Urinary Sodium Excretion Enhances the Effect of Alcohol on Blood Pressure
Abstract
:1. Introduction
2. Materials and Methods
2.1. Ethics Approval
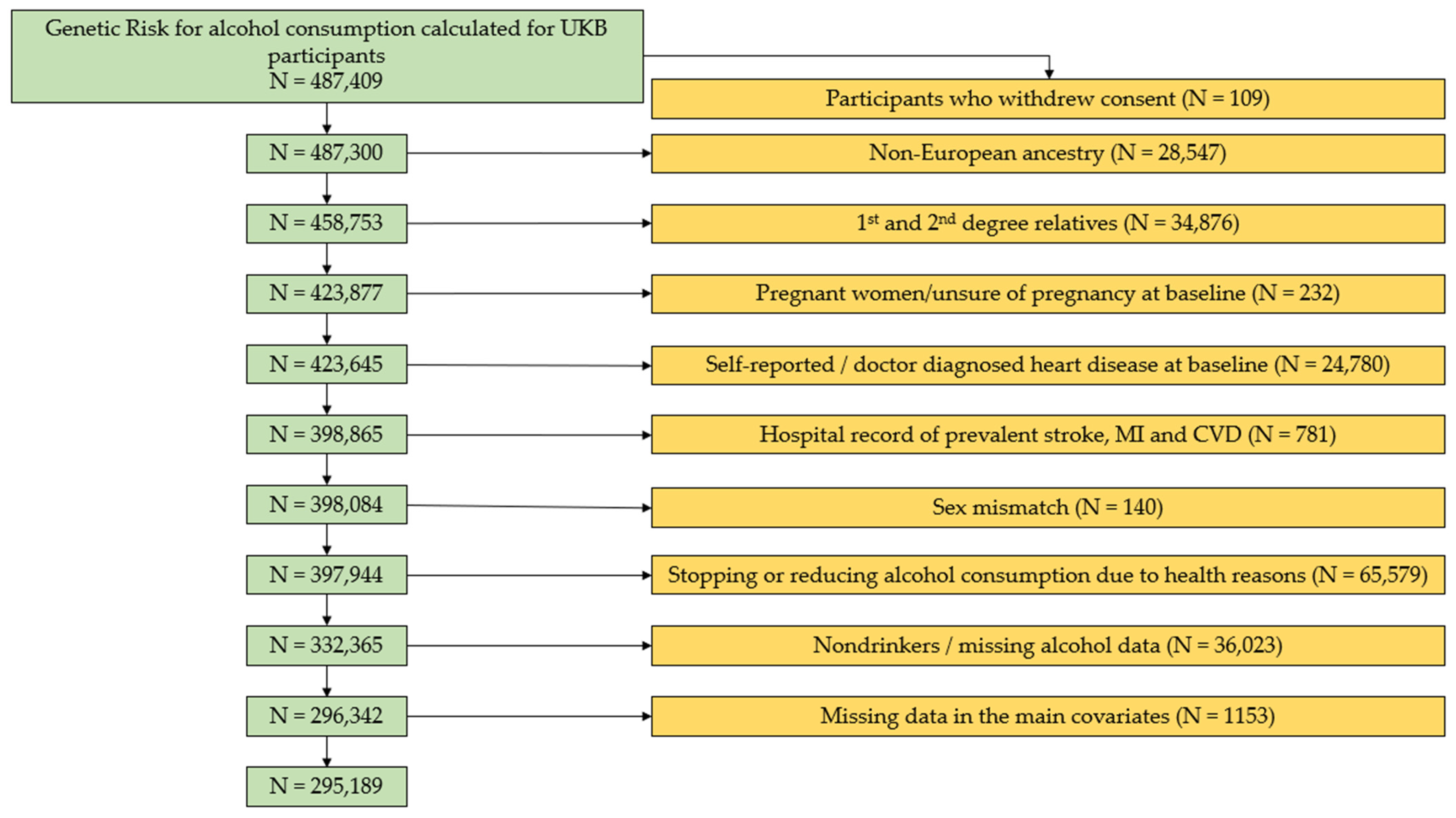
2.2. Study Population and Exclusion Criteria
2.3. Blood Pressure and Definition of Hypertension
2.4. Cardiovascular Diseases
2.5. Assessment of Lifestyle Factors
2.6. Genotyping, Imputation, and Genetic Calculations in the UKB
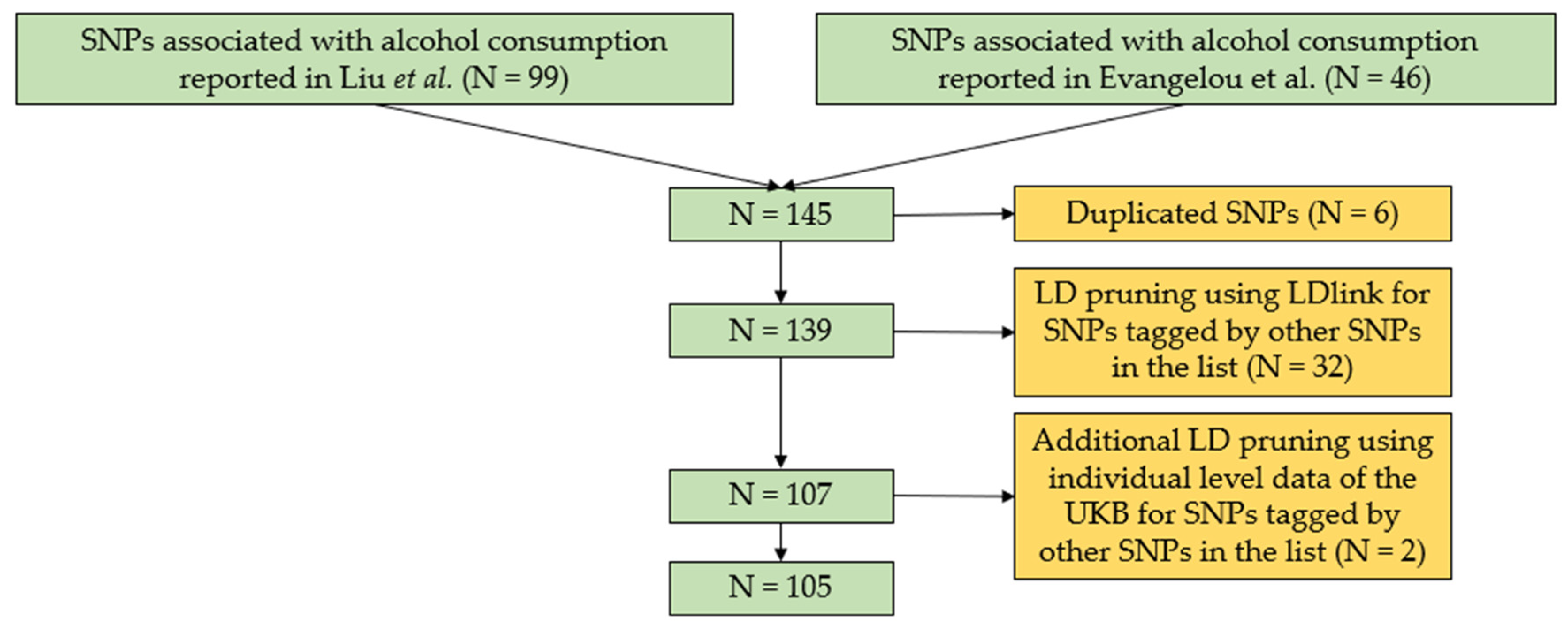
2.7. GRS for Alcohol Consumption
2.8. Secondary Analyses
2.9. Statistical Analysis
2.10. Power Calculation
2.11. Software and Packages
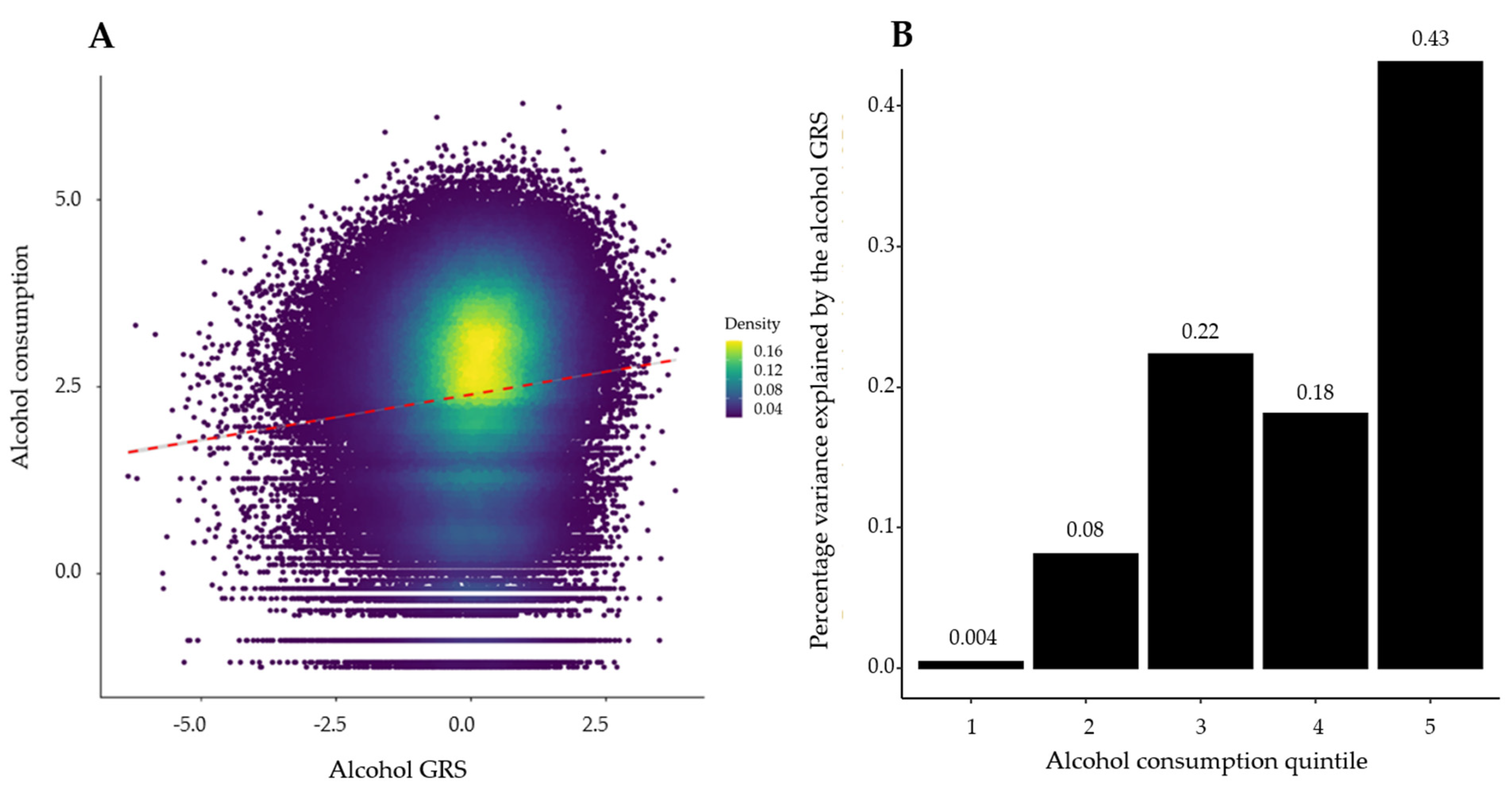
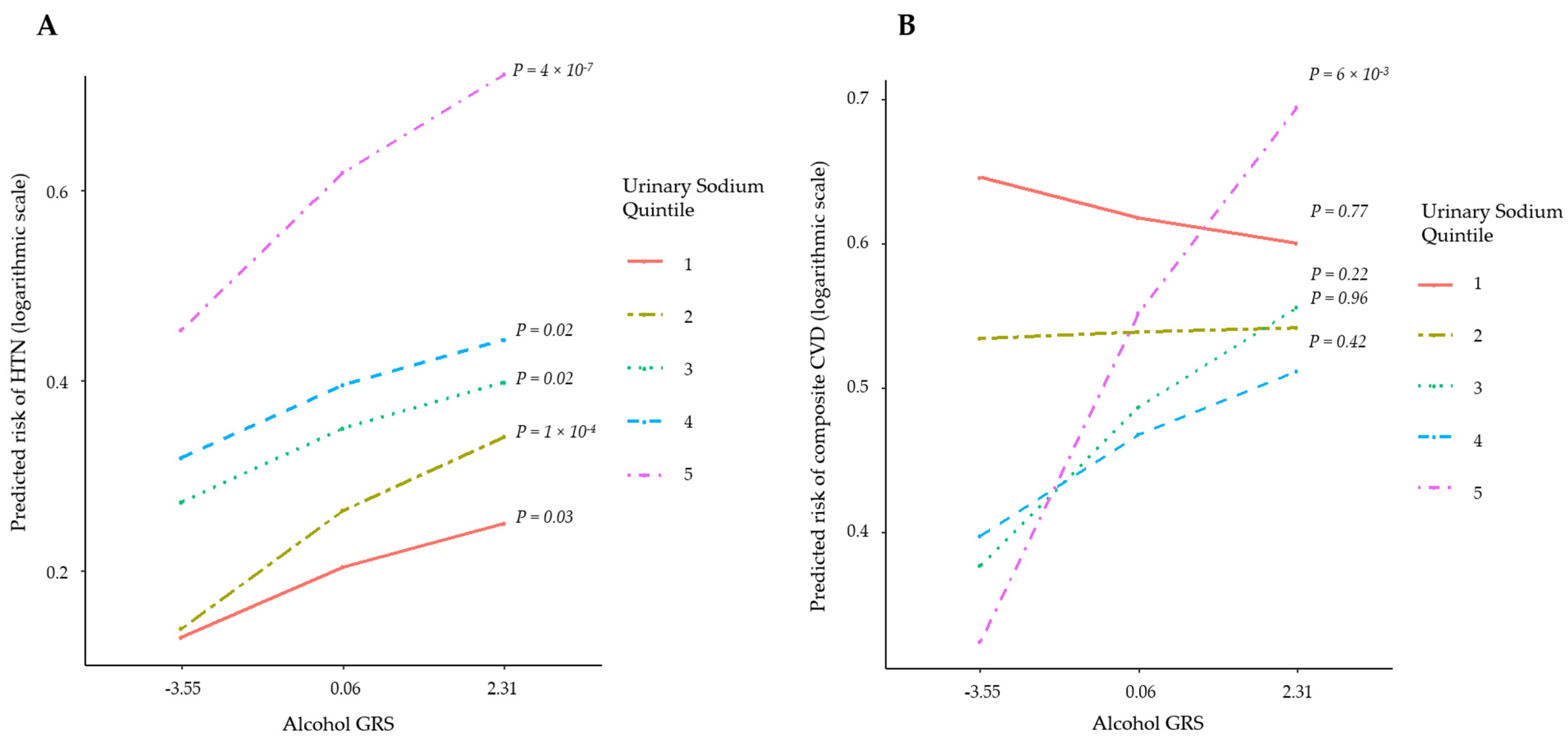
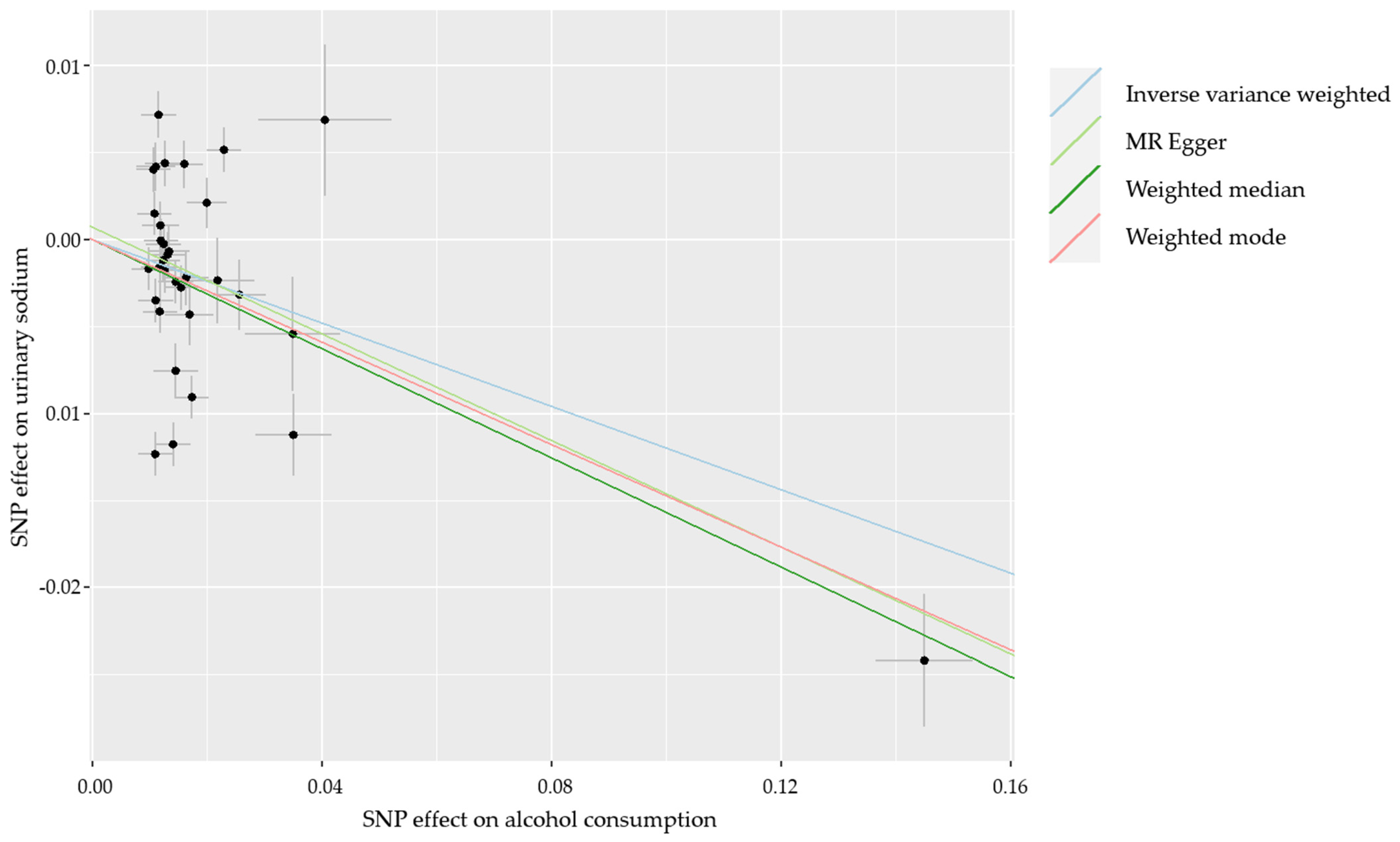
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- The World Health Organization. Cardiovasuclar Diseases (CVDs); The World Health Organization: Geneva, Switzerland, 2017. [Google Scholar]
- Chiva-Blanch, G.; Badimon, L. Benefits and Risks of Moderate Alcohol Consumption on Cardiovascular Disease: Current Findings and Controversies. Nutrients 2020, 12, 108. [Google Scholar]
- Puddey, I.B.; Mori, T.A.; Barden, A.E.; Beilin, L.J. Alcohol and Hypertension—New Insights and Lingering Controversies. Curr. Hypertens. Rep. 2019, 21, 79. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Smith, G.D.; Harbord, R.M.; Lewis, S.J. Alcohol intake and blood pressure: A systematic review implementing a Mendelian randomization approach. PLoS Med. 2008, 5, e52. [Google Scholar]
