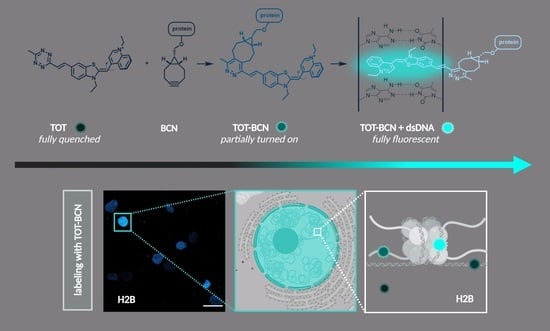
A Bioorthogonal Double Fluorogenic Probe to Visualize Protein–DNA Interaction
Abstract
:1. Introduction
2. Materials and Methods
3. Results and Discussion
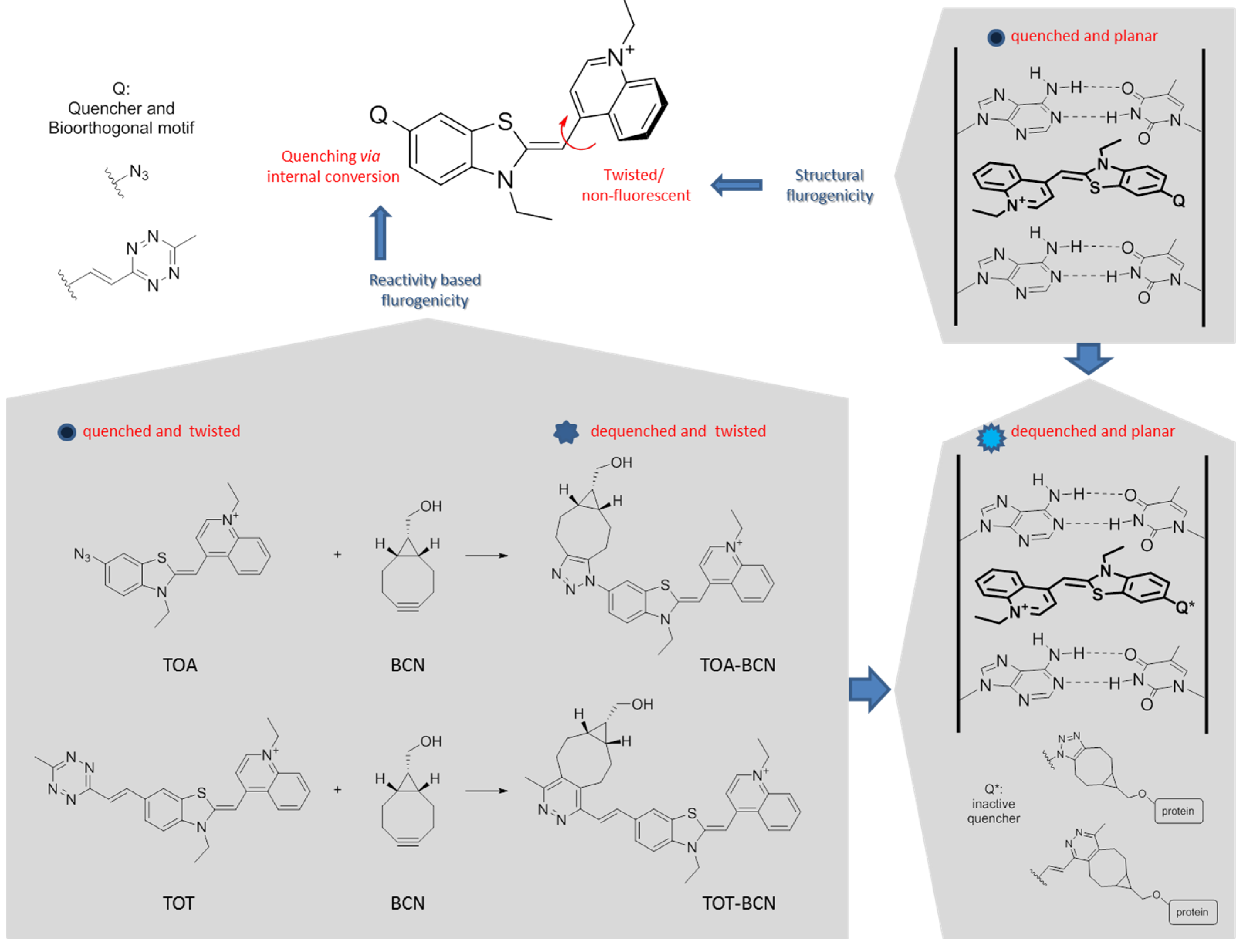
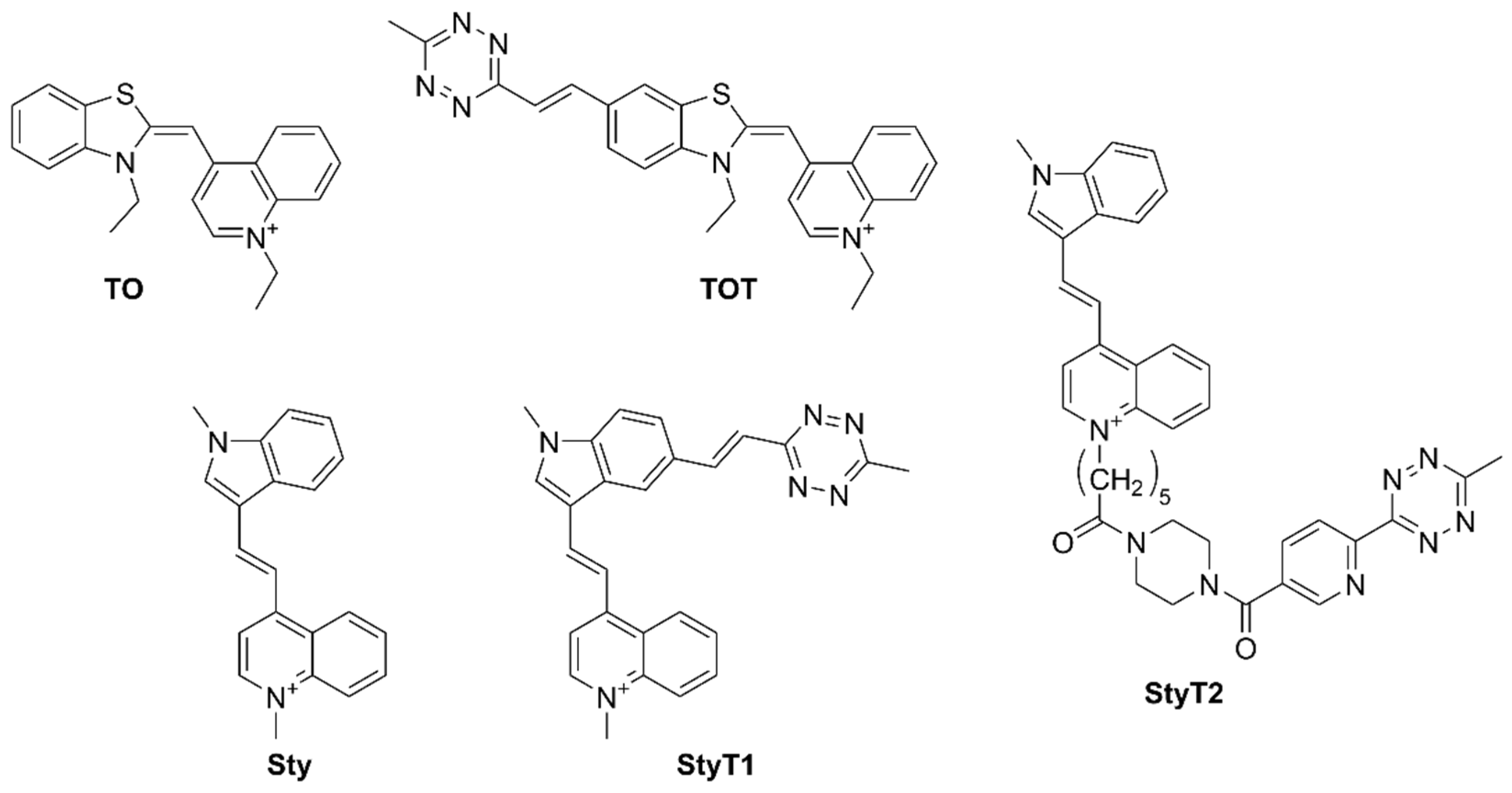
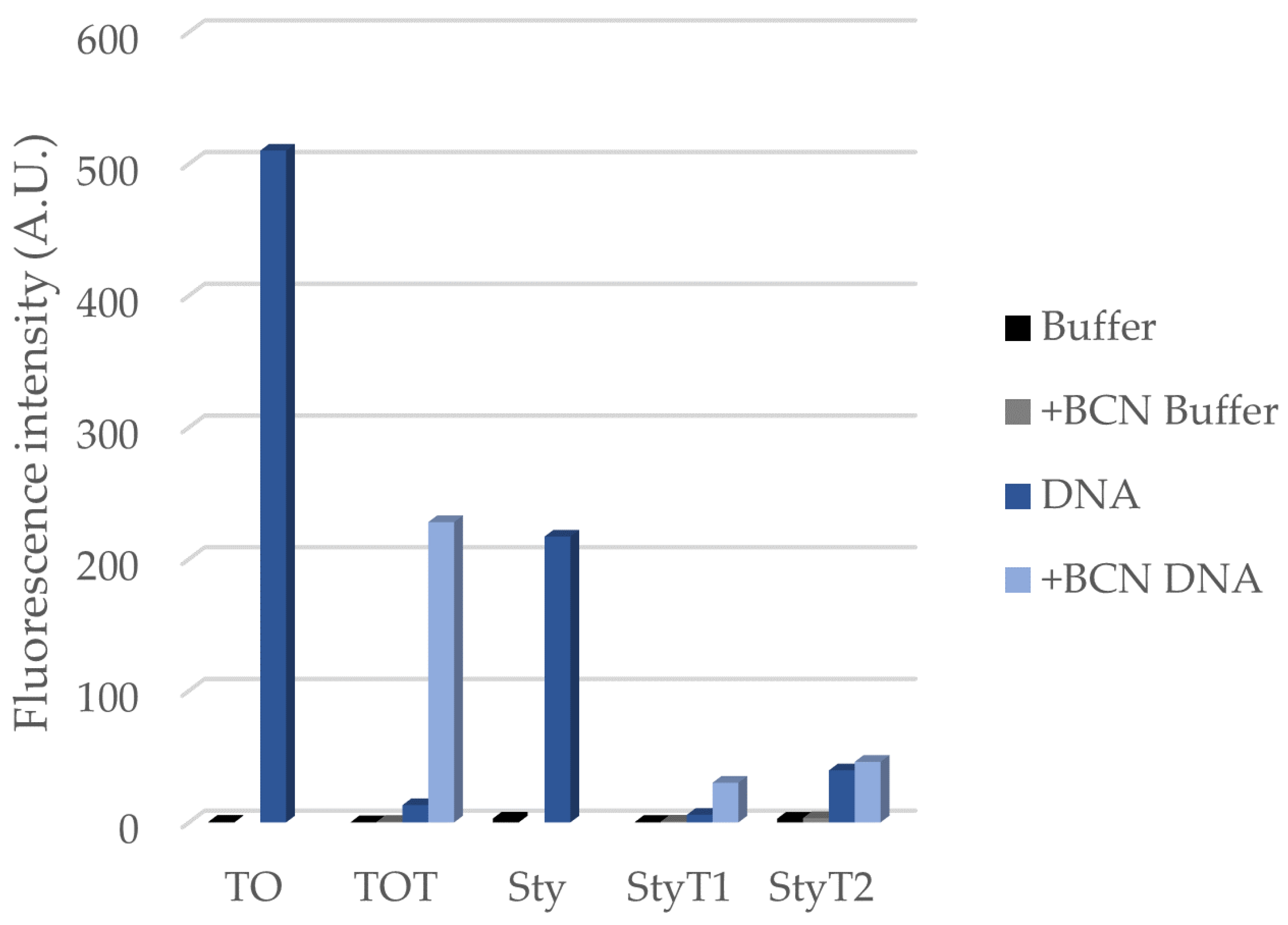
3.1. Synthesis and Fluorogenic Evaluation of Probes
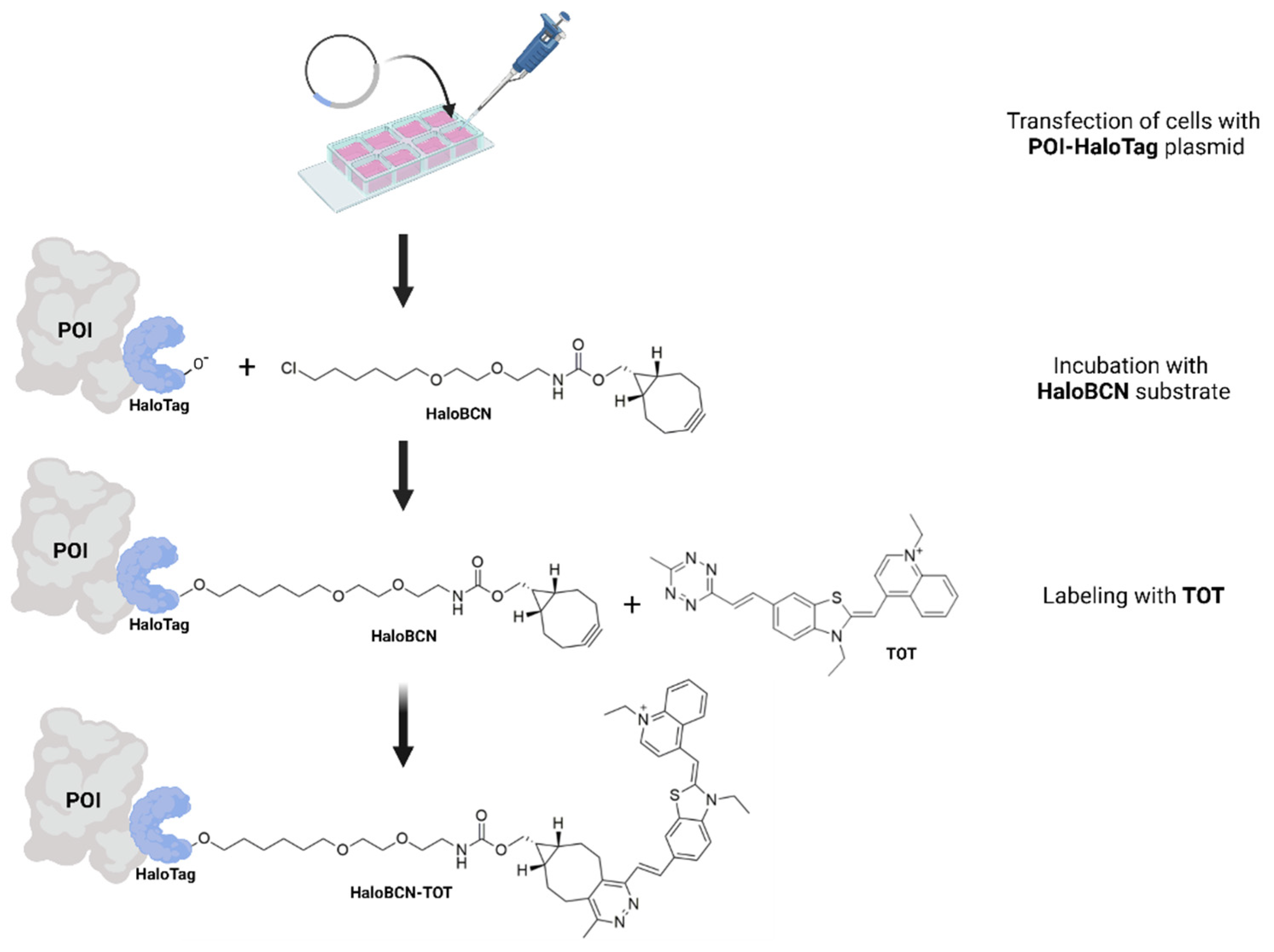
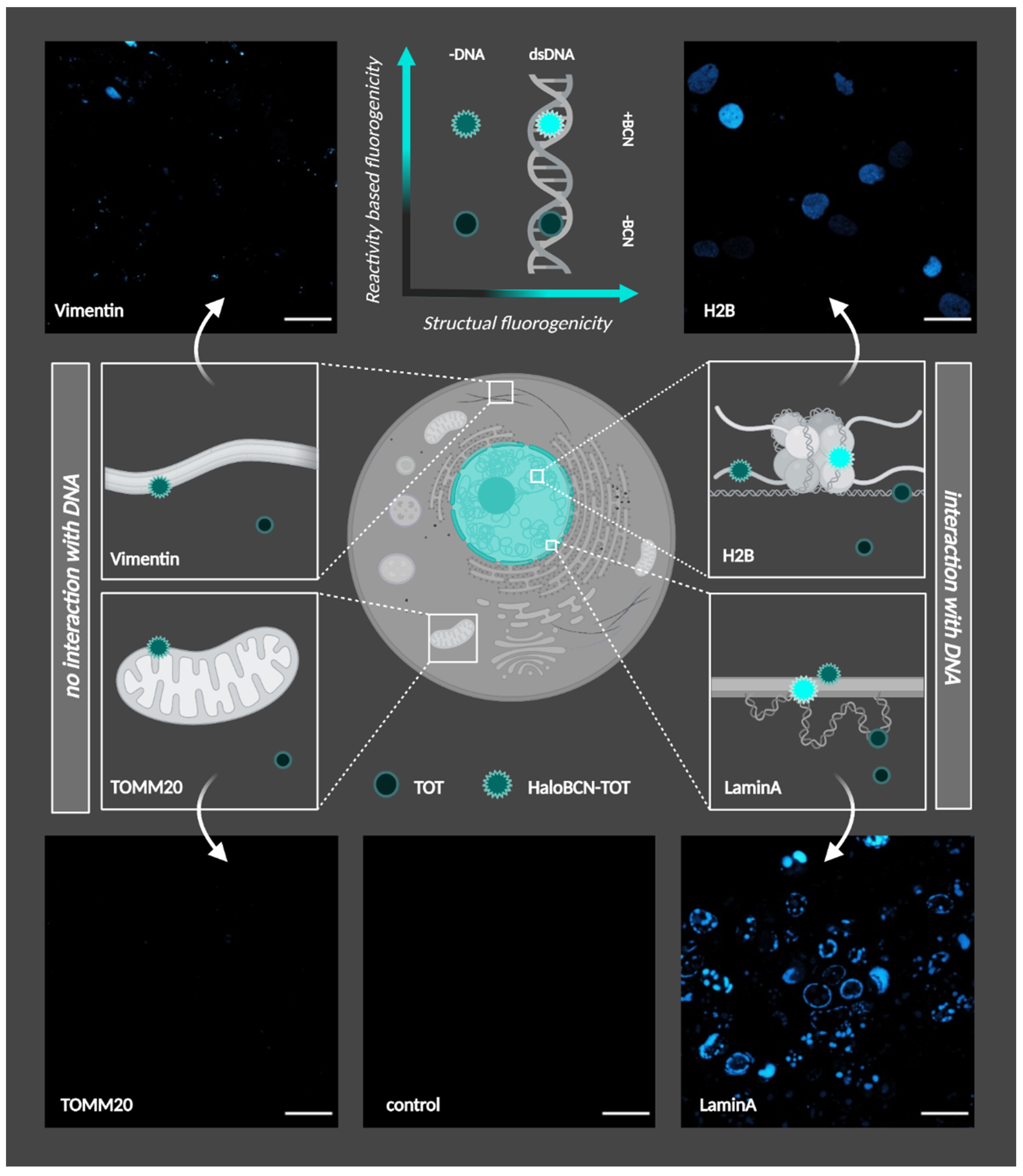
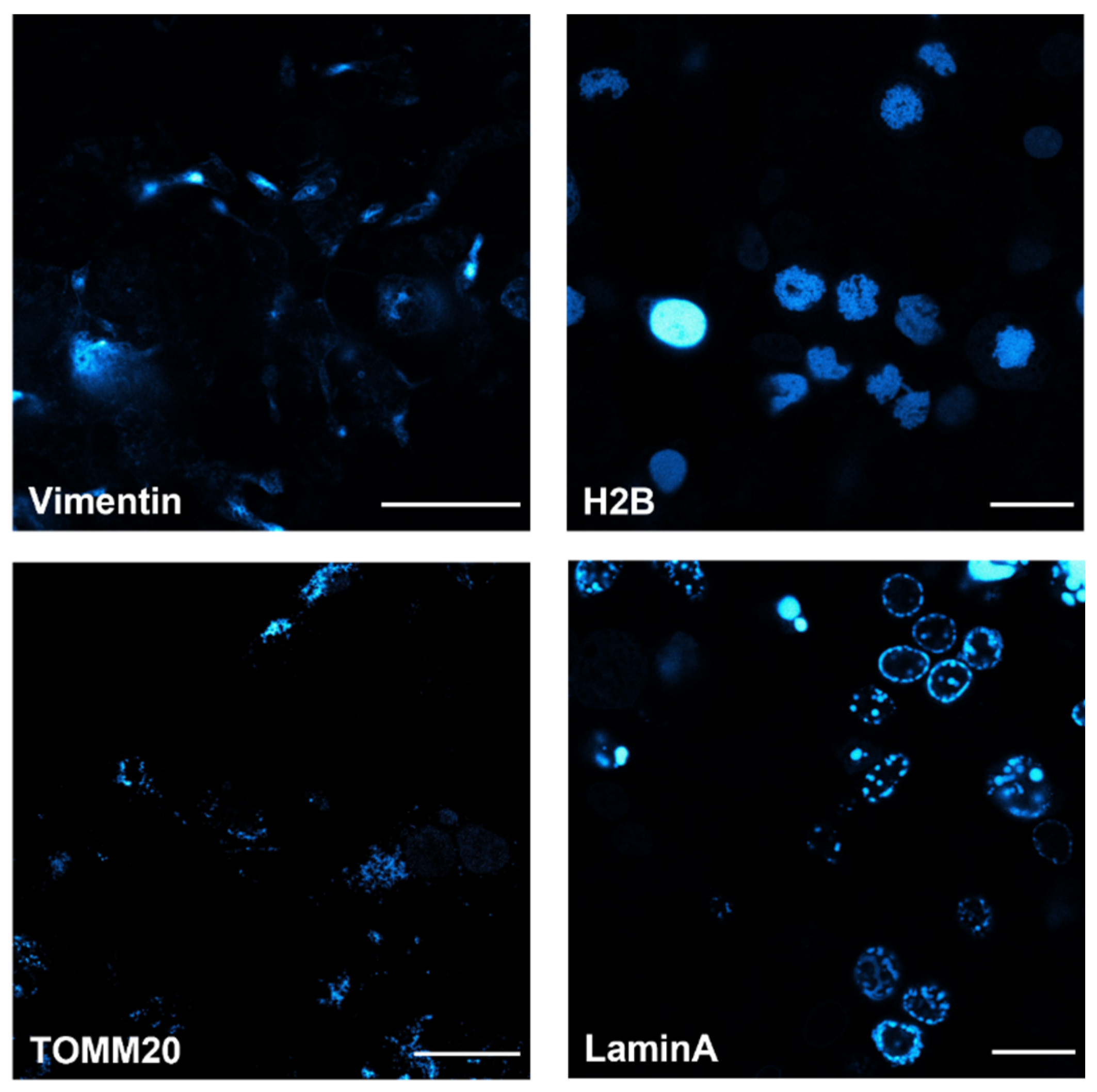
3.2. Cellular Studies
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Furey, T.S. ChIP–seq and beyond: New and improved methodologies to detect and characterize protein–DNA interactions. Nat. Rev. Genet. 2012, 13, 840–852. [Google Scholar] [CrossRef] [Green Version]
- Hellman, L.M.; Fried, M.G. Electrophoretic mobility shift assay (EMSA) for detecting protein–nucleic acid interactions. Nat. Protoc. 2007, 2, 1849–1861. [Google Scholar] [CrossRef] [PubMed]
- Chaparian, R.R.; van Kessel, J.C. Promoter Pull-Down Assay: A Biochemical Screen for DNA-Binding Proteins. In Stem Cell Renewal and Cell-Cell Communication; Turksen, K., Ed.; Springer: New York, NY, USA, 2020; Volume 2346, pp. 165–172. [Google Scholar] [CrossRef]
- Thompson, M.; Woodbury, N.W. Fluorescent and Photochemical Properties of a Single Zinc Finger Conjugated to a Fluorescent DNA-Binding Probe. Biochemistry 2000, 39, 4327–4338. [Google Scholar] [CrossRef] [PubMed]
