Simultaneous Analysis of MLH1, MSH2, MSH6, PMS2 and KRAS in Patients with Gastric and Colon Cancer Using Stochastic Sensors
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals
2.2. Electrochemical Exfoliation of Graphite Rods
2.3. Instruments
2.4. The Stochastic Sensors’ Design
2.5. Procedure of Analysis: The Stochastic Method
2.6. Biological Samples
3. Results and Discussion
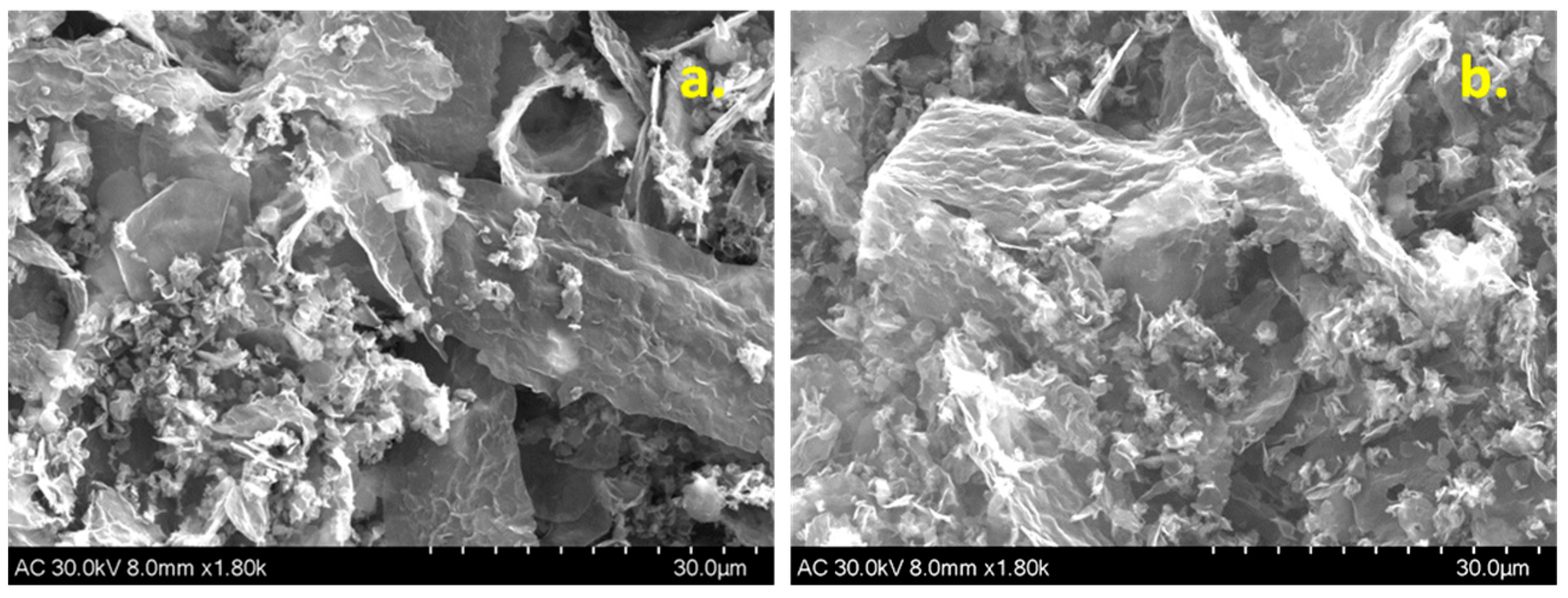
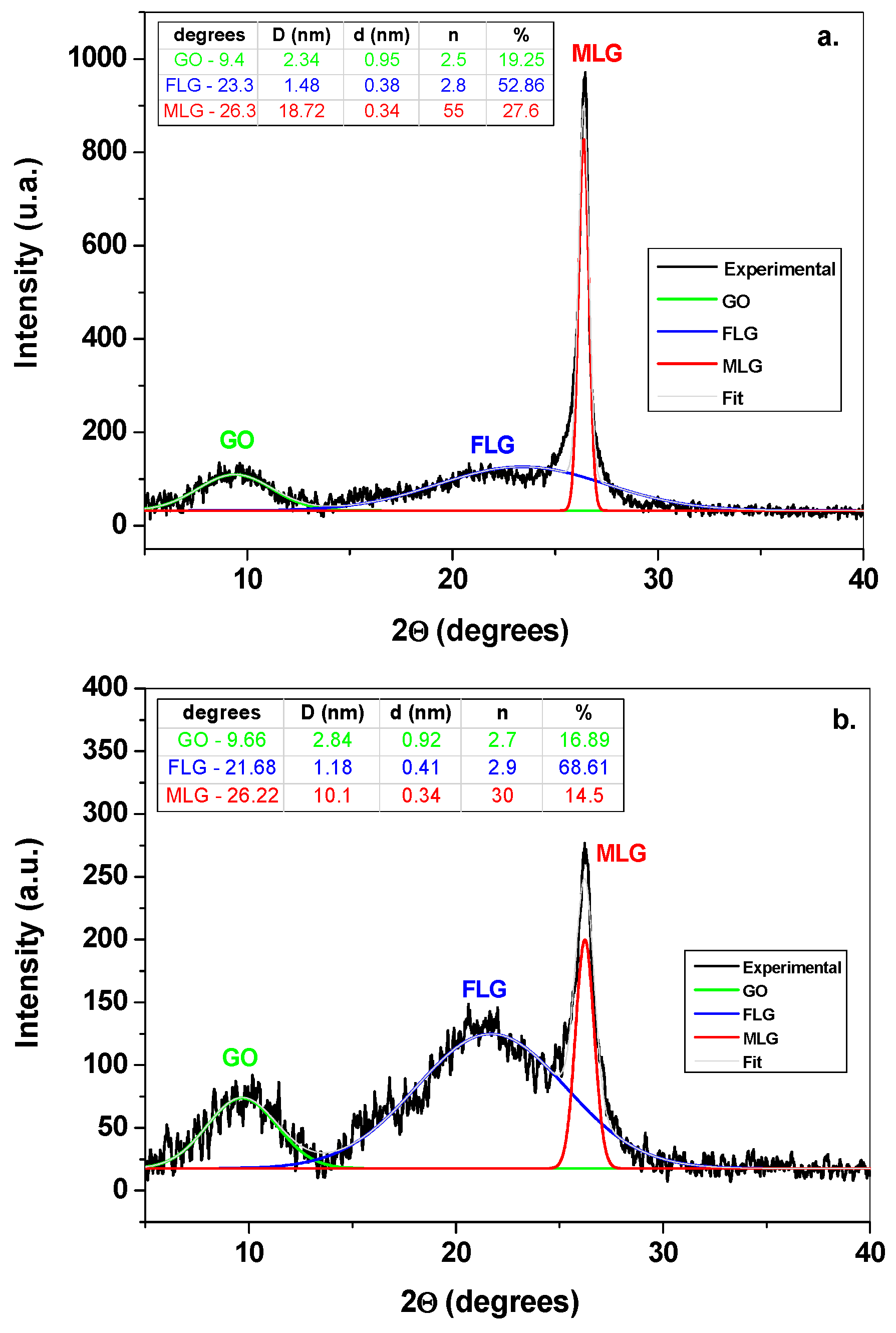
3.1. Morphological and Structural Characterization of Graphene Samples
3.2. Performance Characteristics
3.3. Reproducibility and Stability of the Stochastic Sensors
3.4. Selectivity
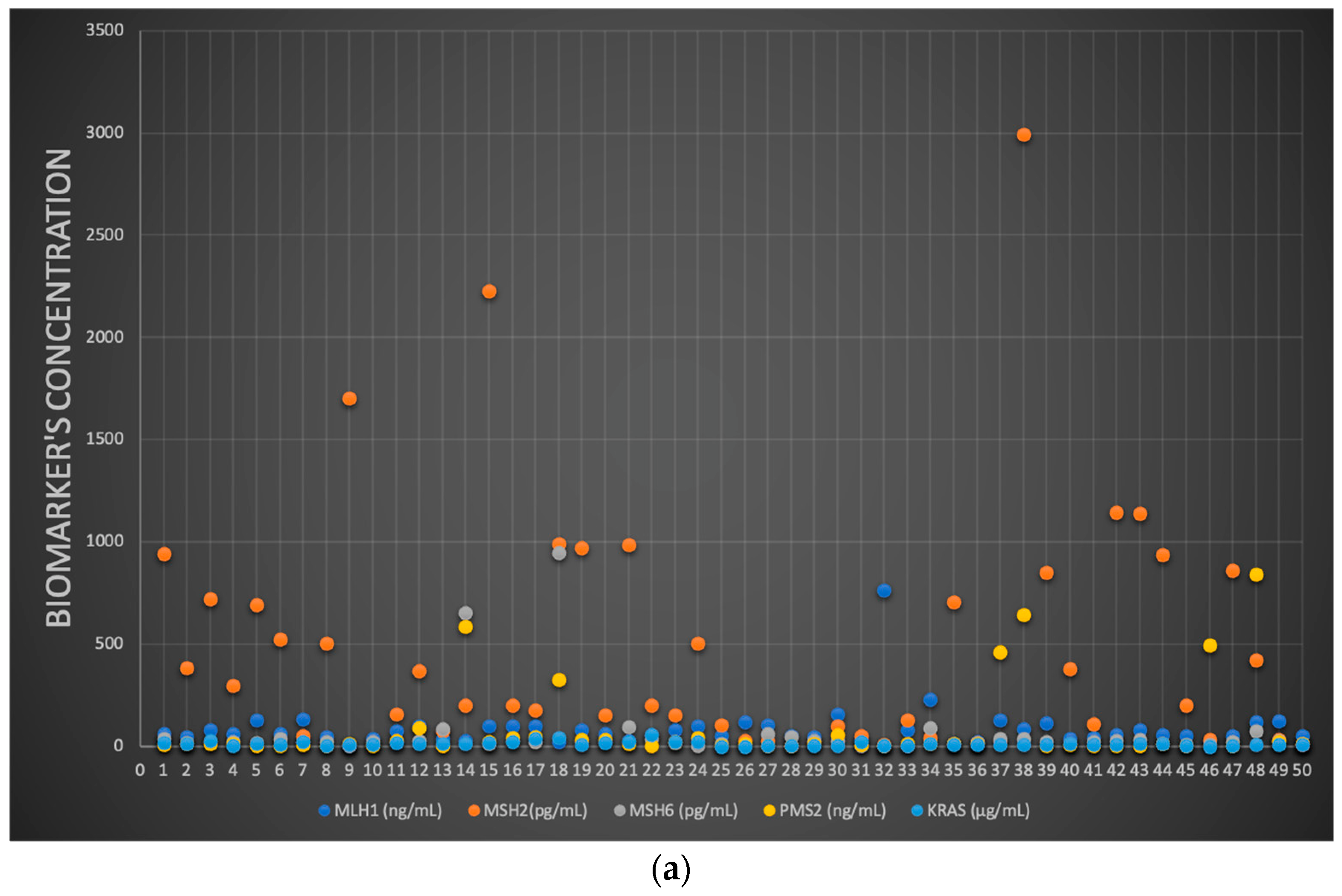
3.5. MLH1, MSH2, MSH6, PMS2 and KRAS: Molecular Identification and Quantification
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Baretti, M.; Le, D.T. DNA mismatch repair in cancer. Pharmacol. Ther. 2018, 189, 45–62. [Google Scholar] [CrossRef]
- Greenson, J.K.; Huang, S.C.; Herron, C.; Moreno, V.; Bonner, J.D.; Tomsho, L.P.; Ben-Izhak, O.; Cohen, H.I.; Trougouboff, P.; Bejhar, J.; et al. Pathologic predictors of microsatellite instability in colorectal cancer. Am. J. Surg. Pathol. 2009, 33, 126–133. [Google Scholar] [CrossRef] [PubMed]
- Hampel, H.; Frankel, W.L.; Martin, E.; Arnold, M.; Khanduja, K.; Kuebler, P.; Clendenning, M.; Sotamaa, K.; Prior, T.; Westman, J.A.; et al. Feasibility of screening for lynch syndrome among patients with colorectal cancer. J. Clin. Oncol. 2008, 26, 5783–5788. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Keijzers, G.; Rasmussen, L.J. DNA mismatch repair and its many roles in eukaryotic cells. Mutat. Res. 2017, 773, 174–187. [Google Scholar] [CrossRef] [PubMed]