- Roerecke, M.; Kaczorowski, J.; Tobe, S.W.; Gmel, G.; Hasan, O.S.M.; Rehm, J. The effect of a reduction in alcohol consump-tion on blood pressure: A systematic review and meta-analysis. Lancet Public Health 2017, 2, e108–e120. [Google Scholar] [CrossRef] [Green Version]
- Huang, L.; Trieu, K.; Yoshimura, S.; Neal, B.; Woodward, M.; Campbell, N.R.C.; Li, Q.; Lackland, D.T.; Leung, A.A.; An-derson, C.A.M.; et al. Effect of dose and duration of reduction in dietary sodium on blood pressure levels: Systematic review and meta-analysis of randomised trials. BMJ 2020, 368, m315. [Google Scholar] [CrossRef] [Green Version]
- Mente, A.; O’Donnell, M.J.; Rangarajan, S.; McQueen, M.J.; Poirier, P.; Wielgosz, A.; Morrison, H.; Li, W.; Wang, X.; Di, C.; et al. Association of Urinary Sodium and Potassium Excretion with Blood Pressure. N. Engl. J. Med. 2014, 371, 601–611. [Google Scholar] [CrossRef] [Green Version]
- Re, F.; Hammami, I.; Littlejohns, T.J.; Arnold, M.; Lewington, S.; Clarke, R.J.; Carter, J.L. Effects of Within-Person Variability in Spot Urinary Sodium Measurements on Associations With Blood Pressure and Cardiovascular Disease. Hypertension 2021, 78, 1628–1636. [Google Scholar] [CrossRef]
