Pd@Pt Nanodendrites as Peroxidase Nanomimics for Enhanced Colorimetric ELISA of Cytokines with Femtomolar Sensitivity
Abstract
:1. Introduction
2. Materials and Methods
2.1. Preparation of 18 nm Pd NCs
2.2. Synthesis of Pd@Pt NDs
2.3. Apparent Steady-State Kinetic Assays
2.4. Preparation of Streptavidin-Conjugated Pd@Pt NDs (Pd@Pt ND-SA Conjugates)
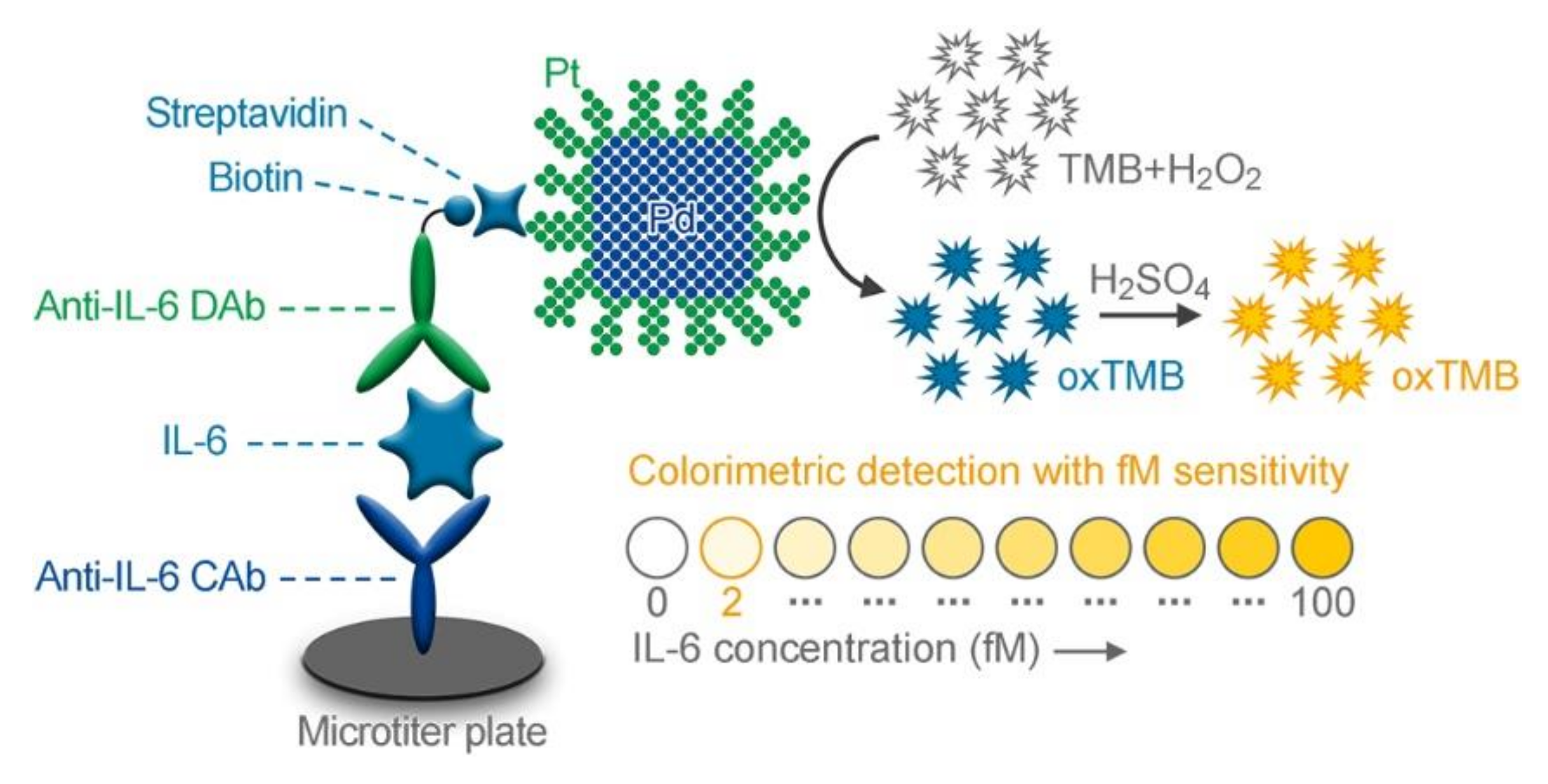
2.5. Colorimetric Pd@Pt ND ELISA of IL-6
3. Results and Discussion
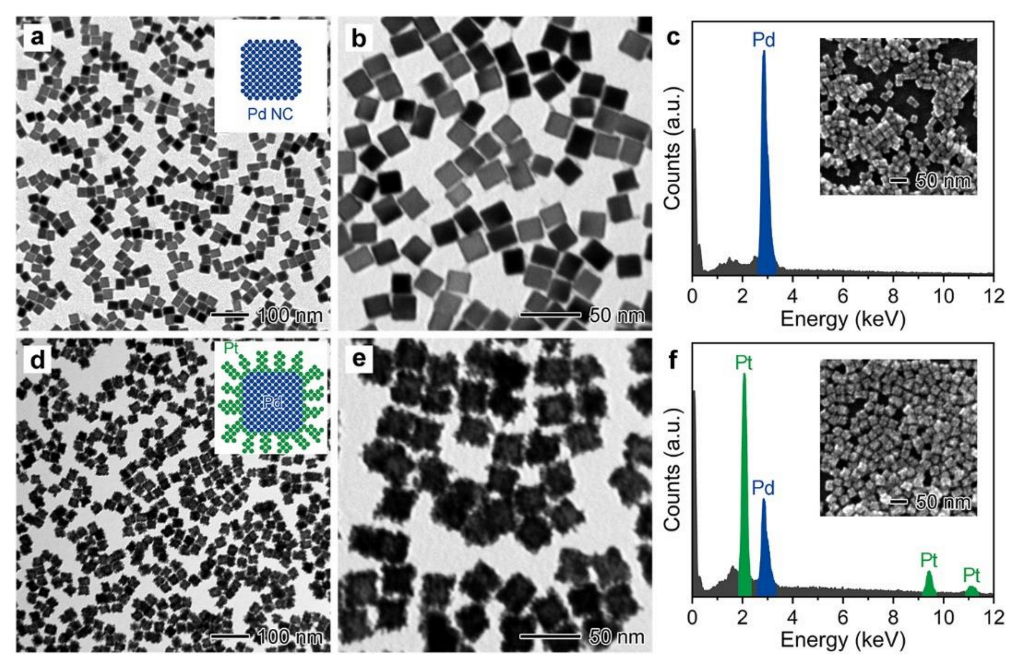
3.1. Synthesis and Characterization of Pd@Pt NDs
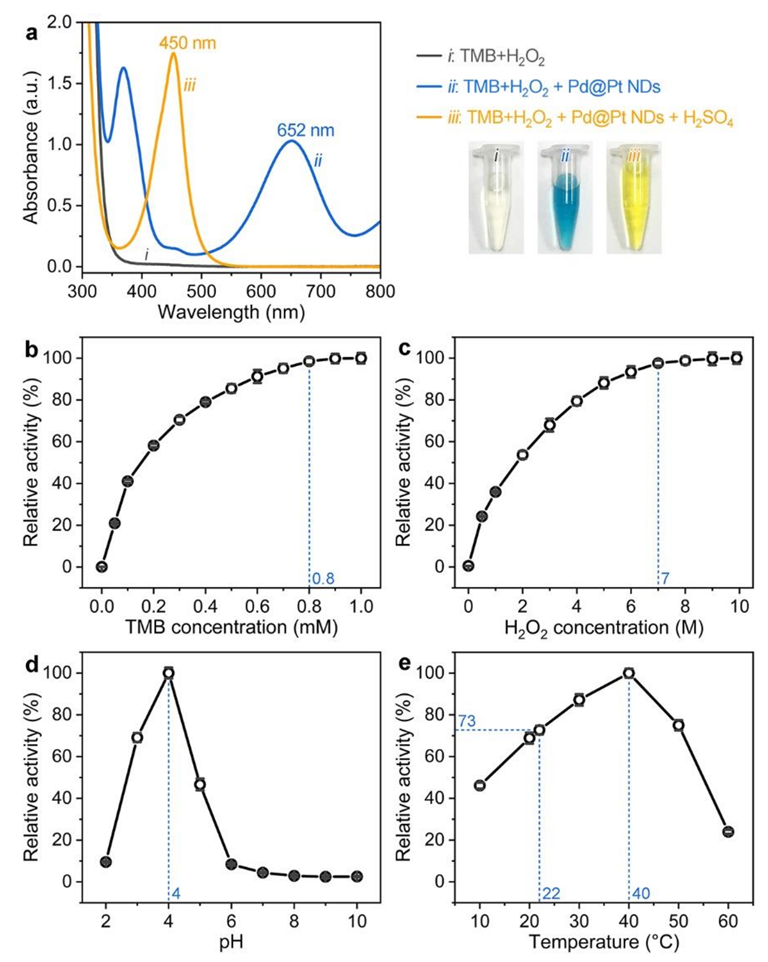
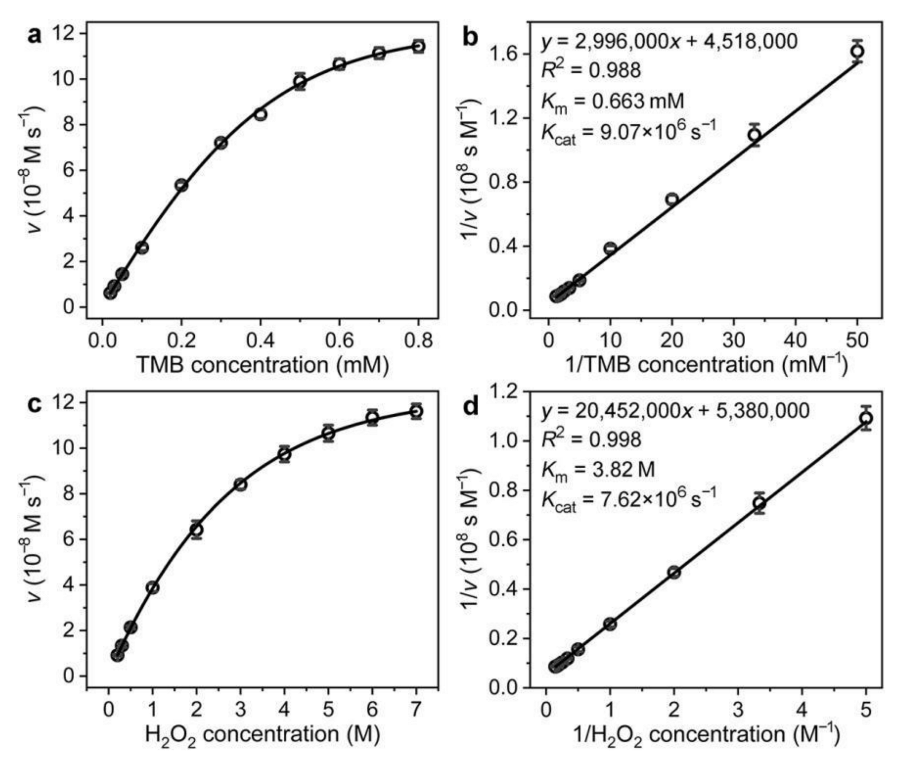
3.2. Peroxidase-Like Catalytic Activity of Pd@Pt NDs
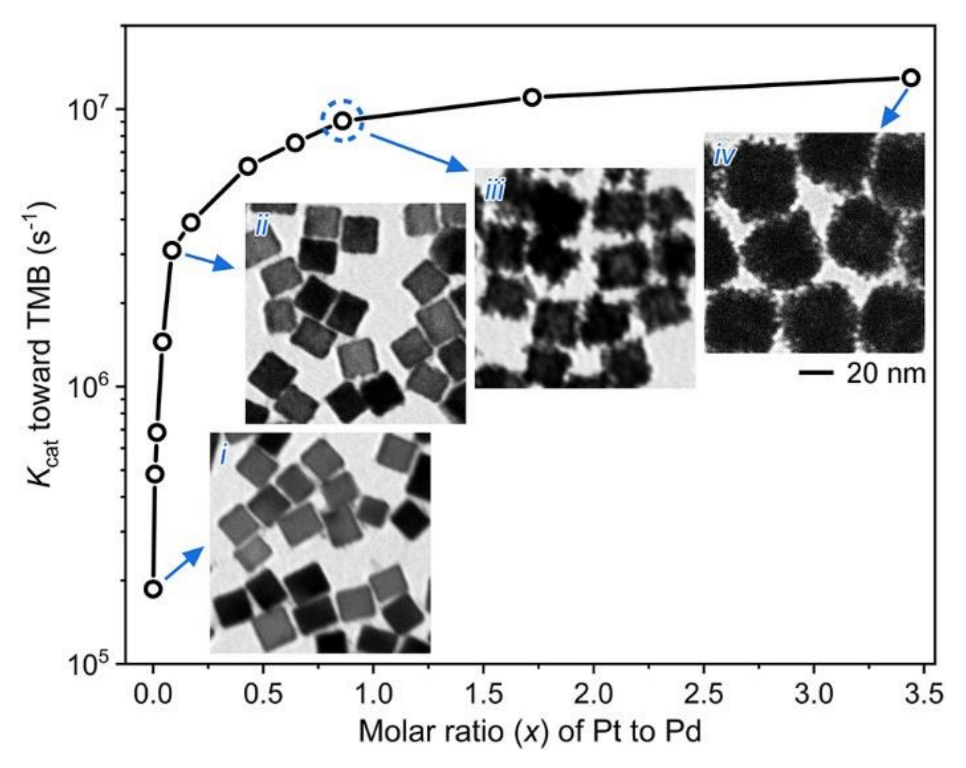
3.3. Effect of Pt Content on the Peroxidase-Like Catalytic Activity of Pd@Pt NDs
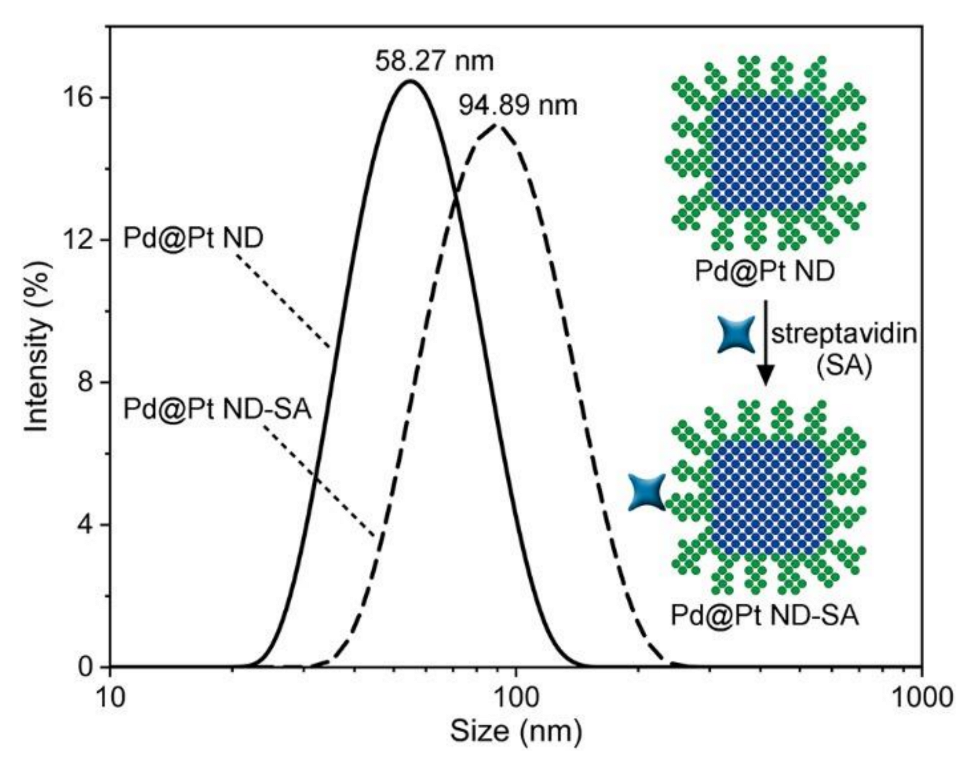
3.4. Preparation and Characterization of Pd@Pt ND-SA Conjugates
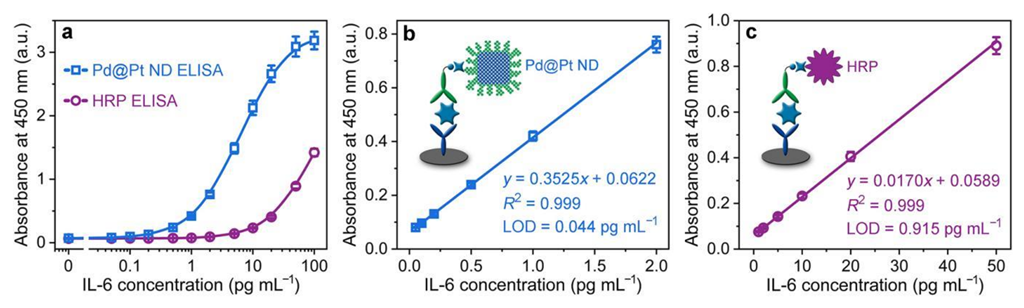
3.5. Analytical Performance of the Colorimetric Pd@Pt ND ELISA for Detection of IL-6
3.6. Analysis of Real Samples and Method Validation
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Thomson, A.W.; Lotze, M.T. The Cytokine Handbook, Two-Volume Set; Elsevier: Amsterdam, The Netherlands, 2003. [Google Scholar]
- Preedy, V.R.; Hunter, R. Cytokines; Taylor & Francis: Abingdon, UK, 2011. [Google Scholar]
- Stenken, J.A.; Poschenrieder, A.J. Bioanalytical chemistry of cytokines—A review. Anal. Chim. Acta 2015, 853, 95–115. [Google Scholar] [CrossRef]
- Liu, C.; Chu, D.; Kalantar-Zadeh, K.; George, J.; Young, H.A.; Liu, G. Cytokines: From clinical significance to quantification. Adv. Sci. 2021, 8, 2004433. [Google Scholar] [CrossRef]
- Stanley, A.C.; Lacy, P. Pathways for cytokine secretion. Physiology 2010, 25, 218–229. [Google Scholar] [CrossRef]
- Altan-Bonnet, G.; Mukherjee, R. Cytokine-mediated communication: A quantitative appraisal of immune complexity. Nat. Rev. Immunol. 2019, 19, 205–217. [Google Scholar] [CrossRef]
- Dembic, Z. The Cytokines of the Immune System: The Role of Cytokines in Disease Related to Immune Response; Academic Press: Cambridge, MA, USA, 2015. [Google Scholar]
- Goldring, M.B.; Goldring, S.R. Cytokines and cell growth control. Crit. Rev. Eukaryot. Gene Expr. 1991, 1, 301–326. [Google Scholar]
- Mizoguchi, I.; Higuchi, K.; Mitobe, K.; Tsunoda, R.; Mizuguchi, J.; Yoshimoto, T. Cytokine Frontiers: Regulation of Immune Responses in Health and Disease; Springer: Berlin/Heidelberg, Germany, 2013. [Google Scholar]
