4.1. Simulated Spectra
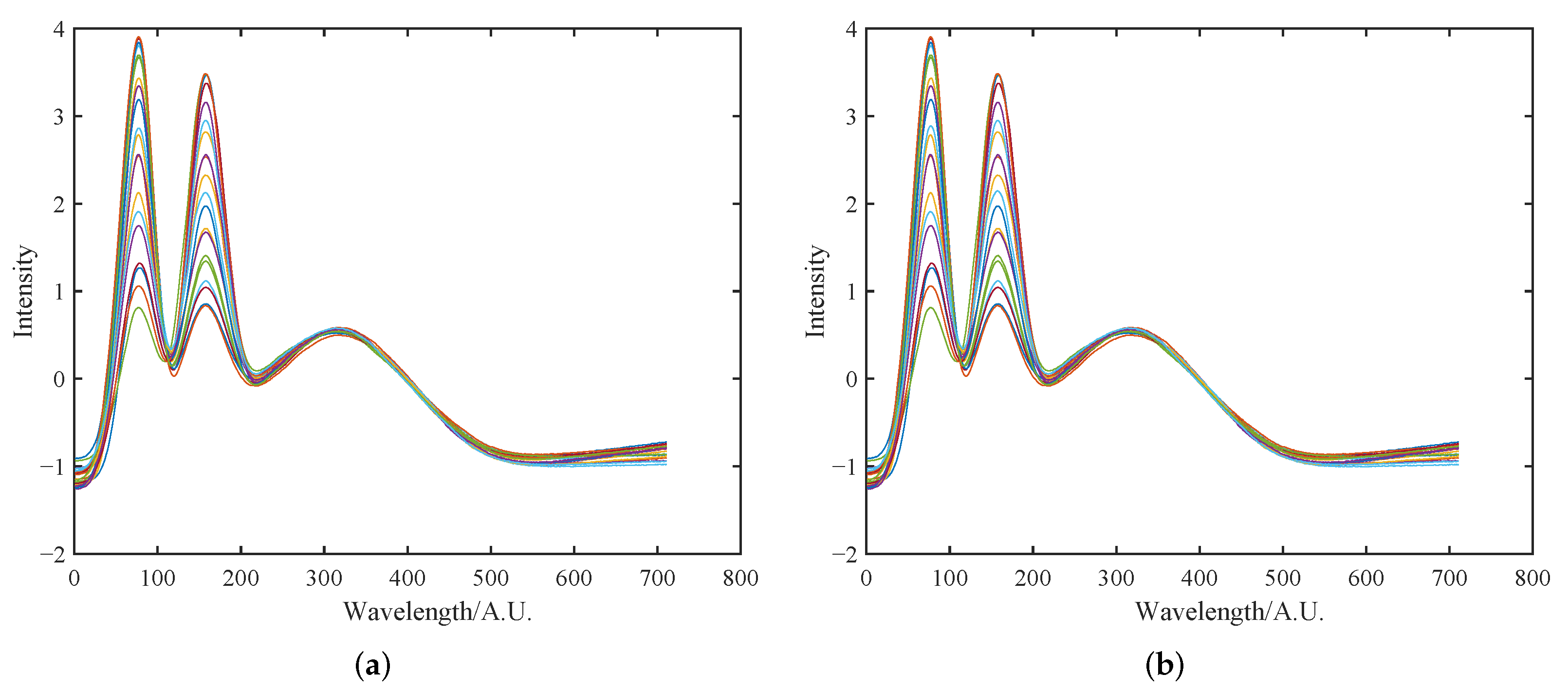
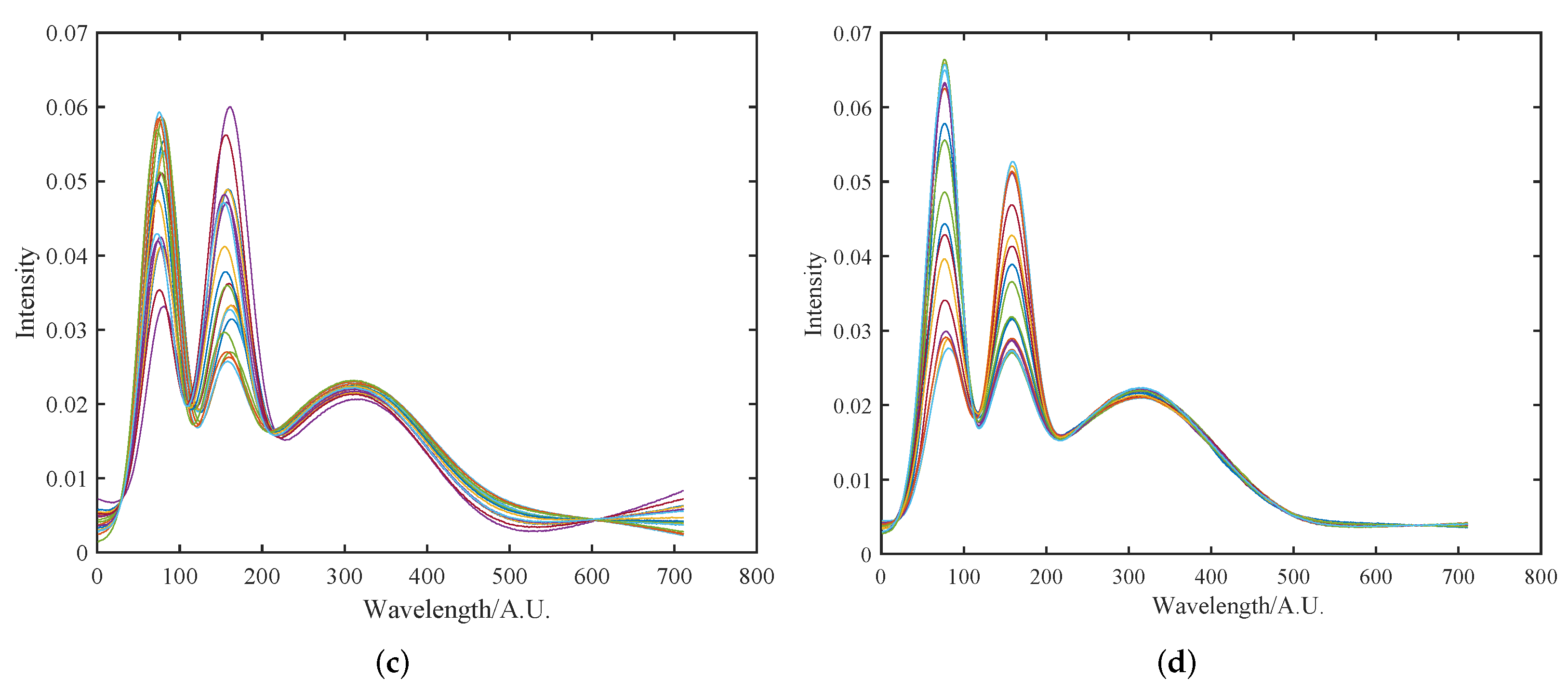
4.1.1. Initial Multiplicative and Additive Effect Correction Results Using SOR Method
4.1.2. Peak Shift Correction and Recalibration Results Using SOR Method
4.1.3. Comparative Analysis of Preprocessing Method Results and Performance
4.2. NIR Spectra of Wood
4.2.1. Initial Multiplicative and Additive Effect Correction Results Using SOR Method
4.2.2. Peak Shift Correction and Recalibration Results Using SOR Method
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| EMSC | Extended Multiplicative Signal Correction |
| COW | Correlation-Optimized Warping |
| NIR | near-infrared |
| LC-MS | liquid chromatography-mass spectrometry |
| GC-MS | gas chromatography-mass spectrometry |
| NIRS | near-infrared spectroscopy |
| MSC | multiplicative signal correction |
| SNV | standard normal variate |
| DTW | Dynamic Time Warping |
| SOR | Spectral Offset Recalibration |
References
- Ríos-Reina, R.; Azcarate, S.M. How chemometrics revives the UV-Vis spectroscopy applications as an analytical sensor for spectralprint (nontargeted) analysis. Chemosensors 2023, 11, 8. [Google Scholar] [CrossRef]
- Sorochan Armstrong, M.D.; de la Mata, A.P.; Harynuk, J.J. Review of variable selection methods for discriminant-type problems in chemometrics. Front. Anal. Sci. 2022, 2, 867938. [Google Scholar] [CrossRef]
- Rajendran, H.K.; Fakrudeen, M.A.D.; Chandrasekar, R.; Silvestri, S.; Sillanpaa, M.; Padmanaban, V.C. A comprehensive review on analytical and equation derived multivariate chemometrics for the accurate interpretation of the degradation of aqueous contaminants. Environ. Technol. Innov. 2022, 28, 102827. [Google Scholar] [CrossRef]
- Dayananda, B.; Owen, S.; Kolobaric, A.; Chapman, J.; Cozzolino, D. Pre-processing applied to instrumental data in analytical chemistry: A brief review of the methods and examples. Crit. Rev. Anal. Chem. 2023. [Google Scholar] [CrossRef] [PubMed]