- Pazoki, R.; Evangelou, E.; Mosen-Ansorena, D.; Pinto, R.C.; Karaman, I.; Blakeley, P.; Gill, D.; Zuber, V.; Elliott, P.; Tzoulaki, I.; et al. GWAS for urinary sodium and potassium excretion highlights pathways shared with cardiovascular traits. Nat. Commun. 2019, 10, 3653. [Google Scholar] [CrossRef] [Green Version]
- Koyama, H.; Suzuki, S.; Satoh, H. Effect of Alcohol Ingestion on Urinary Excretion and Intraerythrocyte Concentrations of Sodium and Potassium. Tohoku J. Exp. Med. 1990, 162, 73–78. [Google Scholar] [CrossRef] [Green Version]
- Nan, X.; Lu, H.; Wu, J.; Xue, M.; Qian, Y.; Wang, W.; Wang, X. The interactive association between sodium intake, alcohol consumption and hypertension among elderly in northern China: A cross-sectional study. BMC Geriatr. 2021, 21, 135. [Google Scholar] [CrossRef]
- Evangelou, E.; Gao, H.; Chu, C.; Ntritsos, G.; Blakeley, P.; Butts, A.R.; Pazoki, R.; Suzuki, H.; Koskeridis, F.; Yiorkas, A.M.; et al. New alcohol-related genes suggest shared genetic mechanisms with neuropsychiatric disorders. Nat. Hum. Behav. 2019, 3, 950–961. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, M.; Jiang, Y.; Wedow, R.; Li, Y.; Brazel, D.M.; Chen, F.; Datta, G.; Davila-Velderrain, J.; McGuire, D.; Tian, C.; et al. Association studies of up to 1.2 million individuals yield new insights into the genetic etiology of tobacco and alcohol use. Nat. Genet. 2019, 51, 237–244. [Google Scholar] [CrossRef] [PubMed]