- Cron, R.Q.; Behrens, E.M. Cytokine Storm Syndrome; Springer: Berlin/Heidelberg, Germany, 2019. [Google Scholar]
- Vancurova, I. Cytokine Bioassays; Springer: Berlin/Heidelberg, Germany, 2016. [Google Scholar]
- Chen, P.; Chung, M.T.; McHugh, W.; Nidetz, R.; Li, Y.; Fu, J.; Cornell, T.T.; Shanley, T.P.; Kurabayashi, K. Multiplex serum cytokine immunoassay using nanoplasmonic biosensor microarrays. ACS Nano 2015, 9, 4173–4181. [Google Scholar] [CrossRef]
- Tanaka, T.; Narazaki, M.; Kishimoto, T. IL-6 in inflammation, immunity, and disease. Cold Spring Harb. Perspect. Biol. 2014, 6, a016295. [Google Scholar] [CrossRef]
- Velazquez-Salinas, L.; Verdugo-Rodriguez, A.; Rodriguez, L.L.; Borca, M.V. The role of interleukin 6 during viral infections. Front. Microbiol. 2019, 10, 1057. [Google Scholar] [CrossRef] [PubMed]
- Schaper, F.; Rose-John, S. Interleukin-6: Biology, signaling and strategies of blockade. Cytokine Growth Factor Rev. 2015, 26, 475–487. [Google Scholar] [CrossRef] [PubMed]
- Hack, C.E.; De Groot, E.R.; Fert-Bersma, R.J.F.; Nuijens, J.H.; Strack Van Schijndel, R.J.M.; Eerenberg-Belmer, A.J.M.; Thijs, L.G.; Aarden, L.A. Increased plasma levels of interleukin-6 in sepsis. Blood 1989, 74, 1704–1710. [Google Scholar] [CrossRef] [PubMed]
- Molano Franco, D.; Arevalo-Rodriguez, I.; Roqué i Figuls, M.; Montero Oleas, N.G.; Nuvials, X.; Zamora, J. Plasma interleukin-6 concentration for the diagnosis of sepsis in critically ill adults. Cochrane Database Syst. Rev. 2019, 4, CD011811. [Google Scholar] [CrossRef] [PubMed]
- Rincon, M. Interleukin-6: From an inflammatory marker to a target for inflammatory diseases. Trends Immunol. 2012, 33, 571–577. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Wu, Z.; Li, J.-W.; Zhao, H.; Wang, G.-Q. Cytokine release syndrome in severe COVID-19: Interleukin-6 receptor antagonist tocilizumab may be the key to reduce mortality. Int. J. Antimicrob. Agents 2020, 55, 105954. [Google Scholar] [CrossRef] [PubMed]
- Shankar, G.; Cohen, D.A. Enhanced cytokine detection by a novel cell culture-based elisa. J. Immunoass. 1997, 18, 371–388. [Google Scholar] [CrossRef]
- Liu, G.; Zhang, K.; Nadort, A.; Hutchinson, M.R.; Goldys, E.M. Sensitive cytokine assay based on optical fiber allowing localized and spatially resolved detection of interleukin-6. ACS Sens. 2017, 2, 218–226. [Google Scholar] [CrossRef]
