Methods and Analysis of Biological Contaminants in the Biomanufacturing Industry
Abstract
1. Introduction
2. Biological Contaminants
2.1. Bacteria
2.2. Viruses
2.3. Fungi
3. Regulatory Requirements for Bioprocess Monitoring
3.1. Regulations in the United States
3.2. Regulations in the European Union (EU)
3.3. Challenges and Trends in Meeting Regulatory Requirements
4. Current Methods
4.1. Process Analytical Technology (PAT)
4.2. Microbial Monitoring
4.3. Polymerase Chain Reaction
4.4. Immunoassay Techniques
4.5. Limulus Amebocyte Lysate
5. Emerging Methods
5.1. Optical
5.2. Electrochemical
6. Challenges of Emerging Methods
7. Conclusions and Future Outlook
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Jarrige, M.; Frank, E.; Herardot, E.; Martineau, S.; Darle, A.; Benabides, M.; Domingues, S.; Chose, O.; Habeler, W.; Lorant, J.; et al. The Future of Regenerative Medicine: Cell Therapy Using Pluripotent Stem Cells and Acellular Therapies Based on Extracellular Vesicles. Cells 2021, 10, 240. [Google Scholar] [CrossRef] [PubMed]
- Govardhanagiri, S.; Bethi, S.; Nagaraju, G.P. Small Molecules and Pancreatic Cancer Trials and Troubles. In Breaking Tolerance to Pancreatic Cancer Unresponsiveness to Chemotherapy; Elsevier: Amsterdam, The Netherlands, 2019; pp. 117–131. [Google Scholar]
- Liu, Y.; Zhang, C.; Chen, J.; Fernandez, J.; Vellala, P.; Kulkarni, T.A.; Aguilar, I.; Ritz, D.; Lan, K.; Patel, P.; et al. A Fully Integrated Online Platform for Real Time Monitoring of Multiple Product Quality Attributes in Bio-Pharmaceutical Processes for Monoclonal Antibody Therapeutics. J. Pharm. Sci. 2022, 111, 358–367. [Google Scholar] [CrossRef] [PubMed]
- Zmievskaya, E.; Valiullina, A.; Ganeeva, I.; Petukhov, A.; Rizvanov, A.; Bulatov, E. Application of CAR-T Cell Therapy beyond Oncology: Autoimmune Diseases and Viral Infections. Biomedicines 2021, 9, 59. [Google Scholar] [CrossRef]
- Morris, C.; Lee, Y.S.; Yoon, S. Adventitious Agent Detection Methods in Bio-Pharmaceutical Applications with a Focus on Viruses, Bacteria, and Mycoplasma. Curr. Opin. Biotechnol. 2021, 71, 105–114. [Google Scholar] [CrossRef] [PubMed]
- Cleveland, M.H.; Anekella, B.; Brewer, M.; Chin, P.-J.; Couch, H.; Delwart, E.; Huggett, J.; Jackson, S.; Martin, J.; Monpoeho, S.; et al. Report of the 2019 NIST-FDA Workshop on Standards for next Generation Sequencing Detection of Viral Adventitious Agents in Biologics and Biomanufacturing. Biologicals 2020, 64, 76–82. [Google Scholar] [CrossRef] [PubMed]
- Gustavsson, R.; Mandenius, C.F.; Löfgren, S.; Scheper, T.; Lindner, P. In Situ Microscopy as Online Tool for Detecting Microbial Contaminations in Cell Culture. J. Biotechnol. 2019, 296, 53–60. [Google Scholar] [CrossRef]
- Valiant, W.G.; Cai, K.; Vallone, P.M. A History of Adventitious Agent Contamination and the Current Methods to Detect and Remove Them from Pharmaceutical Products. Biologicals 2022, 80, 6–17. [Google Scholar] [CrossRef]
- Klug, B.; Robertson, J.S.; Condit, R.C.; Seligman, S.J.; Laderoute, M.P.; Sheets, R.; Williamson, A.-L.; Gurwith, M.; Kochhar, S.; Chapman, L.; et al. Adventitious Agents and Live Viral Vectored Vaccines: Considerations for Archiving Samples of Biological Materials for Retrospective Analysis. Vaccine 2016, 34, 6617–6625. [Google Scholar] [CrossRef]
- Cao, Y.; Yu, M.; Dong, G.; Chen, B.; Zhang, B. Digital PCR as an Emerging Tool for Monitoring of Microbial Biodegradation. Molecules 2020, 25, 706. [Google Scholar] [CrossRef]
- Shmidt, A.; Egorova, T. PCR-Based Analytical Methods for Quantification and Quality Control of Recombinant Adeno-Associated Viral Vector Preparations. Pharmaceuticals 2021, 15, 23. [Google Scholar] [CrossRef]
- Bilitewski, U. Chapter 11 Biosensors for Bioprocess Monitoring. In Comprehensive Analytical Chemistry; Elsevier: Amsterdam, The Netherlands, 2005; Volume 44, pp. 539–578. ISBN 978-0-444-50715-0. [Google Scholar]
- Guidance for Industry: PAT—A Framework for Innovative Pharmaceutical Development, Manufacturing and Quality Assurance; US Department of Health and Human Services, Food and Drug Administration: Silver Spring, MD, USA, 2004.