- Babendure, J.; Liddell, P.A.; Bash, R.; LoVullo, D.; Schiefer, T.K.; Williams, M.; Daniel, D.C.; Thompson, M.; Taguchi, A.K.W.; Lohr, D.; et al. Development of a fluorescent probe for the study of nucleosome assembly and dynamics. Anal. Biochem. 2003, 317, 1–11. [Google Scholar] [CrossRef]
- Neefjes, M.; Housmans, B.A.C.; van den Akker, G.G.H.; van Rhijn, L.W.; Welting, T.J.M.; van der Kraan, P.M. Reporter gene comparison demonstrates interference of complex body fluids with secreted luciferase activity. Sci. Rep. 2021, 11, 1359. [Google Scholar] [CrossRef]
- Valuchova, S.; Fulnecek, J.; Petrov, A.P.; Tripsianes, K.; Riha, K. A rapid method for detecting protein-nucleic acid interactions by protein induced fluorescence enhancement. Sci. Rep. 2016, 6, 39653. [Google Scholar] [CrossRef] [Green Version]
- Song, D.; Graham, T.G.W.; Loparo, J.J. A general approach to visualize protein binding and DNA conformation without protein labelling. Nat. Commun. 2016, 7, 10976. [Google Scholar] [CrossRef] [Green Version]
- Hocek, M. Enzymatic Synthesis of Base-Functionalized Nucleic Acids for Sensing, Cross-linking, and Modulation of Protein–DNA Binding and Transcription. Acc. Chem. Res. 2019, 52, 1730–1737. [Google Scholar] [CrossRef] [Green Version]
- Reisacher, U.; Groitl, B.; Strasser, R.; Cserép, G.B.; Kele, P.; Wagenknecht, H.-A. Triazine-modified 7-deaza-2’-deoxyadenosines are better suited for bioorthogonal labelling of DNA by PCR than 2’-deoxyuridines. Bioconjugate Chem. 2019, 30, 1773–1780. [Google Scholar] [CrossRef]