- Tongdee, M.; Yamanishi, C.; Maeda, M.; Kojima, T.; Dishinger, J.; Chantiwas, R.; Takayama, S. One-incubation one-hour multiplex ELISA enabled by aqueous two-phase systems. Analyst 2020, 145, 3517–3527. [Google Scholar] [CrossRef]
- Schweitzer, B.; Roberts, S.; Grimwade, B.; Shao, W.; Wang, M.; Fu, Q.; Shu, Q.; Laroche, I.; Zhou, Z.; Tchernev, V.T.; et al. Multiplexed protein profiling on microarrays by rolling-circle amplification. Nat. Biotechnol. 2002, 20, 359–365. [Google Scholar] [CrossRef] [PubMed]
- Adalsteinsson, V.; Parajuli, O.; Kepics, S.; Gupta, A.; Reeves, W.B.; Hahm, J.I. Ultrasensitive detection of cytokines enabled by nanoscale zno arrays. Anal. Chem. 2008, 80, 6594–6601. [Google Scholar] [CrossRef]
- Liu, G.; Qi, M.; Hutchinson, M.R.; Yang, G.; Goldys, E.M. Recent advances in cytokine detection by immunosensing. Biosens. Bioelectron. 2016, 79, 810–821. [Google Scholar] [CrossRef]
- Wei, H.; Wang, E. Nanomaterials with enzyme-like characteristics (nanozymes): Next-generation artificial enzymes. Chem. Soc. Rev. 2013, 42, 6060–6093. [Google Scholar] [CrossRef]
- Wu, J.; Wang, X.; Wang, Q.; Lou, Z.; Li, S.; Zhu, Y.; Qin, L.; Wei, H. Nanomaterials with enzyme-like characteristics (nanozymes): Next-generation artificial enzymes (II). Chem. Soc. Rev. 2019, 48, 1004–1076. [Google Scholar] [PubMed]
- Ye, H.; Xi, Z.; Magloire, K.; Xia, X. Noble-metal nanostructures as highly efficient peroxidase mimics. ChemNanoMat 2019, 5, 860–868. [Google Scholar] [CrossRef]
- Liu, X.; Huang, D.; Lai, C.; Qin, L.; Zeng, G.; Xu, P.; Li, B.; Yi, H.; Zhang, M. Peroxidase-like activity of smart nanomaterials and their advanced application in colorimetric glucose biosensors. Small 2019, 15, 1900133. [Google Scholar] [CrossRef] [PubMed]
- Gao, L.; Zhuang, J.; Nie, L.; Zhang, J.; Zhang, Y.; Gu, N.; Wang, T.; Feng, J.; Yang, D.; Perrett, S.; et al. Intrinsic peroxidase-like activity of ferromagnetic nanoparticles. Nat. Nanotechnol. 2007, 2, 577–583. [Google Scholar] [CrossRef]
- Mu, J.; Wang, Y.; Zhao, M.; Zhang, L. Intrinsic peroxidase-like activity and catalase-like activity of Co3O4 nanoparticles. Chem. Comm. 2012, 48, 2540–2542. [Google Scholar] [CrossRef] [PubMed]
- Wan, Y.; Qi, P.; Zhang, D.; Wu, J.; Wang, Y. Manganese oxide nanowire-mediated enzyme-linked immunosorbent assay. Biosens. Bioelectron. 2012, 33, 69–74. [Google Scholar] [CrossRef]