- Bycroft, C.; Freeman, C.; Petkova, D.; Band, G.; Elliott, L.T.; Sharp, K.; Motyer, A. The UK Biobank resource with deep phenotyping and genomic data. Nature 2018, 562, 203–209. [Google Scholar] [CrossRef] [Green Version]
- UK Biobank. Genotyping and Quality Control of UK Biobank, a Large-Scale, Extensively Phenotyped Prospective Resource. 2015. Available online: https://biobank.ctsu.ox.ac.uk/crystal/crystal/docs/genotyping_qc.pdf (accessed on 12 July 2022).
- Pazoki, R.; Dehghan, A.; Evangelou, E.; Warren, H.; Gao, H.; Caulfield, M. Genetic Predisposition to High Blood Pressure and Lifestyle Factors. Circulation 2018, 137, 653–661. [Google Scholar] [CrossRef] [PubMed]
- Tobin, M.D.; Sheehan, N.A.; Scurrah, K.J.; Burton, P.R. Adjusting for treatment effects in studies of quantitative traits: An-tihypertensive therapy and systolic blood pressure. Stat. Med. 2005, 24, 2911–2935. [Google Scholar] [CrossRef]
- Whelton, P.K.; Carey, R.M.; Aronow, W.S.; Casey, D.E.; Collins, K.J.; Dennison Himmelfarb, C.; DePalma, S.M.; Gidding, S.; Jamerson, K.A.; Jones, D.W.; et al. 2017 ACC/AHA/AAPA/ABC/ACPM/AGS/APhA/ASH/ASPC/NMA/PCNA Guideline for the Prevention, Detection, Evaluation, and Management of High Blood Pressure in Adults: Executive Summary: A Report of the American College of Cardiology/American Heart Association Task F. Hypertension 2018, 71, 1269–1324. [Google Scholar] [CrossRef]