- Dziuba, D.; Jurkiewicz, P.; Cebecauer, M.; Hof, M.; Hocek, M. A Rotational BODIPY Nucleotide: An Environment-Sensitive Fluorescence-Lifetime Probe for DNA Interactions and Applications in Live-Cell Microscopy. Angew. Chem. Int. Ed. 2016, 55, 174–178. [Google Scholar] [CrossRef]
- Güixens-Gallardo, P.; Humpolickova, J.; Miclea, S.P.; Pohl, R.; Kraus, T.; Jurkiewicz, P.; Hof, M.; Hocek, M. Thiophene-linked tetramethylbodipy-labeled nucleotide for viscosity-sensitive oligonucleotide probes of hybridization and protein–DNA interactions. Org. Biomol. Chem. 2020, 18, 912–919. [Google Scholar] [CrossRef] [PubMed]
- Matyašovský, J.; Tack, L.; Palágyi, A.; Kuba, M.; Pohl, R.; Kraus, T.; Güixens-Gallardo, P.; Hocek, M. Nucleotides bearing aminophenyl- or aminonaphthyl-3-methoxychromone solvatochromic fluorophores for the enzymatic construction of DNA probes for the detection of protein–DNA binding. Org. Biomol. Chem. 2021, 19, 9966–9974. [Google Scholar] [CrossRef]
- Tokugawa, M.; Masaki, Y.; Canggadibrata, J.C.; Kaneko, K.; Shiozawa, T.; Kanamori, T.; Grøtli, M.; Wilhelmsson, L.M.; Sekine, M.; Seio, K. 7-(Benzofuran-2-yl)-7-deazadeoxyguanosine as a fluorescence turn-ON probe for single-strand DNA binding protein. Chem. Commun. 2016, 52, 3809–3812. [Google Scholar] [CrossRef] [PubMed]
- Schneider, A.F.L.; Hackenberger, C.P.R. Fluorescent labelling in living cells. Curr. Opin. Biotechnol. 2017, 48, 61–68. [Google Scholar] [CrossRef] [PubMed]