- André, R.; Natálio, F.; Humanes, M.; Leppin, J.; Heinze, K.; Wever, R.; Schröder, H.-C.; Müller, W.E.G.; Tremel, W. V2O5 nanowires with an intrinsic peroxidase-like activity. Adv. Funct. Mater. 2011, 21, 501–509. [Google Scholar] [CrossRef]
- Lin, T.; Zhong, L.; Guo, L.; Fu, F.; Chen, G. Seeing diabetes: Visual detection of glucose based on the intrinsic peroxidase-like activity of MoS2 nanosheets. Nanoscale 2014, 6, 11856–11862. [Google Scholar] [CrossRef]
- Lin, T.; Zhong, L.; Song, Z.; Guo, L.; Wu, H.; Guo, Q.; Chen, Y.; Fu, F.; Chen, G. Visual detection of blood glucose based on peroxidase-like activity of WS2 nanosheets. Biosens. Bioelectron. 2014, 62, 302–307. [Google Scholar] [CrossRef]
- Song, Y.; Qu, K.; Zhao, C.; Ren, J.; Qu, X. Graphene oxide: Intrinsic peroxidase catalytic activity and its application to glucose detection. Adv. Mater. 2010, 22, 2206–2210. [Google Scholar] [CrossRef]
- Jv, Y.; Li, B.; Cao, R. Positively-charged gold nanoparticles as peroxidiase mimic and their application in hydrogen peroxide and glucose detection. Chem. Comm. 2010, 46, 8017–8019. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; Chen, Z.; Cao, H.; Huang, Y. Peroxidase-like activity of chitosan stabilized silver nanoparticles for visual and colorimetric detection of glucose. Analyst 2012, 137, 5560–5564. [Google Scholar] [CrossRef]
- Lan, J.; Xu, W.; Wan, Q.; Zhang, X.; Lin, J.; Chen, J.; Chen, J. Colorimetric determination of sarcosine in urine samples of prostatic carcinoma by mimic enzyme palladium nanoparticles. Anal. Chim. Acta 2014, 825, 63–68. [Google Scholar] [CrossRef] [PubMed]
- Ma, M.; Zhang, Y.; Gu, N. Peroxidase-like catalytic activity of cubic Pt nanocrystals. Colloids Surf. A Physicochem. Eng. Asp. 2011, 373, 6–10. [Google Scholar] [CrossRef]
- Su, H.; Liu, D.-D.; Zhao, M.; Hu, W.-L.; Xue, S.-S.; Cao, Q.; Le, X.-Y.; Ji, L.-N.; Mao, Z.-W. Dual-enzyme characteristics of polyvinylpyrrolidone-capped iridium nanoparticles and their cellular protective effect against H2O2-induced oxidative damage. ACS Appl. Mater. Interfaces 2015, 7, 8233–8242. [Google Scholar] [CrossRef] [PubMed]
- Ye, H.; Mohar, J.; Wang, Q.; Catalano, M.; Kim, M.J.; Xia, X. Peroxidase-like properties of ruthenium nanoframes. Sci. Bull. 2016, 61, 1739–1745. [Google Scholar] [CrossRef]
- Jiang, T.; Song, Y.; Wei, T.; Li, H.; Du, D.; Zhu, M.-J.; Lin, Y. Sensitive detection of escherichia coli o157:H7 using Pt–Au bimetal nanoparticles with peroxidase-like amplification. Biosens. Bioelectron. 2016, 77, 687–694. [Google Scholar] [CrossRef]