- Sudlow, C.; Gallacher, J.; Allen, N.; Beral, V.; Burton, P.; Danesh, J. UK biobank: An open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 2015, 12, e1001779. [Google Scholar] [CrossRef] [Green Version]
- Bloch, M.J. The Dietary Approaches to Stop Hypertension (DASH) diet—Promise Unmet. J. Am. Soc. Hypertens. 2017, 11, 323–324. [Google Scholar] [CrossRef]
- Townsend, P.; Phillimore, P.; Beattie, A. Health and Deprivation: Inequality and the North; Croom Helm: Kent, UK, 1988; pp. 1–211. [Google Scholar]
- Machiela, M.J.; Chanock, S.J. LDlink: A web-based application for exploring population-specific haplotype structure and linking correlated alleles of possible functional variants. Bioinformatics 2015, 31, 3555–3557. [Google Scholar] [CrossRef]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.R.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.W.; Daly, M.J.; et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef] [Green Version]
- Smith, G.D.; Ebrahim, S. ‘Mendelian randomization’: Can genetic epidemiology contribute to understanding environmental determinants of disease? Int. J. Epidemiol. 2003, 32, 1–22. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hemani, G.; Zheng, J.; Elsworth, B.; Wade, K.H.; Haberland, V.; Baird, D.; Laurin, C.; Burgess, S.; Bowden, J.; Langdon, R.; et al. The MR-Base platform supports systematic causal inference across the human phenome. eLife 2018, 7, e34408. [Google Scholar] [CrossRef] [PubMed]