- Miceli, R.; An, J.; Di Bartolomeo, M.; Morano, F.; Kim, S.T.; Park, S.H.; Choi, M.G.; Lee, J.H.; Raimondi, A.; Fucá, G.; et al. Prognostic Impact of Microsatellite Instability in Asian Gastric Cancer Patients Enrolled in the ARTIST. Trial. Oncol. 2019, 97, 38–43. [Google Scholar] [CrossRef] [PubMed]
- Di Bartolomeo, M.; Morano, F.; Raimondi, A.; Miceli, R.; Corallo, S.; Tamborini, E.; Perrone, F.; Antista, M.; Niger, M.; Pellegrinelli, A.; et al. Prognostic and Predictive Value of Microsatellite Instability, Inflammatory Reaction and PD-L1 in Gastric Cancer Patients Treated with Either Adjuvant 5-FU/LV or Sequential FOLFIRI Followed by Cisplatin and Docetaxel: A Translational Analysis from the ITA. Oncologist 2020, 25, e460–e468. [Google Scholar] [CrossRef]
- Wright, C.L.; Stewart, I.D. Histopathology and mismatch repair status of 458 consecutive colorectal carcinomas. Am. J. Surg. Pathol. 2003, 27, 1393–1406. [Google Scholar] [CrossRef] [PubMed]
- Jover, R.; Paya, A.; Alenda, C.; Poveda, M.J.; Peiró, G.; Aranda, F.I.; Pérez-Mateo, M. Defective mismatch-repair colorectal cancer: Clinicopathologic characteristics and usefulness of immunohistochemical analysis for diagnosis. Am. J. Clin. Pathol. 2004, 122, 389–394. [Google Scholar] [CrossRef]