- Jiang, T.; Song, Y.; Du, D.; Liu, X.; Lin, Y. Detection of p53 protein based on mesoporous Pt–Pd nanoparticles with enhanced peroxidase-like catalysis. ACS Sens. 2016, 1, 717–724. [Google Scholar] [CrossRef]
- Gao, Z.; Xu, M.; Lu, M.; Chen, G.; Tang, D. Urchin-like (gold core)@(platinum shell) nanohybrids: A highly efficient peroxidase-mimetic system for in situ amplified colorimetric immunoassay. Biosens. Bioelectron. 2015, 70, 194–201. [Google Scholar] [CrossRef]
- Kwon, E.Y.; Ruan, X.; Wang, L.; Lin, Y.; Du, D.; Van Wie, B.J. Mesoporous Pd@Pt nanoparticle-linked immunosorbent assay for detection of atrazine. Anal. Chim. Acta 2020, 1116, 36–44. [Google Scholar] [CrossRef]
- Fu, Z.; Zeng, W.; Cai, S.; Li, H.; Ding, J.; Wang, C.; Chen, Y.; Han, N.; Yang, R. Porous Au@Pt nanoparticles with superior peroxidase-like activity for colorimetric detection of spike protein of SARS-CoV-2. J. Colloid Interface Sci. 2021, 604, 113–121. [Google Scholar] [CrossRef] [PubMed]
- Ge, C.; Wu, R.; Chong, Y.; Fang, G.; Jiang, X.; Pan, Y.; Chen, C.; Yin, J.-J. Synthesis of Pt hollow nanodendrites with enhanced peroxidase-like activity against bacterial infections: Implication for wound healing. Adv. Funct. Mater. 2018, 28, 1801484. [Google Scholar] [CrossRef]
- Jiao, L.; Zhang, L.; Du, W.; Li, H.; Yang, D.; Zhu, C. Au@Pt nanodendrites enhanced multimodal enzyme-linked immunosorbent assay. Nanoscale 2019, 11, 8798–8802. [Google Scholar] [CrossRef] [PubMed]
- Jin, M.; Liu, H.; Zhang, H.; Xie, Z.; Liu, J.; Xia, Y. Synthesis of Pd nanocrystals enclosed by {100} facets and with sizes < 10 nm for application in CO oxidation. Nano Res. 2011, 4, 83–91. [Google Scholar]
- Gao, Z.; Lv, S.; Xu, M.; Tang, D. High-index {hk0} faceted platinum concave nanocubes with enhanced peroxidase-like activity for an ultrasensitive colorimetric immunoassay of the human prostate-specific antigen. Analyst 2017, 142, 911–917. [Google Scholar] [CrossRef] [PubMed]
- Gao, Z.; Ye, H.; Tang, D.; Tao, J.; Habibi, S.; Minerick, A.; Tang, D.; Xia, X. Platinum-decorated gold nanoparticles with dual functionalities for ultrasensitive colorimetric in vitro diagnostics. Nano Lett. 2017, 17, 5572–5579. [Google Scholar] [CrossRef] [PubMed]
- Gao, Z.; Song, Y.; Hsiao, T.Y.; He, J.; Wang, C.; Shen, J.; MacLachlan, A.; Dai, S.; Singer, B.H.; Kurabayashi, K.; et al. Machine-learning-assisted microfluidic nanoplasmonic digital immunoassay for cytokine storm profiling in COVID-19 patients. ACS Nano 2021, 15, 18023–18036. [Google Scholar] [CrossRef]