- Zandieh, M.; Hosseini, S.N.; Vossoughi, M.; Khatami, M.; Abbasian, S.; Moshaii, A. Label-Free and Simple Detection of En-Dotoxins Using a Sensitive LSPR Biosensor Based on Silver Nanocolumns. Anal. Biochem. 2018, 548, 96–101. [Google Scholar] [CrossRef]
- Khansili, N.; Rattu, G.; Krishna, P.M. Label-Free Optical Biosensors for Food and Biological Sensor Applications. Sens. Actuators B Chem. 2018, 265, 35–49. [Google Scholar] [CrossRef]
- Barone, P.W.; Wiebe, M.E.; Leung, J.C.; Hussein, I.T.M.; Keumurian, F.J.; Bouressa, J.; Brussel, A.; Chen, D.; Chong, M.; Dehghani, H.; et al. Viral Contamination in Biologic Manufacture and Implications for Emerging Therapies. Nat. Biotechnol. 2020, 38, 563–572. [Google Scholar] [CrossRef] [PubMed]
- Cheng, D.L.; Ngo, H.H.; Guo, W.S.; Liu, Y.W.; Zhou, J.L.; Chang, S.W.; Nguyen, D.D.; Bui, X.T.; Zhang, X.B. Bioprocessing for Elimination Antibiotics and Hormones from Swine Wastewater. Sci. Total Environ. 2018, 621, 1664–1682. [Google Scholar] [CrossRef]
- Armstrong, S.E.; Mariano, J.A.; Lundin, D.J. The Scope of Mycoplasma Contamination within the Biopharmaceutical Industry. Biol. J. Int. Assoc. Biol. Stand. 2010, 38, 211–213. [Google Scholar] [CrossRef]
- Vijayakumar, R.; Sandle, T. A Review on Fungal Contamination in Pharmaceutical Products and Phenotypicidentification of Contaminants by Conventional Methods. Eur. J. Parenter. Pharm. Sci. 2012, 17, 4–18. [Google Scholar]
- Kumar, P.; Kausar, M.A.; Singh, A.B.; Singh, R. Biological Contaminants in the Indoor Air Environment and Their Impacts on Human Health. Air Qual. Atmos. Health 2021, 14, 1723–1736. [Google Scholar] [CrossRef]
- Merten, O.-W. Virus contaminations of cell cultures—A biotechnological view. Cytotechnology 2002, 39, 91–116. [Google Scholar] [CrossRef]
- Seguel, J.M.; Merrill, R.; Seguel, D.; Campagna, A.C. Indoor Air Quality. Am. J. Lifestyle Med. 2017, 11, 284–295. [Google Scholar] [CrossRef]
- Zhang, C.; Tian, F.; Zhang, M.; Zhang, Z.; Bai, M.; Guo, G.; Zheng, W.; Wang, Q.; Shi, Y.; Wang, L. Endotoxin Contamination, a Potentially Important Inflammation Factor in Water and Wastewater: A Review. Sci. Total Environ. 2019, 681, 365–378. [Google Scholar] [CrossRef]
- Rasuli, L.; Dehghani, M.H.; Aghaei, M.; Mahvi, A.H.; Mubarak, N.M.; Karri, R.R. Occurrence and Fate of Bacterial Endotoxins in the Environment (Air, Water, Wastewater) and Remediation Technologies: An Overview. Chemosphere 2022, 303, 135089. [Google Scholar] [CrossRef] [PubMed]
- Barile, M.F. Mycoplasma Infections of Cell Cultures. Isr. J. Med. Sci. 1981, 17, 555–562. [Google Scholar] [CrossRef] [PubMed]
- Uphoff, C.C.; Drexler, H.G. Elimination of Mycoplasmas from Infected Cell Lines Using Antibiotics. Cancer Cell Cult. Methods Protoc. 2011, 731, 105–114. [Google Scholar]
- Hay, R.J.; Macy, M.L.; Chen, T.R. Mycoplasma Infection of Cultured Cells. Nature 1989, 339, 487–488. [Google Scholar] [CrossRef]
- Drexler, H.G.; Uphoff, C.C. Mycoplasma contamination of cell cultures: Incidence, sources, effects, detection, elimination, prevention. Cytotechnology 2002, 39, 75–90. [Google Scholar] [CrossRef]
- Fratz-Berilla, E.J.; Angart, P.; Graham, R.J.; Powers, D.N.; Mohammad, A.; Kohnhorst, C.; Faison, T.; Velugula-Yellela, S.R.; Trunfio, N.; Agarabi, C. Impacts on Product Quality Attributes of Monoclonal Antibodies Produced in CHO Cell Bioreactor Cul-Tures during Intentional Mycoplasma Contamination Events. Biotechnol. Bioeng. 2020, 117, 2802–2815. [Google Scholar] [CrossRef]
- Nikfarjam, L.; Farzaneh, P. Prevention and detection of Mycoplasma contamination in cell culture. Cell J. 2012, 13, 203–212. [Google Scholar]
- Becherucci, V.; Curini, L.; Ceccantini, R.; Bisin, S.; Gori, V.; Gentile, F.; De Rienzo, E.; Piccini, L.; Bindi, B.; Pavan, P.; et al. A Practical Approach for Gmp-Compliant Validation of Real-Time PCR Method for Mycoplasma Detection in Human Mesenchymal Stromal Cells as Advanced Therapy Medicinal Product. Biologicals 2021, 73, 31–40. [Google Scholar] [CrossRef]