- Burgess, S.; Thompson, S.G. Bias in causal estimates from Mendelian randomization studies with weak instruments. Stat. Med. 2011, 30, 1312–1323. [Google Scholar] [CrossRef] [PubMed]
- Lawlor, D.A.; Harbord, R.M.; Sterne, J.A.; Timpson, N.; Davey Smith, G. Mendelian randomization: Using genes as in-struments for making causal inferences in epidemiology. Stat. Med. 2008, 27, 1133–1163. [Google Scholar] [CrossRef]
- Staiger, D.; Stock, J.H. Instrumental Variables Regression with Weak Instruments. Econometrica 1997, 65, 557–586. [Google Scholar] [CrossRef]
- Burgess, S.; Thompson, S.G. Avoiding bias from weak instruments in Mendelian randomization studies. Int. J. Epidemiol. 2011, 40, 755–764. [Google Scholar] [CrossRef] [Green Version]
- Shim, H.; Chasman, D.I.; Smith, J.D.; Mora, S.; Ridker, P.M.; Nickerson, D.A.; Krauss, R.M.; Stephens, M. A multivariate genome-wide association analysis of 10 LDL subfractions, and their response to statin treatment, in 1868 Caucasians. PLoS ONE 2015, 10, e0120758. [Google Scholar] [CrossRef] [Green Version]
- Burgess, S.; Bowden, J.; Fall, T.; Ingelsson, E.; Thompson, S.G. Sensitivity Analyses for Robust Causal Inference from Mendelian Randomization Analyses with Multiple Genetic Variants. Epidemiology 2017, 28, 30–42. [Google Scholar] [CrossRef] [Green Version]