- Bird, R.E.; Lemmel, S.A.; Yu, X.; Zhou, Q.A. Bioorthogonal Chemistry and Its Applications. Bioconjugate Chem. 2021, 32, 2457–2479. [Google Scholar] [CrossRef] [PubMed]
- Crnković, A.; Vargas-Rodriguez, O.; Söll, D. Plasticity and Constraints of tRNA Aminoacylation Define Directed Evolution of Aminoacyl-tRNA Synthetases. Int. J. Mol. Sci. 2019, 20, 2294. [Google Scholar] [CrossRef] [Green Version]
- Chung, C.Z.; Amikura, K.; Söll, D. Using Genetic Code Expansion for Protein Biochemical Studies. Front. Bioeng. Biotechnol. 2020, 8, 598577. [Google Scholar] [CrossRef] [PubMed]
- Shandell, M.A.; Tan, Z.; Cornish, V.W. Genetic Code Expansion: A Brief History and Perspective. Biochemistry 2021, 60, 3455–3469. [Google Scholar] [CrossRef]
- de la Torre, D.; Chin, J.W. Reprogramming the genetic code. Nat. Rev. Genet. 2021, 22, 169–184. [Google Scholar] [CrossRef]
- Cserép, G.B.; Herner, A.; Kele, P. Bioorthogonal fluorescent labels: A review on combined forces. Methods Appl. Fluoresc. 2015, 3, 042001. [Google Scholar] [CrossRef]
- Kozma, E.; Demeter, O.; Kele, P. Bio-orthogonal Fluorescent Labelling of Biopolymers through Inverse-Electron-Demand Diels-Alder Reactions. ChemBioChem 2017, 18, 486–501. [Google Scholar] [CrossRef] [Green Version]
- Choi, S.-K.; Kim, J.; Kim, E. Overview of Syntheses and Molecular-Design Strategies for Tetrazine-Based Fluorogenic Probes. Molecules 2021, 26, 1868. [Google Scholar] [CrossRef]
