1. Introduction
Animal manure has traditionally been used as a fertilizer to improve soil fertility and carbon sequestration, as well as support crop growth [
1,
2]. It contains essential nutrients including nitrogen (N), phosphorus (P), and potassium (K), which can improve soil quality and, thus, influence agronomic activities such as soil and crop management [
3]. Most chemical nutrients existing in fertilizers are normally present in their oxide form; thus, P is present as phosphorus pentoxide (P
2O
5), K as potassium oxide (K
2O), calcium (Ca) as calcium oxide (CaO), and magnesium (Mg) as magnesium oxide (MgO), among others.
The presence of the aforementioned chemical nutrients has profound effects in plants. Plant organic structure, physiological makeup, biomass synthesis, and distribution, for example, are affected by N and its deficiency hampers the structure and function of photosynthesis, which in turn lowers crop yield. Insufficient P has a similar effect, significantly reducing leaf area, impairing photosynthesis and carbon metabolism, and, thereby, restricting tillering, biomass buildup, and crop yield. On the other hand, K controls membrane protein transport, steady-state enzyme activation, charge balance, stomatal movement, and osmotic regulation [
4]. Other nutrients such as carbon (C), magnesium (Mg), and sulfur (S) are also present in manure. Despite the importance of these chemical nutrients in the proper growth and functioning of plants, built-up and/or excess application of manure containing these nutrients can also be harmful to the environment and poses health risks [
5]. For example, nearly 10% of all direct emissions of greenhouse gases, including methane, nitrous oxide, and carbon dioxide, from agricultural production are caused by direct application of manure to farmlands [
6]. Manure application can also lead to soil/groundwater contamination by heavy metals such as zinc and copper. In the United States, approximately 1.4 billion tons of manure are generated by the 9.8 billion heads of poultry and livestock produced yearly [
7], implying that there is a need for proper manure management processes. To reduce greenhouse gas emissions and effectively employ the nutrients in manure for soil improvement, processing systems need to be based on a thorough analysis of these nutrients [
8].
Various spectroscopic methods have been utilized by researchers to examine manure chemical properties including using inductively coupled plasma-atomic emission spectroscopy (ICP-AES), atomic absorption spectroscopy (AAS) [
9], ultraviolet-visible (UV-Vis) spectroscopy, fluorescence excitation-emission matrix spectroscopy, Fourier-transform infrared spectroscopy, pyrolysis-mass spectrometry, solid state
13C nuclear magnetic resonance (NMR) spectroscopy [
10], solution NMR, X-ray absorption near-edge structure (XANES) spectroscopy [
11], and near-infrared (NIR) spectroscopy [
12]. However, each of these techniques has advantages and disadvantages. Solution
31P NMR, for instance, can provide relevant data on the organic P forms in animal manures but not on the inorganic P solid phases. Organic and mineral P fractions of manure have also been identified using XANES spectroscopy. Since no liquid extraction is required, this method has an advantage over solution NMR [
11]. In comparison to AES, AAS is more specific for some elements, coupled with the relative ease of its use. As for the disadvantage, AAS is not appropriate for P analysis, and only one element can be studied during each run. ICP-AES, on the other hand, enables rapid multi-element analysis. For many elements, its detection limits are comparable to or lower than those of AAS; nevertheless, compared to AAS, the costs associated with its initial acquisition, use, and maintenance are higher [
9]. In general, the use of the abovementioned methods for assessment of various chemical nutrients present in animal manure require significant time and other resources (e.g., training, finances, etc.) to carry out the analyses.
Present analytical laboratories typically employ traditional standard wet chemical analytical techniques such as distillation, Kjeldahl, colorimetry using an auto-analyzer, combustion, and microwave-assisted digestion for the analysis of chemical constituents present in animal manure [
9]. However, such processes are relatively expensive and time-consuming, making them unsuitable for rapid analysis. Because animal manure is heterogeneous and certain chemical constituents in animal manure are volatile, a rapid, low-cost, and accurate analysis at the time of application is required. Physicochemical modeling and NIR spectroscopy have proven to be such rapid evaluation methods [
13,
14]. Animal manure contains two types of chemical constituents that can be accurately predicted using NIR spectroscopy. The first type is made up of chemical constituents that contain chemical bonds that contribute to NIR spectral absorption. Moisture, organic matter, dry matter, C, and N, for example, can be assigned directly to main NIR absorption bands such as N-H, C-H, and O-H. The NIR technique as applied to these constituents in animal manure has produced satisfactory predictions in most previous studies. The second type consists of chemical constituents that are not spectrally active but correlate well with the first type. Due to their correlations with dry matter content, P and Mg without spectral absorption bonds could be quantitatively analyzed using NIR spectroscopy [
15]. Even though NIR spectroscopy has been demonstrated to be a reliable technique in manure nutrient analysis, sensing systems require periodic maintenance because of large variations in manure, which can be costly, time-consuming, and technically challenging. Furthermore, because the NIR analysis of manure compositions is based on the relationship between nutrient concentrations and spectral data, it is unknown whether the changes in the NIR spectral data are driven by direct variation in N concentration or indirect variation in other manure constituents [
14]. As such, the development of a robust and reliable calibration model using chemometrics and various other machine learning techniques is necessary.