- Shia, J. Immunohistochemistry versus microsatellite instability testing for screening colorectal cancer patients at risk for hereditary nonpolyposis colorectal cancer syndrome. Part, I. The utility of immunohistochemistry. J. Mol. Diagn. 2008, 10, 293–300. [Google Scholar] [CrossRef]
- Gremer, L.; Merbitz-Zahradnik, T.; Dvorsky, R.; Cirstea, I.C.; Kratz, C.P.; Zenker, M.; Wittinghofer, A.; Ahmadian, M.R. Germline KRAS mutations cause aberrant biochemical and physical properties leading to developmental disorders. Hum. Mutat. 2011, 32, 33–43. [Google Scholar] [CrossRef] [Green Version]
- Iyer, R.R.; Pluciennik, A.; Burdett, V.; Modrich, P.L. DNA mismatch repair: Functions and mechanisms. Chem. Rev. 2006, 106, 302–323. [Google Scholar] [CrossRef] [PubMed]
- Larrea, A.A.; Lujan, S.A.; Kunkel, T.A. SnapShot: DNA mismatch repair. Cell 2010, 141, 730.e1. [Google Scholar] [CrossRef] [PubMed]
- Stefan-van Staden, R.I.; Popa-Tudor, I. Molecular Recognition of C-Reactive Protein, Adiponectin and Zn2+ in Serum Samples. J. Electrochem. Soc. 2019, 166, B3051. [Google Scholar] [CrossRef]
- Stefan-van Staden, R.I.; Bogea, M.I.; Ilie-Mihai, R.M.; Gheorghe, D.C.; Aboul-Enein, H.Y.; Coros, M.; Pruneanu, S.M. N,S-Decorated graphenes modified with 2,3,7,8,12,13,17,18-octaethyl-21H,23H-porphine manganese(III) chloride-based 3D needle stochastic sensors for enantioanalysis of arginine: A key factor in the metabolomics and early detection of gastric cancer. Anal. Bioanal. Chem. 2022, 414, 6521–6530. [Google Scholar] [CrossRef] [PubMed]