- Trinklein, T.J.; Cain, C.N.; Ochoa, G.S.; Schöneich, S.; Mikaliunaite, L.; Synovec, R.E. Recent advances in GC×GC and chemometrics to address emerging challenges in nontargeted analysis. Anal. Chem. 2023, 95, 264–286. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Peng, S.; Xie, Q.; Han, Q.; Zhang, G.; Sun, H. An improved weighted multiplicative scatter correction algorithm with the use of variable selection: Application to near-infrared spectra. Chemom. Intell. Lab. Syst. 2019, 185, 114–121. [Google Scholar] [CrossRef]
- Mishra, P.; Biancolillo, A.; Roger, J.M.; Marini, F.; Rutledge, D.N. New data preprocessing trends based on ensemble of multiple preprocessing techniques. TrAC Trends Anal. Chem. 2020, 132, 116045. [Google Scholar] [CrossRef]
- Bloemberg, T.G.; Gerretzen, J.; Lunshof, A.; Wehrens, R.; Buydens, L.M. Warping methods for spectroscopic and chromatographic signal alignment: A tutorial. Anal. Chim. Acta 2013, 781, 14–32. [Google Scholar] [CrossRef]
- Zhang, W.; Kasun, L.C.; Wang, Q.J.; Zheng, Y.; Lin, Z. A review of machine learning for near-infrared spectroscopy. Sensors 2022, 22, 9764. [Google Scholar] [CrossRef]
- Li, S.; Viscarra Rossel, R.A.; Webster, R. The cost-effectiveness of reflectance spectroscopy for estimating soil organic carbon. Eur. J. Soil Sci. 2022, 73, e13202. [Google Scholar] [CrossRef]
- Beć, K.B.; Grabska, J.; Huck, C.W. Miniaturized NIR spectroscopy in food analysis and quality control: Promises, challenges, and perspectives. Foods 2022, 11, 1465. [Google Scholar] [CrossRef]
- Kim, J.; Hwang, W.S.; Kim, D.; Kim, D.Y. Fast noniterative data analysis method for frequency-domain near-infrared spectroscopy with the microscopic Beer–Lambert law. Opt. Commun. 2022, 520, 128417. [Google Scholar] [CrossRef]
- Mishra, P.; Roger, J.M.; Rutledge, D.N.; Woltering, E. SPORT pre-processing can improve near-infrared quality prediction models for fresh fruits and agro-materials. Postharvest Biol. Technol. 2020, 168, 111271. [Google Scholar] [CrossRef]
- Yu, H.; Guo, L.; Kharbach, M.; Han, W. Multi-way analysis coupled with near-infrared spectroscopy in food industry: Models and applications. Foods 2021, 10, 802. [Google Scholar] [CrossRef]