- Kahane, I.; Banai, M.; Razin, S.; Feldner, J. Attachment of Mycoplasmas to Host Cell Membranes. Clin. Infect. Dis. 1982, 4, 185–192. [Google Scholar] [CrossRef]
- Benedetti, F.; Curreli, S.; Zella, D. Mycoplasmas–Host Interaction: Mechanisms of Inflammation and Association with Cellular Transformation. Microorganisms 2020, 8, 1351. [Google Scholar] [CrossRef]
- Majdoul, S.; Compton, A.A. Lessons in Self-Defence: Inhibition of Virus Entry by Intrinsic Immunity. Nat. Rev. Immunol. 2022, 22, 339–352. [Google Scholar] [CrossRef] [PubMed]
- Casadevall, A.; Coelho, C.; Alanio, A. Mechanisms of Cryptococcus neoformans-Mediated Host Damage. Front. Immunol. 2018, 9, 855. [Google Scholar] [CrossRef]
- Malik, P.; Mukherjee, S.; Mukherjee, T.K. Microbial Contamination of Mammalian Cell Culture. In Practical Approach to Mammalian Cell and Organ Culture; Mukherjee, T.K., Malik, P., Mukhopadhyay, S., Eds.; Springer Nature: Singapore, 2023; pp. 1–45. [Google Scholar]
- Moineau, S. Bacteriophage. In Brenner’s Encyclopedia of Genetics; Elsevier: Amsterdam, The Netherlands, 2013; pp. 280–283. [Google Scholar]
- Abedon, S.T.; Duffy, S.; Turner, P.E. Bacteriophage Ecology. In Encyclopedia of Microbiology; Elsevier: Amsterdam, The Netherlands, 2009; pp. 42–57. [Google Scholar]
- Ertürk, G.; Lood, R. Bacteriophages as Biorecognition Elements in Capacitive Biosensors: Phage and Host Bacteria Detection. Sens. Actuators B Chem. 2018, 258, 535–543. [Google Scholar] [CrossRef]
- Rao, G.; Moreira, A.; Brorson, K. Disposable Bioprocessing: The Future Has Arrived. Biotechnol. Bioeng. 2009, 102, 348–356. [Google Scholar] [CrossRef]
- Närhi, M.; Nordström, K. National GMP Regulations and Codes and International GMP Guides and Guidelines: Corre-Spondences and Differences. In Pharmaceutical Sciences Encyclopedia; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2010; p. 378. [Google Scholar]
- Haleem, R.M.; Salem, M.Y.; Fatahallah, F.A.; Abdelfattah, L.E. Quality in the Pharmaceutical Industry—A Literature Review. Saudi Pharm. J. 2015, 23, 463–469. [Google Scholar] [CrossRef] [PubMed]
- Rathore, A.S.; Li, Y.; Chhabra, H.; Lohiya, A. FDA Warning Letters: A Retrospective Analysis of Letters Issued to Phar-Maceutical Companies from 2010–2020. J. Pharm. Innov. 2022. [Google Scholar] [CrossRef]
- Bunn, G.P. (Ed.) Good Manufacturing Practices for Pharmaceuticals, 7th ed.; CRC Press, Taylor & Francis Group: Boca Raton, FL, USA, 2019. [Google Scholar]
- Cundell, T. Exclusion of Objectionable Microorganisms from Non-sterile Pharmaceutical Drug Products. In Pharmaceutical Microbiological Quality Assurance and Control; Roesti, D., Goverde, M., Eds.; Wiley: Hoboken, NJ, USA, 2019; pp. 371–400. [Google Scholar]
- Jornitz, M.W. (Ed.) Filtration and Purification in the Biopharmaceutical Industry, 3rd ed.; CRC Press: Boca Raton, FL, USA, 2019. [Google Scholar]
- Seet, W.T.; Afandi, M.A.M.; Shamsuddin, S.A.; Lokanathan, Y.; Ng, M.H.; Maarof, M. Current Good Manufacturing Practice (CGMP) Facility and Production of Stem Cell; Stem Cell Production, Ed.; Springer: Singapore, 2022. [Google Scholar]
- Andriolo, G. GMP-Grade Methods for Cardiac Progenitor Cells: Cell Bank Production and Quality Control. In Stem Cells and Good Manufacturing Practices; Turksen, K., Ed.; Springer: New York, NY, USA, 2020; Volume 2286, pp. 131–166. [Google Scholar]
- EU GMP. The Rules Governing Medicinal Products in the European Union. Volume 4 EU Guidelines to Good Manufacturing PRACTICE—Medicinal Products for Human and Veterinary Use, Annex 1–2008. Available online: https://health.ec.europa.eu/system/files/2022-08/20220825_gmp-an1_en_0.pdf (accessed on 3 March 2023).