- Stefan-van Staden, R.-I.S.; Comnea-Stancu, I.R.; Yanık, H.; Göksel, M.; Alexandru, A.; Durmuş, M. Phthalocyanine-BODIPY Dye: Synthesis, Characterization, and Utilization for Pattern Recognition of CYFRA 21-1 in Whole Blood Samples. Anal. Bioanal. Chem. 2017, 409, 6195–6203. [Google Scholar] [CrossRef] [PubMed]
- Stefan-van Staden, R.I.; Ilie-Mihai, R.M.; Pogacean, F.; Pruneanu, S.M. Needle stochastic sensors for on-site fast recognition and quantification of biomarkers for gastric cancer in biological samples. New J. Chem. 2020, 44, 20203–20211. [Google Scholar] [CrossRef]
- Ilie-Mihai, R.M.; Stefan-van Staden, R.I.; Magerusan, L.; Coros, M.; Pruneanu, S. Enantioanalysis of Tryptophan in Whole Blood Samples Using Stochastic Sensors-A Screening Test for Gastric Cancer. Chirality 2020, 32, 215–222. [Google Scholar] [CrossRef]
- Macchia, E.; Sarcina, L.; Driescher, C.; Gounani, Z.; Tewari, A.; Osterbacka, R.; Palazzo, G.; Tricase, A.; Kovacs Vajna, Z.M.; Viola, F.; et al. Single-molecule bioelectronic label-free assay of both protein and genomic markers of pancreatic mucinous cysts in whole blood serum. Adv. Electron. Mater. 2021, 7, 2100304. [Google Scholar] [CrossRef]
- Stefan-van Staden, R.I.; Ilie-Mihai, R.M.; Gugoasa, L.A.; Stanciu-Gavan, C. Pattern recognition of p53 and KRAS in whole blood samples. J. Electrochem. Soc. 2019, 166, B183–B186. [Google Scholar] [CrossRef]
- Sciammarella, C.; Beneivenga, M.; Mafficini, A.; Piredda, M.; Tsvetkova, V.; Paolino, G.; Mastrosimini, M.; Hetoja, S.; de Manzoni, G.; Mattiolo, P.; et al. Molecular analysis of an intestinal neuroendocrine/non-neuroendocrine neoplasm (MiNEN) reveals MLH1 methylation-driven microsatellite instability and a monoclonal origin: Diagnostic and clinical implications. Appl. Immunohistochem. Molec. Morphol. 2022, 30, 145–152. [Google Scholar] [CrossRef]