- Lu, N.; Wang, J.; Xie, S.; Brink, J.; McIlwrath, K.; Xia, Y.; Kim, M.J. Aberration corrected electron microscopy study of bimetallic Pd–Pt nanocrystal: Core–shell cubic and core–frame concave structures. J. Phys. Chem. C 2014, 118, 28876–28882. [Google Scholar] [CrossRef]
- Shi, Y.; Lyu, Z.; Zhao, M.; Chen, R.; Nguyen, Q.N.; Xia, Y. Noble-metal nanocrystals with controlled shapes for catalytic and electrocatalytic applications. Chem. Rev. 2021, 121, 649–735. [Google Scholar] [CrossRef]
- Wei, Z.; Xi, Z.; Vlasov, S.; Ayala, J.; Xia, X. Nanocrystals of platinum-group metals as peroxidase mimics for in vitro diagnostics. Chem. Commun. 2020, 56, 14962–14975. [Google Scholar] [CrossRef]
- Khoris, I.M.; Takemura, K.; Lee, J.; Hara, T.; Abe, F.; Suzuki, T.; Park, E.Y. Enhanced colorimetric detection of norovirus using in-situ growth of Ag shell on Au NPs. Biosens. Bioelectron. 2019, 126, 425–432. [Google Scholar] [CrossRef] [PubMed]
- Choleva, T.G.; Gatselou, V.A.; Tsogas, G.Z.; Giokas, D.L. Intrinsic peroxidase-like activity of rhodium nanoparticles, and their application to the colorimetric determination of hydrogen peroxide and glucose. Microchim. Acta 2018, 185, 22. [Google Scholar] [CrossRef] [PubMed]
- Xie, S.; Choi, S.-I.; Lu, N.; Roling, L.T.; Herron, J.A.; Zhang, L.; Park, J.; Wang, J.; Kim, M.J.; Xie, Z.; et al. Atomic layer-by-layer deposition of Pt on Pd nanocubes for catalysts with enhanced activity and durability toward oxygen reduction. Nano Lett. 2014, 14, 3570–3576. [Google Scholar] [CrossRef] [PubMed]
- Jiang, M.; Lim, B.; Tao, J.; Camargo, P.H.C.; Ma, C.; Zhu, Y.; Xia, Y. Epitaxial overgrowth of platinum on palladium nanocrystals. Nanoscale 2010, 2, 2406–2411. [Google Scholar] [CrossRef]
- Fang, P.-P.; Duan, S.; Lin, X.-D.; Anema, J.R.; Li, J.-F.; Buriez, O.; Ding, Y.; Fan, F.-R.; Wu, D.-Y.; Ren, B.; et al. Tailoring Au-core Pd-shell Pt-cluster nanoparticles for enhanced electrocatalytic activity. Chem. Sci. 2011, 2, 531–539. [Google Scholar] [CrossRef]
- Gao, Z.; Xu, M.; Hou, L.; Chen, G.; Tang, D. Irregular-shaped platinum nanoparticles as peroxidase mimics for highly efficient colorimetric immunoassay. Anal. Chim. Acta 2013, 776, 79–86. [Google Scholar] [CrossRef]
- He, W.; Liu, Y.; Yuan, J.; Yin, J.-J.; Wu, X.; Hu, X.; Zhang, K.; Liu, J.; Chen, C.; Ji, Y.; et al. Au@Pt nanostructures as oxidase and peroxidase mimetics for use in immunoassays. Biomaterials 2011, 32, 1139–1147. [Google Scholar] [CrossRef]