NIR spectroscopy and chemometrics have been established as reliable techniques for qualitative and quantitative investigations in a variety of industries, including agriculture, food, and oil, among others. NIR spectroscopy is efficient, affordable, and non-destructive, and has become popular because of advancements in computers and the development of new mathematical methods allowing data processing. However, deciphering NIR spectra can be challenging, and chemometrics is useful for extracting and aiding with the interpretation of the acquired data [
16]. The most commonly used chemometric techniques include principal component analysis for qualitative analysis of NIR spectral data and partial least squares (PLS) regression for quantitative prediction of sample parameters of interest [
17]. Least absolute shrinkage and selection operator (LASSO) regression, ridge regression (RIDGE), least angle regression, random forest (RF), and forward stagewise or stepwise regression are other rarely used regression techniques for processing spectral data [
12]. In addition to these machine learning techniques, various signal preprocessing methods such as mean centering, multiplicative scatter correction, Savitzky–Golay smoothing with first and second derivation, and their combinations were also used in processing NIR spectra data prior to the implementation of the aforementioned machine learning techniques [
14]. Other authors have also reported the use of other preprocessing techniques such as standard normal variable (SNV) and de-trending (DT), as well as SNV-DT ratio [
18,
19,
20]
The majority of the existing literature investigated spectral data from various NIR systems to analyze only a single nutrient’s concentration in manure. For example, NIR-sensing systems with reflectance and transflectance modes coupled with PLS were employed in the prediction of N speciation in dairy cow manure using a spiking method [
14]. In another study, Devianti et al. also applied principal component regression and NIR spectroscopy to exclusively determine N content as a quality parameter of organic fertilizer [
19] and P content in organic fertilizer [
20]. The metal nutrient contents of animal manure compost produced in China on fresh and dried weight basis, on the other hand, was previously investigated using PLS and NIR [