- Saliani, M.; Mirzaiebadizi, A.; Javadmanesh, A.; Siavoshi, A.; Ahmadian, M.R. KRAS-related long noncoding RNAs in human cancers. Cancer Gene Ther. 2022, 29, 418–427. [Google Scholar] [CrossRef] [PubMed]
- Ulreich, K.; Firnau, M.B.; Tagscherer, N.; Beyer, S.; Ackermann, A.; Plotz, G.; Brieger, A. High expression of casein kinase2 alpha is responsible for enhanced phosphorylation of DNA mismatch repair protein MLH1 and increased tumor mutation rates in colorectal cancer. Cancers 2022, 14, 1553. [Google Scholar] [CrossRef] [PubMed]
- Padin-Iruegas, E.; Chamorro-Petromacci, C.M.; Sines-Cajade, I.; Lorezo-Pouso, A.I.; Blanco-Carrion, A.; Perez-Jardon, A.; Gaudara-Vila, P.; Perez-Sayans, M. DNA methylation by bisulfite next-generation sequencing for MLH1 and MGMT in oral squamos cell carcinomas and potentially malignant disorders: An integrative analysis towards field cancerization. Medicina 2022, 58, 878. [Google Scholar] [CrossRef] [PubMed]
- Ijsselsteijn, R.; van Hees, S.; Drost, M.; Jansen, J.G.; de Wind, N. Induction of mismatch repair deficiency, compromised DNA damage signaling and compound hypermutagenesis by a dietary mutagen in a cell-based model for Lynch syndrome. Carcinogenesis 2022, 43, 160–169. [Google Scholar] [CrossRef] [PubMed]