- Thygesen, L.G.; Lundqvist, S.O. NIR measurement of moisture content in wood under unstable temperature conditions. Part 2. Handling temperature fluctuations. J. Near Infrared Spectrosc. 2000, 8, 191–199. [Google Scholar] [CrossRef]
- Watari, M.; Ozaki, Y. Calibration models for the vinyl acetate concentration in ethylene-vinyl acetate copolymers and its on-Line monitoring by near-infrared spectroscopy and chemometrics: Use of band shifts associated with variations in the vinyl acetate concentration to improve the models. Appl. Spectrosc. 2005, 59, 912–919. [Google Scholar] [CrossRef]
- Kelly, J.J.; Kelly, K.A.; Barlow, C.H. Tissue temperature by near-infrared spectroscopy. In Proceedings of the Optical Tomography, Photon Migration, and Spectroscopy of Tissue and Model Media: Theory, Human Studies, and Instrumentation; Chance, B., Alfano, R.R., Eds.; International Society for Optics and Photonics: Bellingham, WA, USA, 1995; Volume 2389, pp. 818–828. [Google Scholar] [CrossRef]
- Khosravi, M.; Soleimanmeigouni, I.; Ahmadi, A.; Nissen, A. Reducing the positional errors of railway track geometry measurements using alignment methods: A comparative case study. Measurement 2021, 178, 109383. [Google Scholar] [CrossRef]
- Khosravi, M.; Soleimanmeigouni, I.; Ahmadi, A.; Nissen, A.; Xiao, X. Modification of correlation optimized warping method for position alignment of condition measurements of linear assets. Measurement 2022, 201, 111707. [Google Scholar] [CrossRef]
- Skov, T.; van den Berg, F.; Tomasi, G.; Bro, R. Automated alignment of chromatographic data. J. Chemom. 2006, 20, 484–497. [Google Scholar] [CrossRef]
- Wang, S.; Yao, J.; Liu, J.; Petrick, N.; Van Uitert, R.L.; Periaswamy, S.; Summers, R.M. Registration of prone and supine CT colonography scans using correlation optimized warping and canonical correlation analysis. Med. Phys. 2009, 36, 5595–5603. [Google Scholar] [CrossRef]
- Li, L.; Peng, Y.; Li, Y.; Wang, F. A new scattering correction method of different spectroscopic analysis for assessing complex mixtures. Anal. Chim. Acta 2019, 1087, 20–28. [Google Scholar] [CrossRef] [PubMed]