- Whitford, W.G. Bioprocess Intensification: Technologies and Goals. In Process Control, Intensification, and Digitalisation in Continuous Biomanufacturing; Subramanian, G., Ed.; Wiley: Hoboken, NJ, USA, 2022; pp. 93–136. [Google Scholar]
- Gerzon, G.; Sheng, Y.; Kirkitadze, M. Process Analytical Technologies—Advances in Bioprocess Integration and Future Perspectives. J. Pharm. Biomed. Anal. 2022, 207, 114379. [Google Scholar] [CrossRef]
- Rathore, A.S.; Mishra, S.; Nikita, S.; Priyanka, P. Bioprocess Control: Current Progress and Future Perspectives. Life 2021, 11, 557. [Google Scholar] [CrossRef]
- Gomes, J.; Chopda, V.R.; Rathore, A.S. Integrating systems analysis and control for implementing process analytical technology in bioprocess development: Integrating systems analysis and control. J. Chem. Technol. Biotechnol. 2015, 90, 583–589. [Google Scholar] [CrossRef]
- Vashist, S.K.; Luong, J.H.T. Immunoassays. In Handbook of Immunoassay Technologies; Elsevier: Amsterdam, The Netherlands, 2018; pp. 1–18. [Google Scholar]
- Rathore, A.S.; Bhambure, R.; Ghare, V. Process analytical technology (PAT) for biopharmaceutical products. Anal. Bioanal. Chem. 2010, 398, 137–154. [Google Scholar] [CrossRef]
- Shintani, H. Validation Studies for Microbial Contamination and Control of Contaminants. Biocontrol Sci. 2015, 20, 161–170. [Google Scholar] [CrossRef] [PubMed]
- Abu-Absi, N.R. Real Time Monitoring of Multiple Parameters in Mammalian Cell Culture Bioreactors Using an In-Line Raman Spectroscopy Probe. Biotechnol. Bioeng. 2011, 108, 1215–1221. [Google Scholar] [CrossRef] [PubMed]
- Read, E.K.; Shah, R.B.; Riley, B.S.; Park, J.T.; Brorson, K.A.; Rathore, A.S. Process Analytical Technology (PAT) for Bio-Pharmaceutical Products: Part II. Concepts and Applications. Biotechnol. Bioeng. 2010, 105, 285–295. [Google Scholar] [CrossRef]
- Dervisevic, E.; Dervisevic, M.; Ang, B.; Carthew, J.; Tuck, K.L.; Voelcker, N.H.; Cadarso, V.J. Integrated Microfluidic Device to Monitor Unseen Escherichia Coli Contamination in Mammalian Cell Cul-Ture. Sens. Actuators B Chem. 2022, 359, 131522. [Google Scholar] [CrossRef]
- Wehbe, K.; Vezzalini, M.; Cinque, G. Detection of Mycoplasma in Contaminated Mammalian Cell Culture Using FTIR Mi-Crospectroscopy. Anal. Bioanal. Chem. 2018, 410, 3003–3016. [Google Scholar] [CrossRef]
- Lowder, J.N.; Whelton, P. Microbial Contamination of Cellular Products for Hematolymphoid Transplantation Therapy: Assessment of the Problem and Strategies to Minimize the Clinical Impact. Cytotherapy 2003, 5, 377–390. [Google Scholar] [CrossRef]
- Bustin, S.A. The MIQE Guidelines: Minimum Information for Publication of Quantitative Real-Time PCR Experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef]
- Kleinschmidt, K.; Wilkens, E.; Glaeser, S.P.; Kaempfer, P.; Staerk, A.; Roesti, D. Development of a Qualitative Real-Time PCR for Microbiological Quality Control Testing in Mammalian Cell Culture Production. J. Appl. Microbiol. 2017, 122, 997–1008. [Google Scholar] [CrossRef]
- Davies, C. Immunoassay Performance Measures 1 1This Chapter Is from the First Edition of The Immunoassay Handbook, with Few Changes, Because of the High Regard in Which Chris Davies’ Original Material Is Held. The Theory Still Applies but Some of the Examples given Are Dated. In The Immunoassay Handbook; Elsevier: Amsterdam, The Netherlands, 2013; pp. 11–26. [Google Scholar]
- Findlay, J.W.A. Validation of Immunoassays for Bioanalysis: A Pharmaceutical Industry Perspective. J. Pharm. Biomed. Anal. 2000, 21, 1249–1273. [Google Scholar] [CrossRef]
- Novitsky, T.J. Biomedical Applications of Limulus Amebocyte Lysate. In Biology and Conservation of Horseshoe Crabs; Tanacredi, J.T., Botton, M.L., Smith, D., Eds.; Springer: Boston, MA, USA, 2009; pp. 315–329. [Google Scholar]
- Brown, J.; Bir, A.; Bain, R.E.S. Novel Methods for Global Water Safety Monitoring: Comparative Analysis of Low-Cost, Field-Ready E. Coli Assays. npj Clean. Water 2020, 3, 9. [Google Scholar] [CrossRef]
- Arnaout, R.; Lee, R.A.; Lee, G.R.; Callahan, C.; Yen, C.F.; Smith, K.P.; Arora, R.; Kirby, J.E. SARS-CoV-2 Testing: The Limit of Detection Matters. bioRxiv 2020. [Google Scholar] [CrossRef]
- Maturin, L. Aerobic Plate Count. In Bacteriological Analytical Manual Online. 2001. Available online: https://cir.nii.ac.jp/all?q=http://www.cfsan.fda.gov/%E3%80%9Cebam/bam-3.html (accessed on 3 March 2023).