- Lipunova, G.N.; Nosova, E.V.; Zyryanov, G.V.; Charushin, V.N.; Chupakhin, O.N. 1,2,4,5-Tetrazine derivatives as components and precursors of photo- and electroactive materials. Org. Chem. Front. 2021, 8, 5182–5205. [Google Scholar] [CrossRef]
- Armitage, B.A. Cyanine Dye–DNA Interactions: Intercalation, Groove Binding, and Aggregation. In DNA Binders and Related Subjects; Waring, M.J., Chaires, J.B., Eds.; Topics in Current Chemistry; Springer: Berlin/Heidelberg, Germany, 2005; pp. 55–76. ISBN 9783540314639. [Google Scholar]
- Lartia, R.; Asseline, U. New Cyanine–Oligonucleotide Conjugates: Relationships between Chemical Structures and Properties. Chem. Eur. J. 2006, 12, 2270–2281. [Google Scholar] [CrossRef] [PubMed]
- Ikeda, S.; Kubota, T.; Yuki, M.; Okamoto, A. Exciton-Controlled Hybridization-Sensitive Fluorescent Probes: Multicolor Detection of Nucleic Acids. Angew. Chem. Int. Ed. 2009, 48, 6480–6484. [Google Scholar] [CrossRef] [PubMed]
- Suss, O.; Motiei, L.; Margulies, D. Broad Applications of Thiazole Orange in Fluorescent Sensing of Biomolecules and Ions. Molecules 2021, 26, 2828. [Google Scholar] [CrossRef]
- Nguyen, S.S.; Prescher, J.A. Developing bioorthogonal probes to span a spectrum of reactivities. Nat. Rev. Chem. 2020, 4, 476–489. [Google Scholar] [CrossRef]
- Meimetis, L.G.; Carlson, J.C.T.; Giedt, R.J.; Kohler, R.H.; Weissleder, R. Ultrafluorogenic Coumarin-Tetrazine Probes for Real-Time Biological Imaging. Angew. Chem. Int. Ed. 2014, 53, 7531–7534. [Google Scholar] [CrossRef] [PubMed]