- Salari, S.; Ghadyani, M.; Karimi, M.; Mortezazadeh, M.; Vahedifard, F. Immunohistochemical expression pattern of MLH1, MSH2, MSH6, and PMS2 in tumor specimen of Iranian gastric carcinoma patients. J. Gastroint. Cancer 2022, 53, 192–196. [Google Scholar] [CrossRef]
- Herz, A.L.; Wisser, S.; Kohlruss, M.; Slotta-Huspenina, J.; Jesinghaus, M.; Grosser, B.; Steiger, K.; Novotni, A.; Hapfelmeier, A.; Schmidt, T.; et al. Elevated microsatellite instability at selected tetranucleotide (EMAST) repeats in gastric cancer: A distinct microsatellite instability type with potential clinical impact? J. Pathol. Clin. Res. 2022, 8, 233–244. [Google Scholar] [CrossRef]
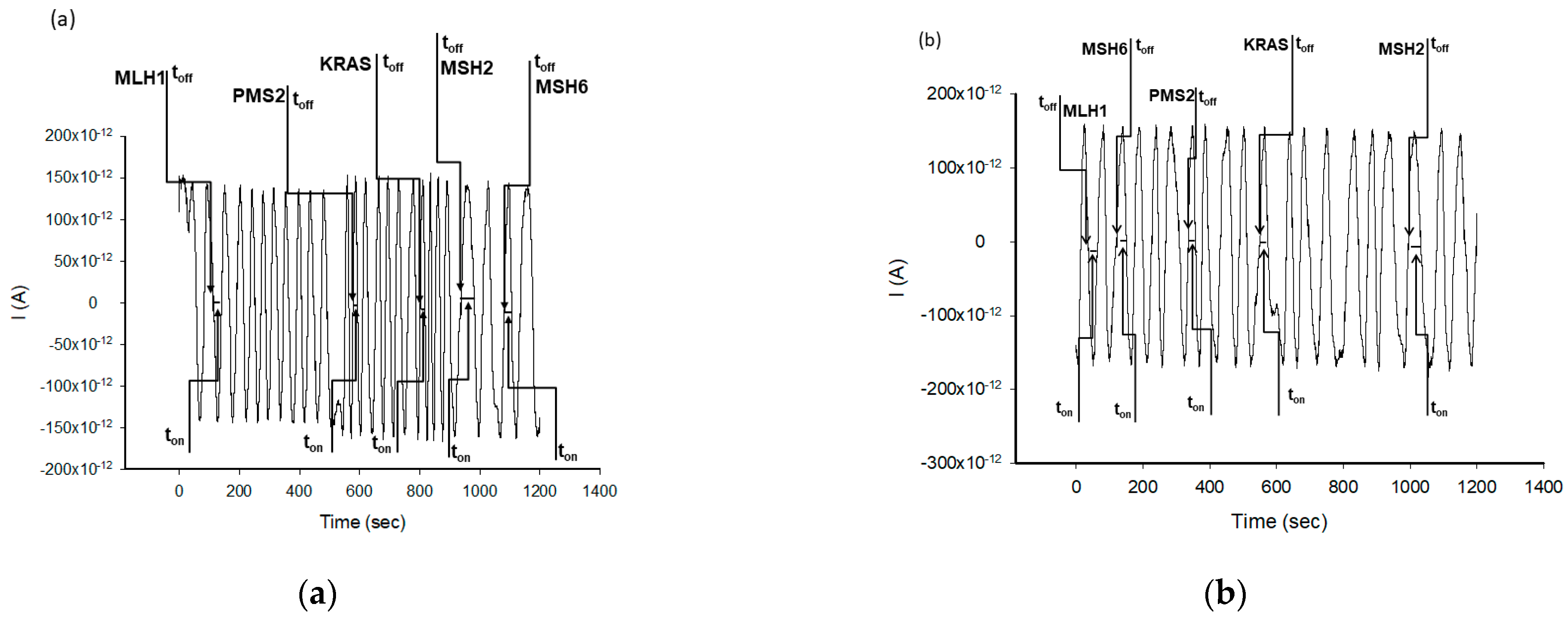


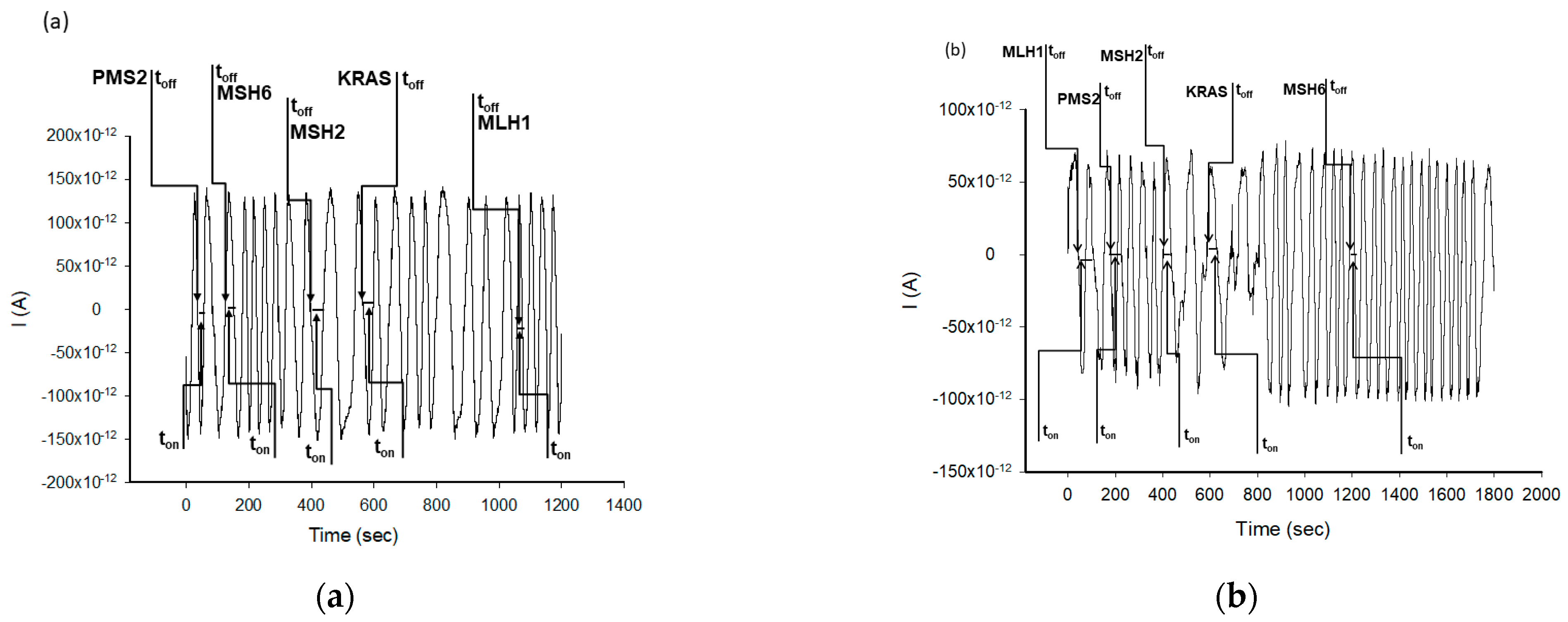



| Stochastic Sensors | Working Concentration Range (g mL−1) | Calibration Equation and r 1 | Signature, toff (s) | Sensitivity (s−1/μg mL−1) | Limit of Determination (g mL−1) |
|---|---|---|---|---|---|
| MLH1 | |||||
| MD/EGR-1 | 3.2 × 10−16−3.2 × 10−8 | 1/ton = 0.03 + 5.31 × 105 C r = 0.9999 | 4.2 | 5.31 × 105 | 3.2 × 10−16 |
| MD/EGR-2 | 3.2 × 10−12−3.2 × 10−9 | 1/ton = 0.03 + 1.21 × 102 C r = 0.9994 | 1.0 | 1.21 × 102 | 3.2 × 10−12 |
| MSH2 | |||||
| MD/EGR-1 | 1.0 × 10−12−1.0 × 10−8 | 1/ton = 0.03 + 2.11 × 10 C r = 0.9998 | 3.3 | 2.11 × 10 | 1.0 × 10−12 |
| MD/EGR-2 | 1.0 × 10−12−1.0 × 10−8 | 1/ton = 0.04 + 4.57 × 10 C r = 0.9999 | 1.2 | 4.57 × 10 | 1.0 × 10−12 |
| MSH6 | |||||
| MD/EGR-1 | 2.3−10−13−2.3 × 10−5 | 1/ton = 0.01 + 743.16 C r = 0.9999 | 2.6 | 743.16 | 2.3 × 10−13 |