- Xiong, X.L.; Wang, S.M.; Zhang, Y.; Chen, L.C. Detection of Endotoxin Concentration Using Piezoelectric Based Biosensor System. AMM 2012, 195–196, 874–878. [Google Scholar] [CrossRef]
- Feng, P.; Weagant, S.; Grant, M. Enumeration of Escherichia coli and the Coliform Bacteria. In Bacteriological Analytical Manual, 8th ed.; FDA/Center for Food Safety & Applied Nutrition: College Park, MD, USA. Available online: http://www.cfsan.fda.gov/~ebam/bam4.html (accessed on 3 March 2023).
- Manoharan, H.; Kalita, P.; Gupta, S.; Sai, V.V.R. Plasmonic biosensors for bacterial endotoxin detection on biomimetic C-18 supported fiber optic probes. Biosens. Bioelectron. 2019, 129, 79–86. [Google Scholar] [CrossRef] [PubMed]
- Numnuam, A.; Kanatharana, P.; Mattiasson, B.; Asawatreratanakul, P.; Wongkittisuksa, B.; Limsakul, C.; Thavarungkul, P. Capacitive Biosensor for Quantification of Trace Amounts of DNA. Biosens. Bioelectron. 2009, 24, 2559–2565. [Google Scholar] [CrossRef]
- Kim, S.-E.; Su, W.; Cho, M.; Lee, Y.; Choe, W.-S. Harnessing Aptamers for Electrochemical Detection of Endotoxin. Anal. Biochem. 2012, 424, 12–20. [Google Scholar] [CrossRef] [PubMed]
- Damborský, P.; Švitel, J.; Katrlík, J. Optical biosensors. Essays Biochem. 2016, 60, 91–100. [Google Scholar] [CrossRef]