- Bowden, J.; Davey Smith, G.; Burgess, S. Mendelian randomization with invalid instruments: Effect estimation and bias detection through Egger regression. Int. J. Epidemiol. 2015, 44, 512–525. [Google Scholar] [CrossRef] [Green Version]
- Bowden, J.; Davey Smith, G.; Haycock, P.C.; Burgess, S. Consistent Estimation in Mendelian Randomization with Some Invalid Instruments Using a Weighted Median Estimator. Genet. Epidemiol. 2016, 40, 304–314. [Google Scholar] [CrossRef] [Green Version]
- Hartwig, F.P.; Davey Smith, G.; Bowden, J. Robust inference in summary data Mendelian randomization via the zero modal pleiotropy assumption. Int. J. Epidemiol. 2017, 46, 1985–1998. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schoenfeld, D. Partial Residuals for The Proportional Hazards Regression Model. Biometrika 1982, 69, 239–241. [Google Scholar] [CrossRef]
- Gauderman, W.; Morrison, J.M.; Gauderman, W.J.; Morrison, J.M.; Morrison, W.G.J. QUANTO 1.1: A Computer Program for Power and Sample Size Calculations for Genetic-Epidemiology Studies; University of Southern California: Los Angeles, CA, USA, 2006; Available online: https://www.scienceopen.com/document?vid=2944f68a-3b3d-4e86-90a3-3d3c1fab3ffa (accessed on 12 July 2022).