- Gao, Z.; Tang, D.; Tang, D.; Niessner, R.; Knopp, D. Target-induced nanocatalyst deactivation facilitated by core@shell nanostructures for signal-amplified headspace-colorimetric assay of dissolved hydrogen sulfide. Anal. Chem. 2015, 87, 10153–10160. [Google Scholar] [CrossRef]
- Xia, X.; Zhang, J.; Lu, N.; Kim, M.J.; Ghale, K.; Xu, Y.; McKenzie, E.; Liu, J.; Ye, H. Pd–Ir core–shell nanocubes: A type of highly efficient and versatile peroxidase mimic. ACS Nano 2015, 9, 9994–10004. [Google Scholar] [CrossRef]
- Liu, X.; Wang, Q.; Zhao, H.; Zhang, L.; Su, Y.; Lv, Y. BSA-templated MnO2 nanoparticles as both peroxidase and oxidase mimics. Analyst 2012, 137, 4552–4558. [Google Scholar] [CrossRef]
- Wan, S.; Wang, Q.; Ye, H.; Kim, M.J.; Xia, X. Pd–Ru bimetallic nanocrystals with a porous structure and their enhanced catalytic properties. Part. Part. Syst. Charact. 2018, 35, 1700386. [Google Scholar] [CrossRef]
- Ye, H.; Liu, Y.; Chhabra, A.; Lilla, E.; Xia, X. Polyvinylpyrrolidone (PVP)-capped Pt nanocubes with superior peroxidase-like activity. ChemNanoMat 2017, 3, 33–38. [Google Scholar] [CrossRef]
- Davidson, E.; Xi, Z.; Gao, Z.; Xia, X. Ultrafast and sensitive colorimetric detection of ascorbic acid with Pd-Pt core-shell nanostructure as peroxidase mimic. Sens. Int. 2020, 1, 100031. [Google Scholar] [CrossRef]
- Lim, B.; Jiang, M.; Camargo, P.H.C.; Cho, E.C.; Tao, J.; Lu, X.; Zhu, Y.; Xia, Y. Pd-Pt bimetallic nanodendrites with high activity for oxygen reduction. Science 2009, 324, 1302–1305. [Google Scholar] [CrossRef] [PubMed]
- Avvakumova, S.; Colombo, M.; Galbiati, E.; Mazzucchelli, S.; Rotem, R.; Prosperi, D. Chapter 6—Bioengineered approaches for site orientation of peptide-based ligands of nanomaterials. In Biomedical Applications of Functionalized Nanomaterials; Sarmento, B., das Neves, J., Eds.; Elsevier: Amsterdam, The Netherlands, 2018; pp. 139–169. [Google Scholar]
- Kuzuya, A.; Numajiri, K.; Kimura, M.; Komiyama, M. Single-molecule accommodation of streptavidin in nanometer-scale wells formed in DNA nanostructures. Nucleic Acids Symp. Ser. 2008, 52, 681–682. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharjee, S. DLS and zeta potential—What they are and what they are not? J. Control. Release 2016, 235, 337–351. [Google Scholar] [CrossRef]