- Singh, P. Surface Plasmon Resonance: A Boon for Viral Diagnostics. In Reference Module in Life Sciences; Elsevier: Amsterdam, The Netherlands, 2017; p. 9780128096338123000. [Google Scholar]
- Balbinot, S.; Srivastav, A.M.; Vidic, J.; Abdulhalim, I.; Manzano, M. Plasmonic biosensors for food control. Trends Food Sci. Technol. 2021, 111, 128–140. [Google Scholar] [CrossRef]
- Hamza, M.E.; Othman, M.A.; Swillam, M.A. Plasmonic Biosensors: Review. Biology 2022, 11, 621. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, K.R.; Awasthi, S.; Mishra, P.K.; Srivastava, P.K. Biosensors/Molecular Tools for Detection of Waterborne Pathogens. In Waterborne Pathogens; Elsevier: Amsterdam, The Netherlands, 2020; pp. 237–277. [Google Scholar]
- Mulchandani, A.; Bassi, A.S. Principles and Applications of Biosensors for Bioprocess Monitoring and Control. Crit. Rev. Biotechnol. 1995, 15, 105–124. [Google Scholar] [CrossRef]
- Carpenter, A.; Paulsen, I.; Williams, T. Blueprints for Biosensors: Design, Limitations, and Applications. Genes 2018, 9, 375. [Google Scholar] [CrossRef]
- Gavrilaș, S.; Ursachi, C.Ș.; Perța-Crișan, S.; Munteanu, F.-D. Recent Trends in Biosensors for Environmental Quality Monitoring. Sensors 2022, 22, 1513. [Google Scholar] [CrossRef]