- Devaraj, N.K.; Hilderbrand, S.; Upadhyay, R.; Mazitschek, R.; Weissleder, R. Bioorthogonal Turn-On Probes for Imaging Small Molecules inside Living Cells. Angew. Chem. Int. Ed. 2010, 49, 2869–2872. [Google Scholar] [CrossRef]
- Lee, Y.; Cho, W.; Sung, J.; Kim, E.; Park, S.B. Monochromophoric Design Strategy for Tetrazine-Based Colorful Bioorthogonal Probes with a Single Fluorescent Core Skeleton. J. Am. Chem. Soc. 2018, 140, 974–983. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bojtár, M.; Németh, K.; Domahidy, F.; Knorr, G.; Verkman, A.; Kállay, M.; Kele, P. Conditionally Activatable Visible-Light Photocages. J. Am. Chem. Soc. 2020, 142, 15164–15171. [Google Scholar] [CrossRef]
- Bohländer, P.R.; Wagenknecht, H.-A. Bright and photostable cyanine-styryl chromophores with green and red fluorescence colour for DNA staining. Methods Appl. Fluoresc. 2015, 3, 044003. [Google Scholar] [CrossRef]
- Dittmer, T.A.; Misteli, T. The lamin protein family. Genome Biol. 2011, 12, 222. [Google Scholar] [CrossRef] [Green Version]
- Li, B.X.; Chen, J.; Chao, B.; Zheng, Y.; Xiao, X. A Lamin-Binding Ligand Inhibits Homologous Recombination Repair of DNA Double-Strand Breaks. ACS Cent. Sci. 2018, 4, 1201–1210. [Google Scholar] [CrossRef] [PubMed]
- Kornberg, R.D.; Lorch, Y. Primary Role of the Nucleosome. Mol. Cell 2020, 79, 371–375. [Google Scholar] [CrossRef]
- Kurumizaka, H.; Kujirai, T.; Takizawa, Y. Contributions of Histone Variants in Nucleosome Structure and Function. J. Mol. Biol. 2021, 433, 166678. [Google Scholar] [CrossRef] [PubMed]