- Martens, H.; Nielsen, J.P.; Engelsen, S.B. Light scattering and light absorbance separated by extended multiplicative signal correction. Application to near-infrared transmission analysis of powder mixtures. Anal. Chem. 2003, 75, 394–404. [Google Scholar] [CrossRef]
- Afseth, N.K.; Kohler, A. Extended multiplicative signal correction in vibrational spectroscopy—A tutorial. Chemom. Intell. Lab. Syst. 2012, 117, 92–99. [Google Scholar] [CrossRef]
- Rabatel, G.; Marini, F.; Walczak, B.; Roger, J.M. VSN: Variable sorting for normalization. J. Chemom. 2020, 34, e3164. [Google Scholar] [CrossRef]
- Kucha, C.T.; Liu, L.; Ngadi, M.; Gariépy, C. Prediction and visualization of fat content in polythene-packed meat using near-infrared hyperspectral imaging and chemometrics. J. Food Compos. Anal. 2022, 111, 104633. [Google Scholar] [CrossRef]
- Haddad, F.; Boudet, S.; Peyrodie, L.; Vandenbroucke, N.; Poupart, J.; Hautecoeur, P.; Chieux, V.; Forzy, G. Oligoclonal band straightening based on optimized hierarchical warping for multiple sclerosis diagnosis. Sensors 2022, 22, 724. [Google Scholar] [CrossRef]
- Solheim, J.H.; Zimmermann, B.; Tafintseva, V.; Dzurendová, S.; Shapaval, V.; Kohler, A. The use of constituent spectra and weighting in extended multiplicative signal correction in infrared spectroscopy. Molecules 2022, 27, 1900. [Google Scholar] [CrossRef]
- Khodabakhshian, R.; Seyedalibeyk Lavasani, H.; Weller, P. A methodological approach to preprocessing FTIR spectra of adulterated sesame oil. Food Chem. 2023, 419, 136055. [Google Scholar] [CrossRef]
- Joshi, P.; Pahariya, P.; Al-Ani, M.F.; Choudhary, R. Monitoring and prediction of sensory shelf-life in strawberry with ultraviolet-visible-near-infrared (UV-VIS-NIR) spectroscopy. Appl. Food Res. 2022, 2, 100123. [Google Scholar] [CrossRef]
- Wang, Y.; Ren, Z.; Li, M.; Lu, C.; Deng, W.W.; Zhang, Z.; Ning, J. From lab to factory: A calibration transfer strategy from HSI to online NIR optimized for quality control of green tea fixation. J. Food Eng. 2023, 339, 111284. [Google Scholar] [CrossRef]