18]. In another recent study, particle swarm optimization and two multiple stacked generalizations were used to assess the quantity of N and organic matter in manure using Vis-NIR spectroscopy [
21].
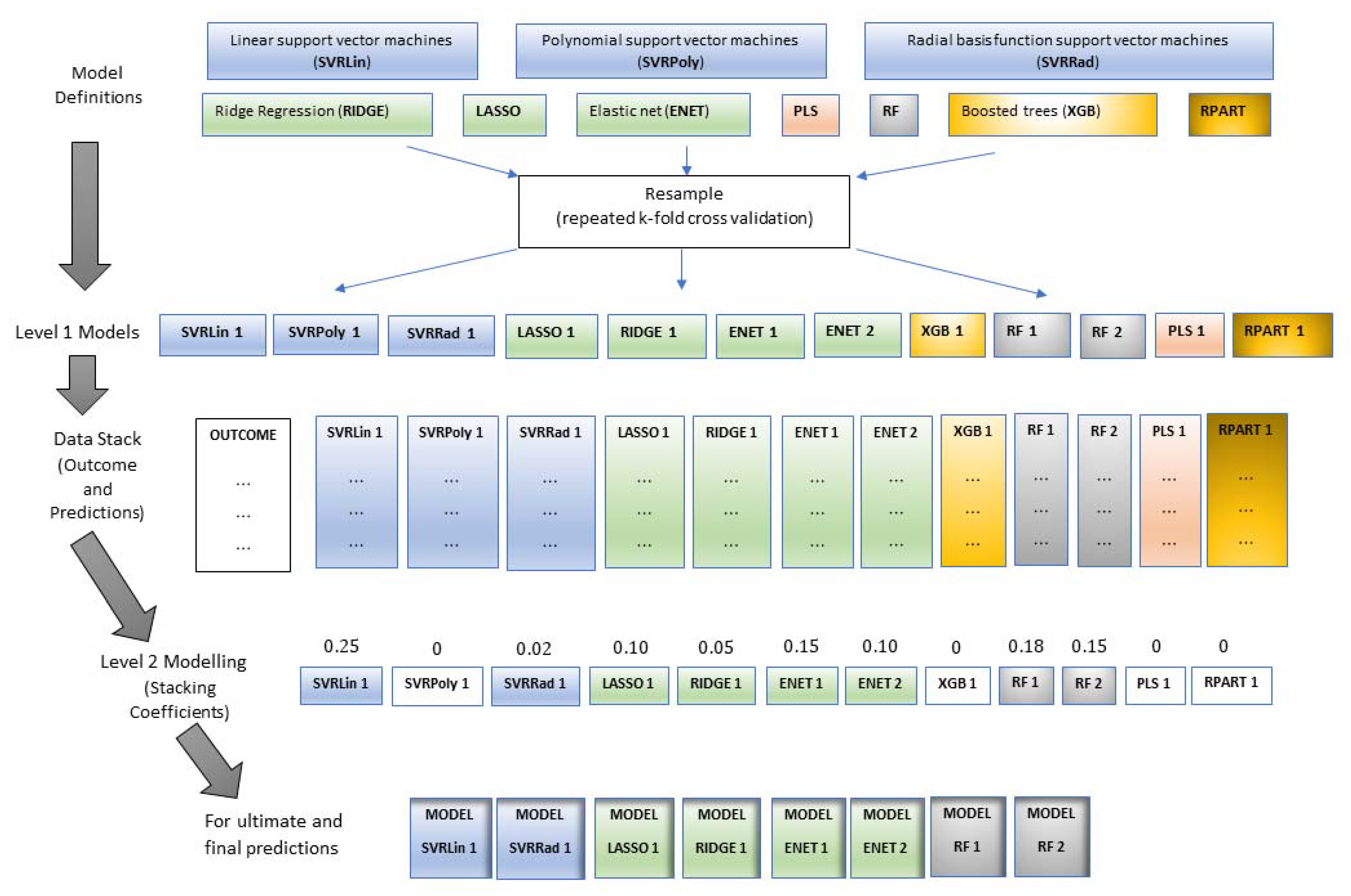
This study is the first to comprehensively compare and test a wide array of machine learning techniques including support vector regression with linear (SVRLin), polynomial (SVRPoly), and radial (SVRRad) basis kernels, LASSO, (RIDGE, elastic net regression (ENET), PLS, RF, recursive partitioning and regression trees (RPART), and boosted trees (XGB) using NIR spectral data, for the simultaneous prediction of seven chemical constituents present in animal manure. These components include dry matter (DM), total ammonium nitrogen (NH4) (designated as NH4 in this study), total N (designated as N in this study), P2O5, CaO, MgO, and K2O. In so doing, we determined the most suitable techniques for the simultaneous determination of the abovementioned chemical components. Results of this study show that stacked regression that collated the performance of the various abovementioned machine learning techniques appears to be a robust machine algorithm for the simultaneous quantification of the seven chemical components in fresh cattle and poultry manure.
4. Discussion
No studies have comprehensively investigated and compared the role of various machine learning techniques including stacked regression for the simultaneous quantification of DM, NH
4, N, P
2O
5, CaO, MgO, and K
2O contents in both cattle and poultry manure collected from livestock production. While previous studies have traditionally utilized a PLS approach for the analysis of the abovementioned chemical constituents using NIR systems [
14,
60,
61], alternative machine learning algorithms may provide better and more accurate results.
Since the optimization of results for this study is highly dependent on the choice of the hyperparameters used, a rigorous and exhaustive grid search approach was used covering an extensive range of values (
Table S1). Once hyperparameter values were optimized (
Table S2), we then tested and compared the performance of various machine learning techniques.