- Szatmári, Á.; Cserép, G.B.; Molnár, T.Á.; Söveges, B.; Biró, A.; Várady, G.; Szabó, E.; Németh, K.; Kele, P. A Genetically Encoded Isonitrile Lysine for Orthogonal Bioorthogonal Labeling Schemes. Molecules 2021, 26, 4988. [Google Scholar] [CrossRef] [PubMed]
- Danielsson, F.; Peterson, M.; Caldeira Araújo, H.; Lautenschläger, F.; Gad, A. Vimentin Diversity in Health and Disease. Cells 2018, 7, 147. [Google Scholar] [CrossRef] [Green Version]
- Endo, T.; Kohda, D. Functions of outer membrane receptors in mitochondrial protein import. Biochim. Biophys. Acta Mol. Cell Res. 2002, 1592, 3–14. [Google Scholar] [CrossRef] [Green Version]
- Wurm, C.A.; Neumann, D.; Lauterbach, M.A.; Harke, B.; Egner, A.; Hell, S.W.; Jakobs, S. Nanoscale distribution of mitochondrial import receptor Tom20 is adjusted to cellular conditions and exhibits an inner-cellular gradient. Proc. Natl. Acad. Sci. USA 2011, 108, 13546–13551. [Google Scholar] [CrossRef] [Green Version]
- Kozma, E.; Estrada Girona, G.; Paci, G.; Lemke, E.A.; Kele, P. Bioorthogonal Double-Fluorogenic Siliconrhodamine Probes for Intracellular Superresolution Microscopy. Chem. Commun. 2017, 53, 6696–6699. [Google Scholar] [CrossRef] [PubMed] [Green Version]
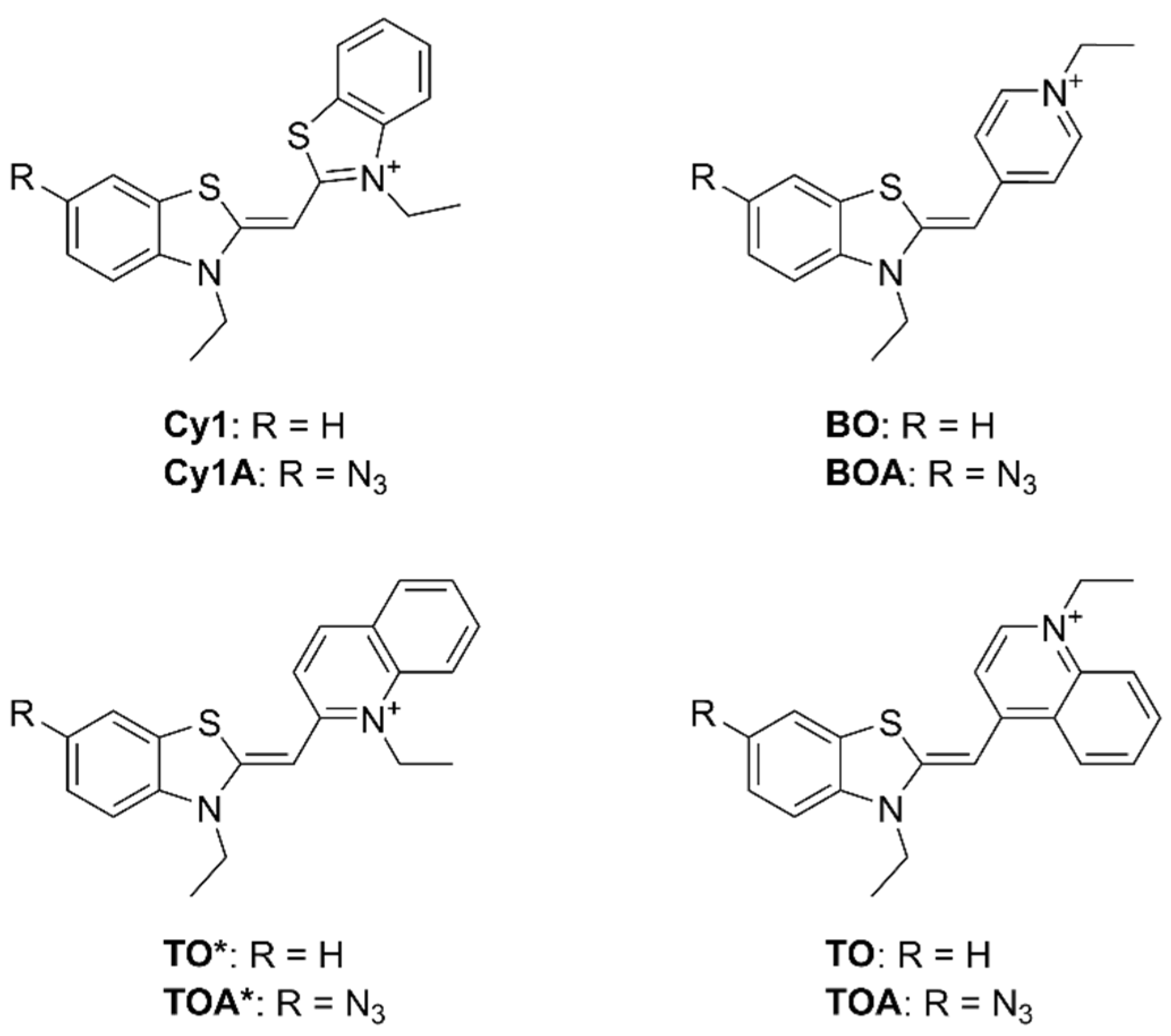
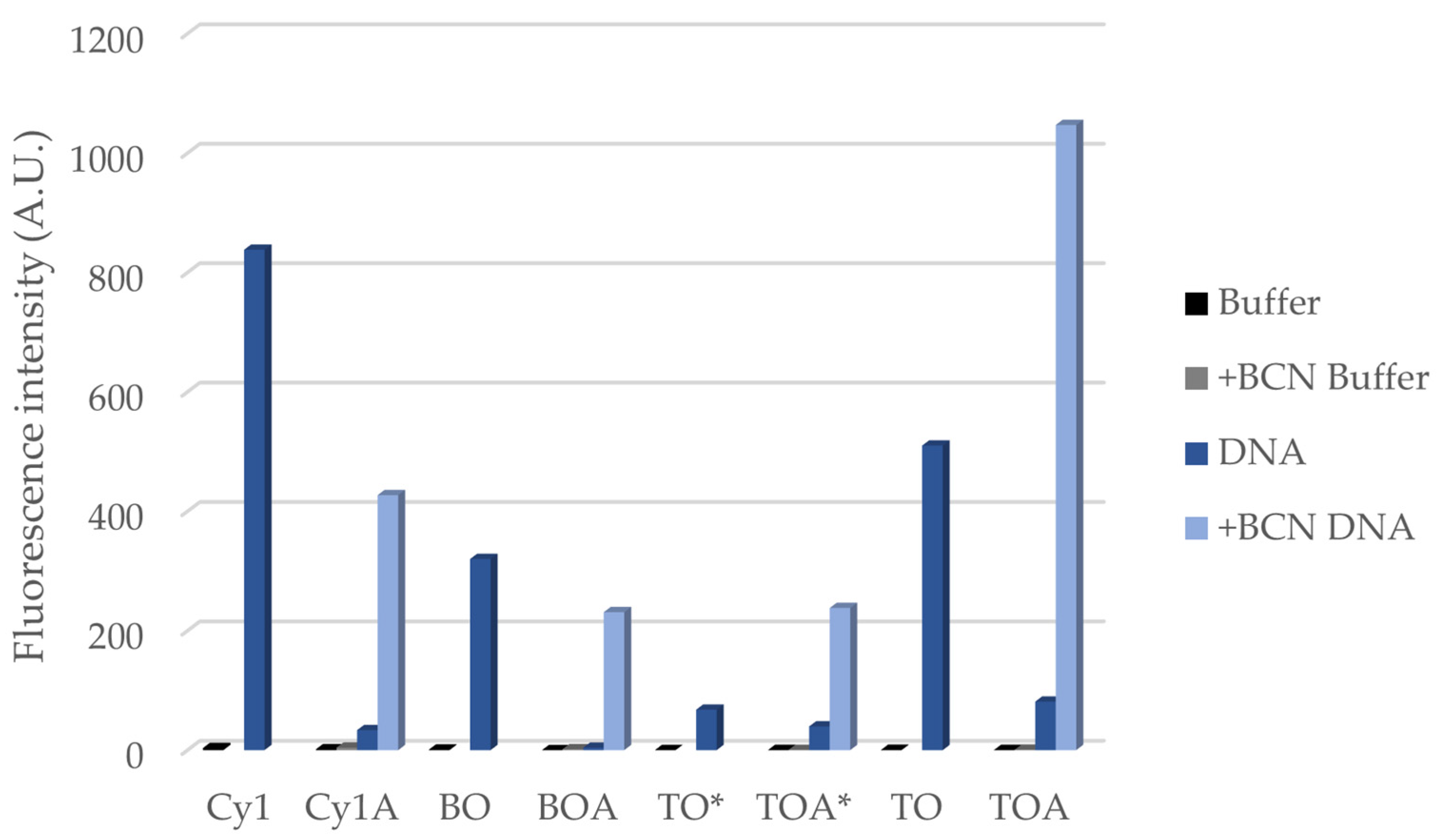
| ΦPBS | +BCN ΦPBS | ΦDNA | +BCN ΦDNA | |
|---|---|---|---|---|
| Cy1 | 0.0028 | - | 0.41 | - |
| Cy1A | 0.0014 | 0.0028 | 0.047 | 0.19 |
| BO | 0.00053 | - | 0.24 | - |
| BOA | N.D. a | 0.0010 | 0.017 | 0.094 |