- Yuan, H.; Liu, C.; Wang, H.; Wang, L.; Dai, L. PLS-DA and Vis-NIR spectroscopy based discrimination of abdominal tissues of female rabbits. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2022, 271, 120887. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; He, H.; Wang, L.; Xu, Y.; Song, Z.; Wang, X.; Wang, X. Assessment of total glycerol core aldehyde and monomer content based on NIR and PLS. J. Food Compos. Anal. 2023, 123, 105526. [Google Scholar] [CrossRef]
- Malvandi, A.; Kapoor, R.; Feng, H.; Kamruzzaman, M. Non-destructive measurement and real-time monitoring of apple hardness during ultrasonic contact drying via portable NIR spectroscopy and machine learning. Infrared Phys. Technol. 2022, 122, 104077. [Google Scholar] [CrossRef]
- Scholkmann, F.; Kleiser, S.; Metz, A.J.; Zimmermann, R.; Mata Pavia, J.; Wolf, U.; Wolf, M. A review on continuous wave functional near-infrared spectroscopy and imaging instrumentation and methodology. NeuroImage 2014, 85, 6–27. [Google Scholar] [CrossRef]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jiang, D.; Zhang, Y.; Ge, Y.; Wang, K. Fusion Recalibration Method for Addressing Multiplicative and Additive Effects and Peak Shifts in Analytical Chemistry. Chemosensors 2023, 11, 472. https://doi.org/10.3390/chemosensors11090472
Jiang D, Zhang Y, Ge Y, Wang K. Fusion Recalibration Method for Addressing Multiplicative and Additive Effects and Peak Shifts in Analytical Chemistry. Chemosensors. 2023; 11(9):472. https://doi.org/10.3390/chemosensors11090472
Chicago/Turabian StyleJiang, Dapeng, Yizhuo Zhang, Yilin Ge, and Keqi Wang. 2023. "Fusion Recalibration Method for Addressing Multiplicative and Additive Effects and Peak Shifts in Analytical Chemistry" Chemosensors 11, no. 9: 472. https://doi.org/10.3390/chemosensors11090472
APA StyleJiang, D., Zhang, Y., Ge, Y., & Wang, K. (2023). Fusion Recalibration Method for Addressing Multiplicative and Additive Effects and Peak Shifts in Analytical Chemistry. Chemosensors, 11(9), 472. https://doi.org/10.3390/chemosensors11090472