| MD/EGR-2 | 2.3−10−15−2.3 × 10−9 | 1/ton = 0.02 + 2.19 × 105 C r = 0.9999 | 4.6 | 2.19 × 105 | 2.3 × 10−15 |
| PMS2 | |||||
| MD/EGR-1 | 2.7−10−10−2.7 × 10−8 | 1/ton = 0.04 + 8.85 × 10−1 C r = 0.9999 | 1.3 | 8.85 × 10−1 | 2.7 × 10−10 |
| MD/EGR-2 | 2.7−10−12−2.7 × 10−5 | 1/ton = 0.02 + 1.35 × 102 C r = 0.9991 | 2.0 | 1.35 × 102 | 2.7 × 10−12 |
| KRAS | |||||
| MD/EGR-1 | 2.2 × 10−8−2.2 × 10−5 | 1/ton = 0.03 + 2.46 × 10−3 C r = 0.9999 | 2.0 | 2.46 × 10−3 | 2.2 × 10−8 |
| MD/EGR-2 | 2.2 × 10−9−2.2 × 10−6 | 1/ton = 0.04 + 2.39 × 10−2 C r = 0.9992 | 1.4 | 2.39 × 10−2 | 2.2 × 10−9 |
| Stochastic Sensors Modified with MD and Based on | MLH1 Signature (s) | MSH2 Signature (s) | MSH6 Signature (s) | PMS2 Signature (s) | KRAS Signature (s) | CA19-9 Signature (s) | p53 Signature (s) | Glutamine Signature (s) | CA72-4 Signature (s) |
|---|---|---|---|---|---|---|---|---|---|
| EGR-1 | 4.2 | 3.3 | 2.6 | 1.3 | 2.0 | 0.5 | 3.0 | 1.7 | 2.8 |
| EGR-2 | 1.0 | 1.2 | 4.6 | 2.0 | 1.4 | 0.7 | 2.6 | 0.7 | 3.3 |
| Stochastic Sensors Modified with MD and Based on | Recovery % | ||||
|---|---|---|---|---|---|
| MLH-1 | MSH-2 | MSH-6 | PMS-2 | KRAS | |
| Whole blood | |||||
| EGR-1 | 99.21 ± 0.02 | 99.15 ± 0.03 | 99.73 ± 0.01 | 99.77 ± 0.05 | 98.99 ± 0.02 |
| EGR-2 | 99.13 ± 0.01 | 99.98 ± 0.02 | 99.03 ± 0.02 | 99.10 ± 0.02 | 99.87 ± 0.03 |
| Saliva | |||||
| EGR-1 | 99.15 ± 0.01 | 99.24 ± 0.02 | 99.12 ± 0.01 | 99.20 ± 0.02 | 99.03 ± 0.05 |
| EGR-2 | 99.43 ± 0.03 | 99.57 ± 0.02 | 99.70 ± 0.08 | 98.59 ± 0.07 | 98.98 ± 0.02 |
| Urine | |||||