| TO* | 0.00013 | - | 0.050 | - |
| TOA* | 0.00021 | 0.00046 | 0.030 | 0.16 |
| TO | 0.00029 | - | 0.30 | - |
| TOA | 0.00033 | 0.00061 | 0.083 | 0.53 |
| ΦPBS | +BCN ΦPBS | ΦDNA | +BCN ΦDNA | |
|---|---|---|---|---|
| TO | 0.00029 | - | 0.30 | - |
| TOT | N.D. a | 0.0016 | 0.014 | 0.24 |
| Sty | 0.0031 | - | 0.19 | - |
| StyT1 | 0.00093 | 0.0024 | 0.014 | 0.072 |
| StyT2 | 0.014 | 0.013 | 0.15 | 0.15 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kormos, A.; Egyed, A.; Olvany, J.M.; Szatmári, Á.; Biró, A.; Csorba, Z.; Kele, P.; Németh, K. A Bioorthogonal Double Fluorogenic Probe to Visualize Protein–DNA Interaction. Chemosensors 2022, 10, 37. https://doi.org/10.3390/chemosensors10010037
Kormos A, Egyed A, Olvany JM, Szatmári Á, Biró A, Csorba Z, Kele P, Németh K. A Bioorthogonal Double Fluorogenic Probe to Visualize Protein–DNA Interaction. Chemosensors. 2022; 10(1):37. https://doi.org/10.3390/chemosensors10010037
Chicago/Turabian StyleKormos, Attila, Alexandra Egyed, Jasmine M. Olvany, Ágnes Szatmári, Adrienn Biró, Zsóka Csorba, Péter Kele, and Krisztina Németh. 2022. "A Bioorthogonal Double Fluorogenic Probe to Visualize Protein–DNA Interaction" Chemosensors 10, no. 1: 37. https://doi.org/10.3390/chemosensors10010037
APA StyleKormos, A., Egyed, A., Olvany, J. M., Szatmári, Á., Biró, A., Csorba, Z., Kele, P., & Németh, K. (2022). A Bioorthogonal Double Fluorogenic Probe to Visualize Protein–DNA Interaction. Chemosensors, 10(1), 37. https://doi.org/10.3390/chemosensors10010037