- Mukhopadhyay, A.; Joshi, N.; Chattopadhyay, K.; De, G. A facile synthesis of PEG-coated magnetite (Fe3O4) nanoparticles and their prevention of the reduction of cytochrome C. ACS Appl. Mater. Interfaces 2012, 4, 142–149. [Google Scholar] [CrossRef] [PubMed]
- Leopold, L.F.; Tódor, I.S.; Diaconeasa, Z.; Rugină, D.; Ştefancu, A.; Leopold, N.; Coman, C. Assessment of PEG and BSA-PEG gold nanoparticles cellular interaction. Colloids Surf. A 2017, 532, 70–76. [Google Scholar] [CrossRef]
- Wang, C.; Huang, C.; Gao, Z.; Shen, J.; He, J.; MacLachlan, A.; Ma, C.; Chang, Y.; Yang, W.; Cai, Y.; et al. Nanoplasmonic sandwich immunoassay for tumor-derived exosome detection and exosomal PD-L1 profiling. ACS Sens. 2021, 6, 3308–3319. [Google Scholar] [CrossRef]
| Catalyst | [E] (M) | Substance | Km (M) | Vmax (M s−1) | Kcat (s−1) |
|---|---|---|---|---|---|
| Pd@Pt NDs | 2.44 × 10−14 | TMB | 6.63 × 10−4 | 2.21 × 10−7 | 9.06 × 106 |
| 2.44 × 10−14 | H2O2 | 3.82 × 100 | 1.86 × 10−7 | 7.62 × 106 | |
| HRP [30] | 2.5 × 10−11 | TMB | 4.3 × 10−4 | 1.0 × 10−7 | 4.0 × 103 |
| 2.5 × 10−11 | H2O2 | 3.7 × 10−3 | 8.7 × 10−8 | 3.5 × 103 |
| Sample No. | IL-6 Level Spiked (pg mL−1) | IL-6 Level Meaured (pg mL−1) | CV (%, n = 12) | Recovery (%) |
|---|---|---|---|---|
| 1 | 0.2 | 0.19 | 9.96 | 94.69 |
| 2 | 0.5 | 0.47 | 7.55 | 93.65 |
| 3 | 1 | 0.98 | 5.60 | 97.55 |
| 4 | 2 | 1.92 | 6.70 | 96.12 |
| 5 | 5 | 5.28 | 8.86 | 105.53 |
| 6 | 10 | 9.97 | 8.34 | 99.74 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gao, Z.; Wang, C.; He, J.; Chen, P. Pd@Pt Nanodendrites as Peroxidase Nanomimics for Enhanced Colorimetric ELISA of Cytokines with Femtomolar Sensitivity. Chemosensors 2022, 10, 359. https://doi.org/10.3390/chemosensors10090359
Gao Z, Wang C, He J, Chen P. Pd@Pt Nanodendrites as Peroxidase Nanomimics for Enhanced Colorimetric ELISA of Cytokines with Femtomolar Sensitivity. Chemosensors. 2022; 10(9):359. https://doi.org/10.3390/chemosensors10090359
Chicago/Turabian StyleGao, Zhuangqiang, Chuanyu Wang, Jiacheng He, and Pengyu Chen. 2022. "Pd@Pt Nanodendrites as Peroxidase Nanomimics for Enhanced Colorimetric ELISA of Cytokines with Femtomolar Sensitivity" Chemosensors 10, no. 9: 359. https://doi.org/10.3390/chemosensors10090359
APA StyleGao, Z., Wang, C., He, J., & Chen, P. (2022). Pd@Pt Nanodendrites as Peroxidase Nanomimics for Enhanced Colorimetric ELISA of Cytokines with Femtomolar Sensitivity. Chemosensors, 10(9), 359. https://doi.org/10.3390/chemosensors10090359