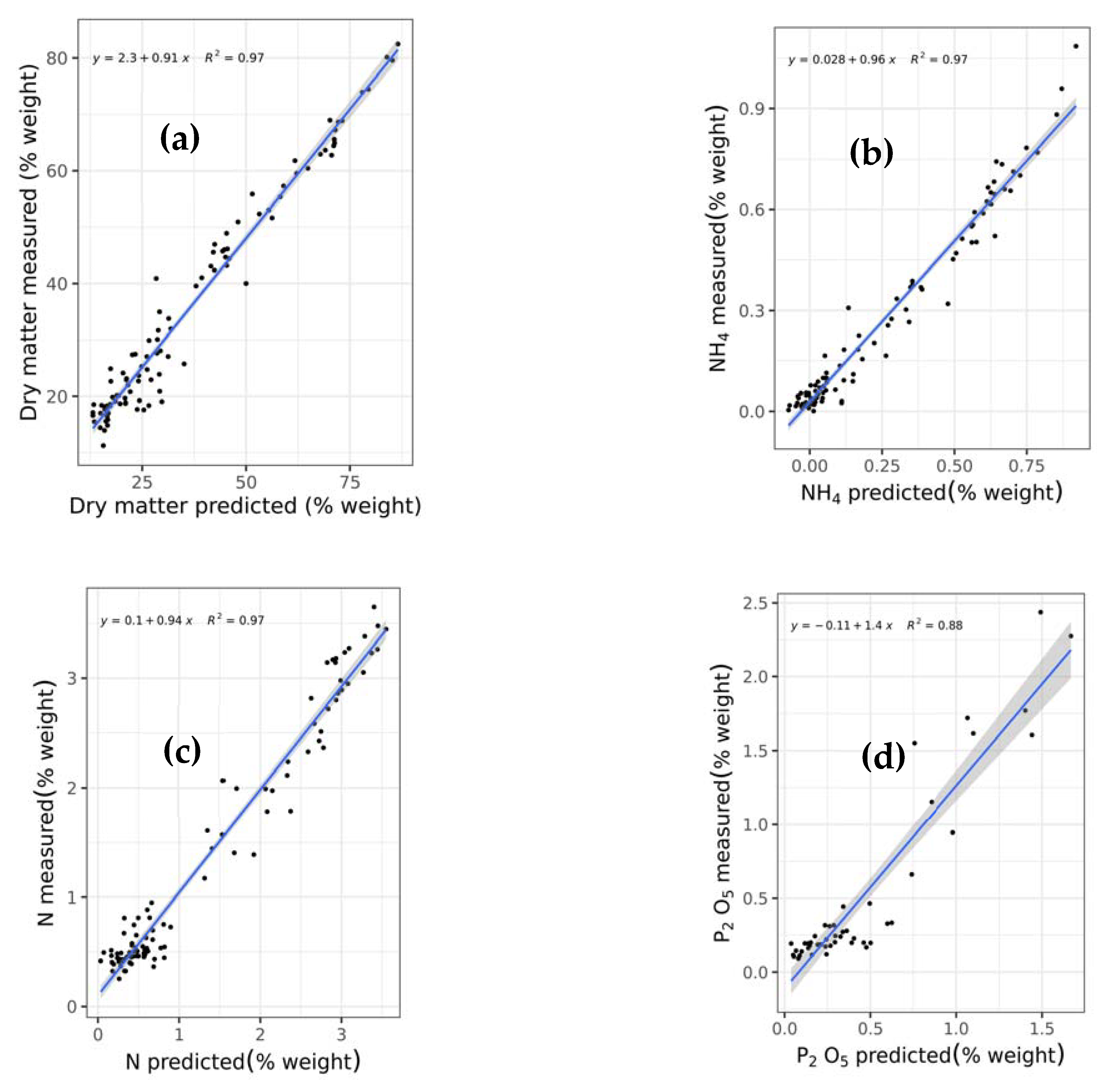
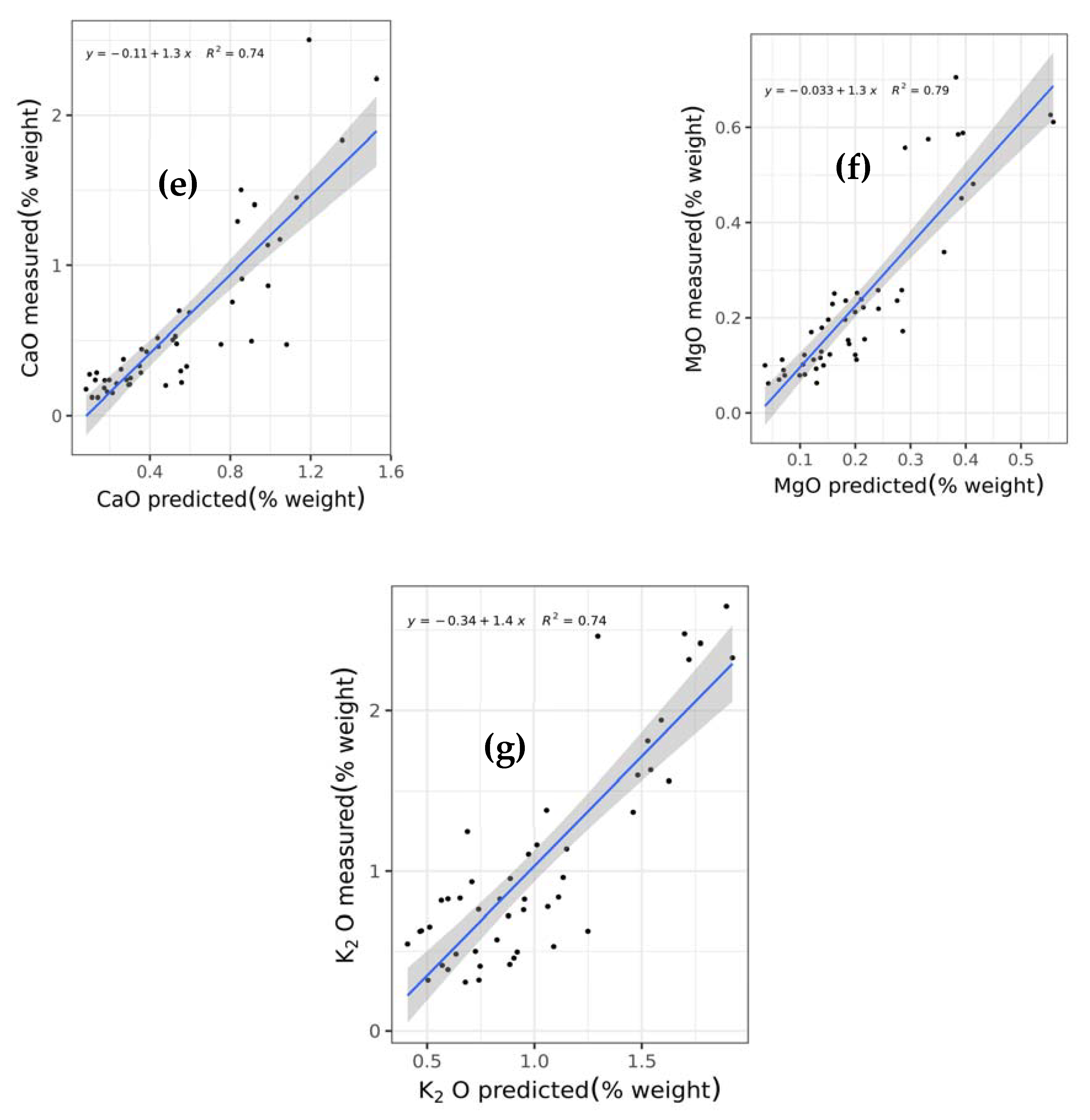
As shown in this study, PLS, the traditionally used technique in NIR analysis, did not perform fairly well as compared to stacking various machine learning techniques (
Table 6,
Table 7 and
Table 8). While PLS offers several advantages including the ability to handle missing data and intercorrelated predictors, as well as having a robust calibration model, it offers several disadvantages such as difficulty in interpreting loadings of independent variables, as well as that the learned projections are the results of linear combinations of all variables in the independent and dependent datasets, making it challenging to interpret the solutions [
62,
63,
64].
In general, a large number of samples are required to develop a robust calibration model in PLS [
65]. In one study, however, a sample size of 100 may be sufficient to achieve acceptable power for moderate effect sizes [
66]. While PLS generated an excellent model reliability across all seven chemical constituents (
RPDaverage = 2.073) (
Table 8), its performance is less superior as compared to the stacked regression technique.
Models with different hyperparameters or configurations were ranked on the basis of their optimum
R2 and
RMSE values. That is, models (i.e., machine learning algorithms) with the lowest
RMSE and highest
R2 values were highly ranked (
Figures S1–S7). As was evident, PLS was not the top-performing algorithm for each of the chemical components in the workflow rank. The top-performing models were not guaranteed to be included in the Level 2 modeling.
In the stacking procedure implemented in this study, all these models with their corresponding prediction values were stacked together, and a Level 2 model via elastic net was fitted on each of the Level 1 model that became the predictors; stacking coefficients or weights were then determined for each stack member where the only non-zero coefficients were retained to be used for final prediction on the test set (
Table S3).
Stacked regression is a very powerful approach that has been successfully applied to a wide array of fields including anticancer drug response prediction, prediction of photosynthetic capacities, image quality assessment, and mortality forecasting, among others [
67,
68,
69,
70]. The aforementioned technique has also shown its superior performance in several agricultural applications such as in the estimation of the leaf area index, wild blueberry yield prediction, and crop yield prediction, among others [
71,
72,
73]. However, its particular application in the simultaneous prediction of these seven important chemical components in fresh cattle and poultry manure has not been studied. This study has shown the robust performance of this approach as compared to PLS and other traditionally used machine learning techniques such as SVR (linear, polynomial, and radial), LASSO, RIDGE, ENET, RF, RPART, and XGB. Future studies may consider the effects of various signal preprocessing techniques, as well as wavelength selection strategies for each algorithm prior to stacked regression.