| EGR-1 | 98.21 ± 0.03 | 99.01 ± 0.02 | 98.42 ± 0.03 | 98.21 ± 0.02 | 98.32 ± 0.03 |
| EGR-2 | 98.18 ± 0.05 | 98.55 ± 0.03 | 99.00 ± 0.02 | 98.11 ± 0.02 | 98.14 ± 0.02 |
| Tumoral tissue | |||||
| EGR-1 | 99.99 ± 0.02 | 99.95 ± 0.03 | 99.51 ± 0.02 | 99.87 ± 0.01 | 99.90 ± 0.02 |
| EGR-2 | 99.91 ± 0.02 | 99.00 ± 0.01 | 99.44 ± 0.03 | 99.00 ± 0.02 | 99.69 ± 0.07 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gheorghe, D.-C.; Stefan-van Staden, R.-I.; Pogacean, F.; Pruneanu, S. Simultaneous Analysis of MLH1, MSH2, MSH6, PMS2 and KRAS in Patients with Gastric and Colon Cancer Using Stochastic Sensors. Chemosensors 2022, 10, 380. https://doi.org/10.3390/chemosensors10100380
Gheorghe D-C, Stefan-van Staden R-I, Pogacean F, Pruneanu S. Simultaneous Analysis of MLH1, MSH2, MSH6, PMS2 and KRAS in Patients with Gastric and Colon Cancer Using Stochastic Sensors. Chemosensors. 2022; 10(10):380. https://doi.org/10.3390/chemosensors10100380
Chicago/Turabian StyleGheorghe, Damaris-Cristina, Raluca-Ioana Stefan-van Staden, Florina Pogacean, and Stela Pruneanu. 2022. "Simultaneous Analysis of MLH1, MSH2, MSH6, PMS2 and KRAS in Patients with Gastric and Colon Cancer Using Stochastic Sensors" Chemosensors 10, no. 10: 380. https://doi.org/10.3390/chemosensors10100380
APA StyleGheorghe, D.-C., Stefan-van Staden, R.-I., Pogacean, F., & Pruneanu, S. (2022). Simultaneous Analysis of MLH1, MSH2, MSH6, PMS2 and KRAS in Patients with Gastric and Colon Cancer Using Stochastic Sensors. Chemosensors, 10(10), 380. https://doi.org/10.3390/chemosensors10100380