- Reardon, K.F. Practical Monitoring Technologies for Cells and Substrates in Biomanufacturing. Curr. Opin. Biotechnol. 2021, 71, 225–230. [Google Scholar] [CrossRef] [PubMed]
- Liu, C. Acoustic Ejection Mass Spectrometry: Fundamentals and Applications in High-Throughput Drug Discovery. Expert Opin. Drug Discov. 2022, 17, 775–787. [Google Scholar] [CrossRef] [PubMed]

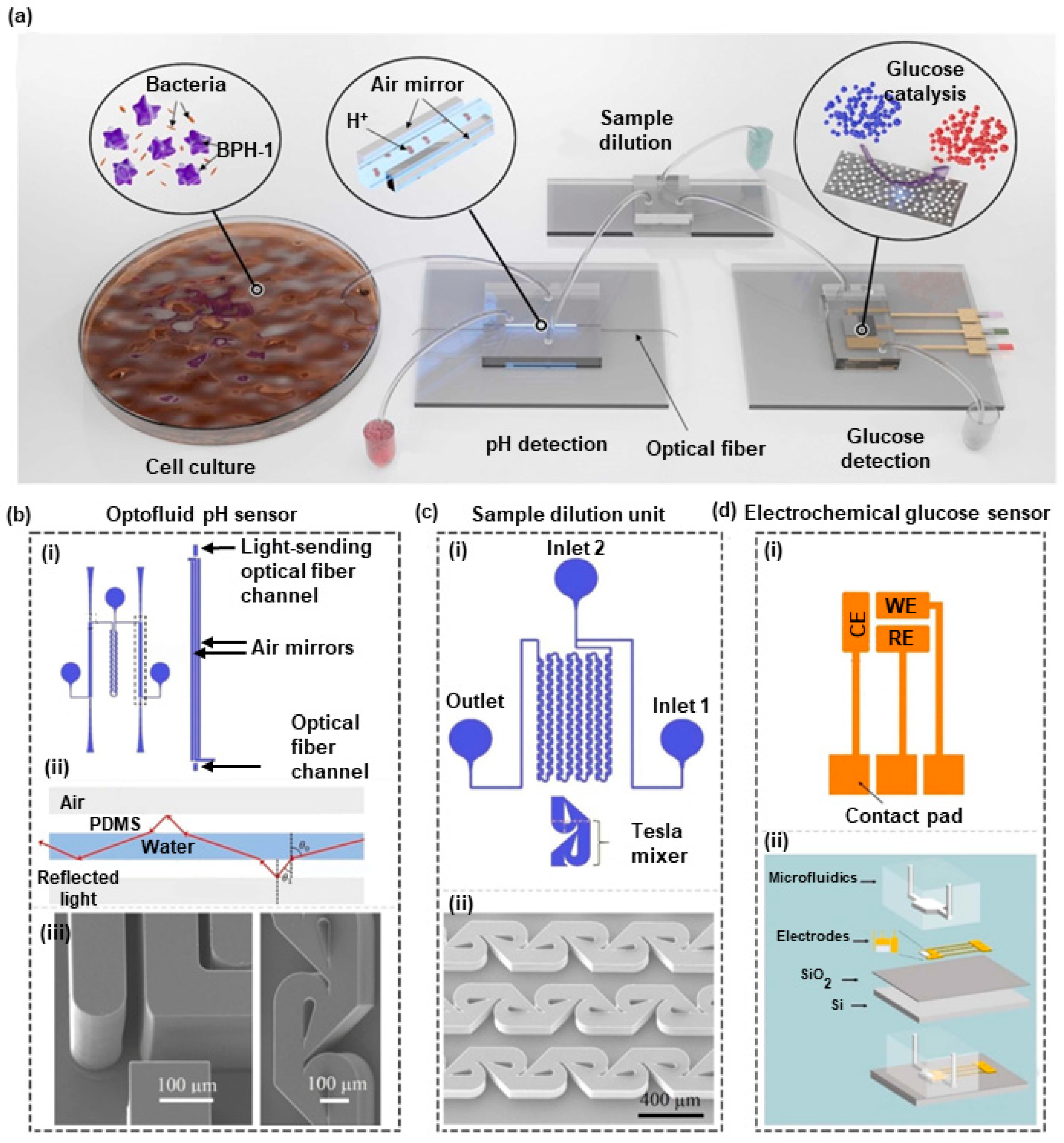
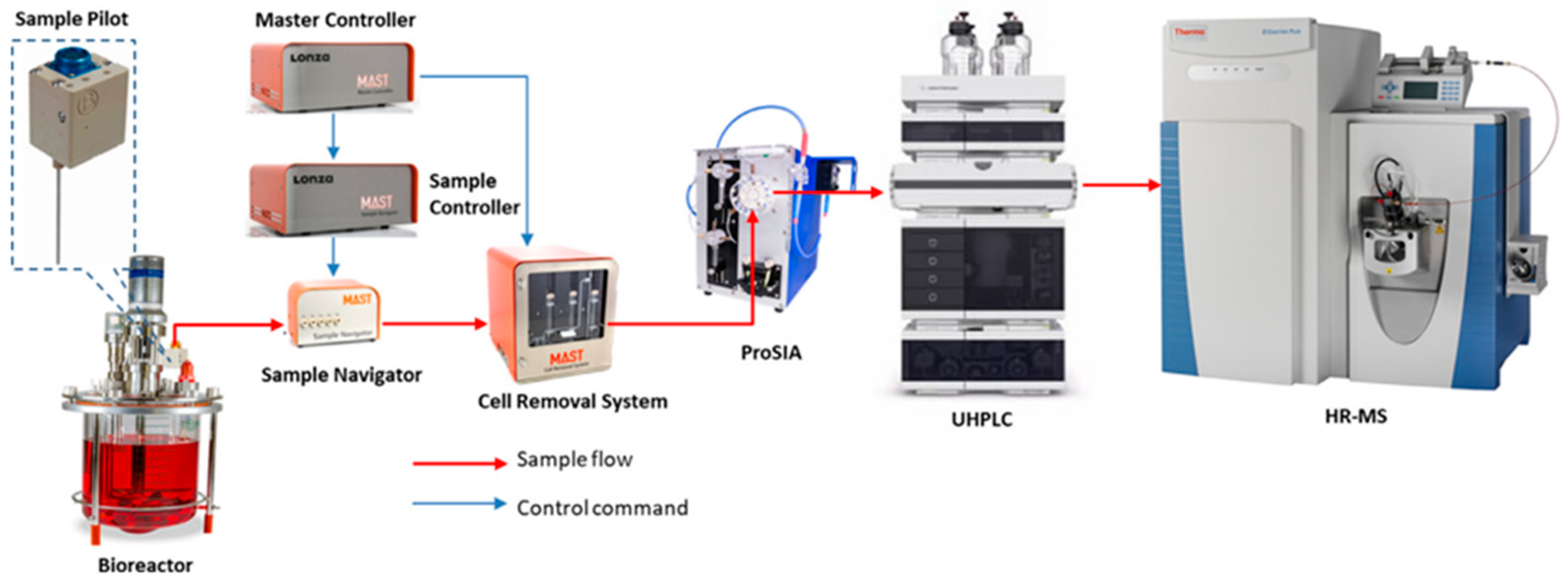
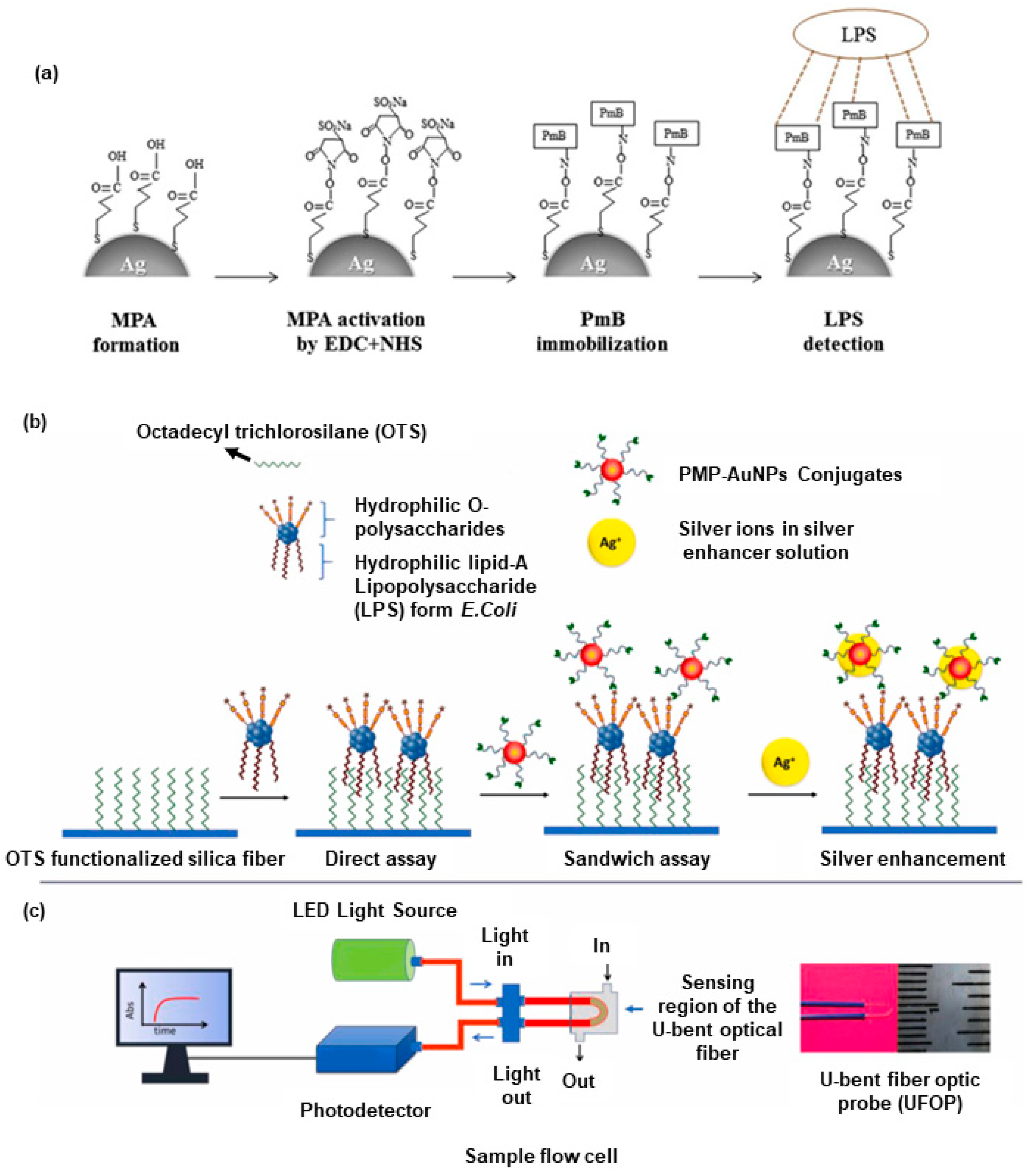
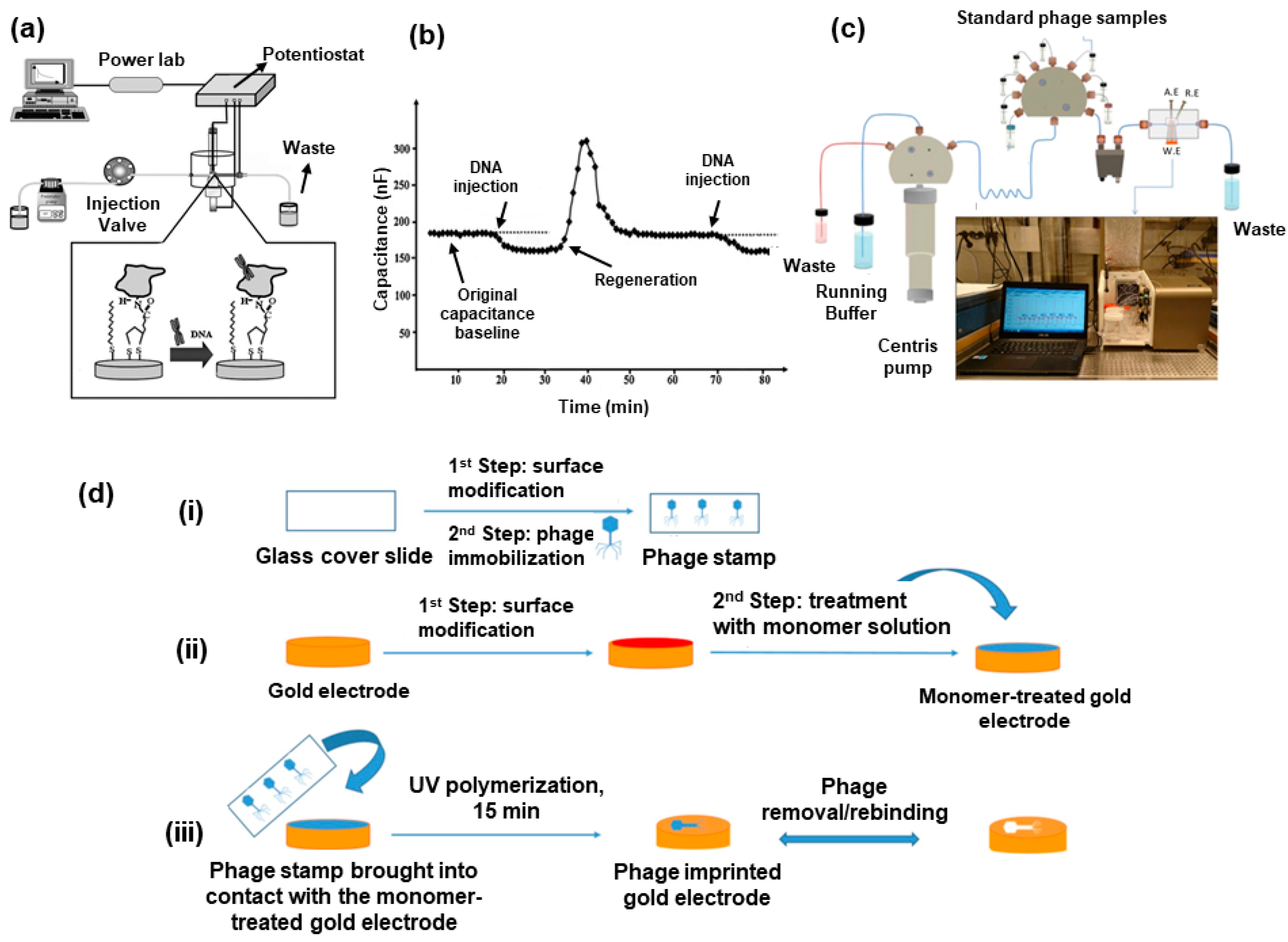
| Method Type | Target Biocontaminant | Time Required to Do the Test | LOD | Specificity | Reference |
|---|---|---|---|---|---|
| LSPR | Endotoxin | 1 to 24 h | 340 pg/mL | Selectively detects endotoxin | [14] |
| UFOPs | Endotoxin | 25 min | 0.4 ng/mL | Highly specific | [72] |
| Capacitive | Residual DNA | 14 min | 10−5 ng/L | Specific to DNA regardless of source | [73] |
| MIP/Capacitive | E. coli phage E. coli | Real-time | 10 pfu/mL 1.0 × 102 cfu/mL | Selective for E. coli phage in river water Selective for E. coli but recognized other bacteria | [39] |
| Impedance-based aptasensor | Endotoxin | 15 min | 0.01 ng/mL | Minimal response to other media components (pDNA, RNA, proteins, saccharides, and lipids) | [74] |
| Integrated microfluidic device | E. coli | Less than 8 h | 27.7 ± 1.3 µM of glucose | Highly selective toward glucose | [59] |
| FTIR microspectroscopy | Mycoplasma bovis | 6 min (data acquisition) | NA | Not validate for specificity | [60] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Janghorban, M.; Kazemi, S.; Tormon, R.; Ngaju, P.; Pandey, R. Methods and Analysis of Biological Contaminants in the Biomanufacturing Industry. Chemosensors 2023, 11, 298. https://doi.org/10.3390/chemosensors11050298
Janghorban M, Kazemi S, Tormon R, Ngaju P, Pandey R. Methods and Analysis of Biological Contaminants in the Biomanufacturing Industry. Chemosensors. 2023; 11(5):298. https://doi.org/10.3390/chemosensors11050298
Chicago/Turabian StyleJanghorban, Mohammad, Sara Kazemi, Rigel Tormon, Philippa Ngaju, and Richa Pandey. 2023. "Methods and Analysis of Biological Contaminants in the Biomanufacturing Industry" Chemosensors 11, no. 5: 298. https://doi.org/10.3390/chemosensors11050298
APA StyleJanghorban, M., Kazemi, S., Tormon, R., Ngaju, P., & Pandey, R. (2023). Methods and Analysis of Biological Contaminants in the Biomanufacturing Industry. Chemosensors, 11(5), 298. https://doi.org/10.3390/chemosensors11050298