- Gauderman, W.J. Sample size requirements for matched case-control studies of gene-environment interaction. Stat. Med. 2002, 21, 35–50. [Google Scholar] [CrossRef]
- Roerecke, M.; Tobe, S.W.; Kaczorowski, J.; Bacon, S.L.; Vafaei, A.; Hasan, O.S.M.; Krishnan, R.J.; Raifu, A.O.; Rehm, J. Sex-Specific Associations Between Alcohol Consumption and Incidence of Hypertension: A Systematic Review and Me-ta-Analysis of Cohort Studies. J. Am. Heart Assoc. 2018, 7, e008202. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wood, A.M.; Kaptoge, S.; Butterworth, A.S.; Willeit, P.; Warnakula, S.; Bolton, T.; Paige, E.; Paul, D.S.; Sweeting, M.; Burgess, S.; et al. Risk thresholds for alcohol consumption: Combined analysis of individual-participant data for 599,912 current drinkers in 83 prospective studies. Lancet 2018, 391, 1513–1523. [Google Scholar] [CrossRef] [Green Version]
- Millwood, I.Y.; Walters, R.G.; Mei, X.W.; Guo, Y.; Yang, L.; Bian, Z.; Bennett, D.A.; Chen, Y.; Dong, C.; Hu, R.; et al. Con-ventional and genetic evidence on alcohol and vascular disease aetiology: A prospective study of 500,000 men and women in China. Lancet 2019, 393, 1831–1842. [Google Scholar] [CrossRef] [Green Version]
- Chen, I.C.; Kuo, P.H.; Yang, A.C.; Tsai, S.J.; Liu, T.H.; Liu, H.J.; Lan, T.H.; Chen, H.M.; Huang, H.N.; Chung, R.H.; et al. CUX2, BRAP and ALDH2 are associated with metabolic traits in people with excessive alcohol consumption. Sci. Rep. 2020, 10, 18118. [Google Scholar] [CrossRef] [PubMed]
- Biddinger, K.J.; Emdin, C.A.; Haas, M.E.; Wang, M.; Hindy, G.; Ellinor, P.T.; Kathiresan, S.; Khera, A.V.; Aragam, K.G. Association of Habitual Alcohol Intake With Risk of Cardiovascular Disease. JAMA Netw. Open 2022, 5, e223849. [Google Scholar] [CrossRef]
- Bulik-Sullivan, B.; Finucane, H.K.; Anttila, V.; Gusev, A.; Day, F.R.; Loh, P.R.; Duncan, L.; Perry, J.R.; Patterson, N.; Robinson, E.B.; et al. An atlas of genetic correlations across human diseases and traits. Nat. Genet. 2015, 47, 1236–1241. [Google Scholar] [CrossRef] [Green Version]
- Musabayane, C.T.; Brimble, M.J.; Balment, R.J. Renal sodium retention and vasopressin induced kaliuresis in ethanol anaesthetised rats. Acta Endocrinol. 1985, 110, 214–220. [Google Scholar] [CrossRef]
- Kawamura, M.; Kawasaki, T. Clinical application of the second morning urine method for estimating salt intake in patients with hypertension. Clin. Exp. Hypertens. 2015, 37, 89–96. [Google Scholar] [CrossRef]
- Cappuccio, F.P.; Beer, M.; Strazzullo, P. Population dietary salt reduction and the risk of cardiovascular disease. A scientific statement from the European Salt Action Network. Nutr. Metab. Cardiovasc. Dis. NMCD 2018, 29, 107–114. [Google Scholar] [CrossRef] [Green Version]
- Cherpitel, C.J.; Ye, Y.; Stockwell, T.; Vallance, K.; Chow, C. Recall bias across 7 days in self-reported alcohol consumption prior to injury among emergency department patients. Drug Alcohol Rev. 2018, 37, 382–388. [Google Scholar]
- Tan, M.; Wang, C.; Song, J.; He, F.J.; MacGregor, G.A. Spot urinary sodium to monitor relative changes in population salt intake during the UK salt reduction programme. J. Hypertens. 2022, 40, 1406–1410. [Google Scholar] [CrossRef]
| Overall (N = 295,189) | Males (N = 134,169) | Females (N = 161,020) | |
|---|---|---|---|
| Age at recruitment, mean (SD), years | 56.3 (8) | 56.4 (8.1) | 56.2 (7.9) |
| Males, N (%) | 134,169 (45.5) | NA | NA |
| Smoking, N (%) | |||
| Current | 30,388 (10.3) | 16,341 (12.2) | 14,047 (8.7) |
| Past | 148,209 (50.2) | 70,575 (52.6) | 77,634 (48.2) |
| Never | 116,592 (39.5) | 47,253 (35.2) | 69,339 (43.1) |
| Healthy diet score (DASH), mean (SD) | 2.7 (1) | 2.4 (1) | 2.9 (1) |
| Sedentary lifestyle, median [IQR), hours/day | 4 (3, 6) | 5 (3, 6) | 4 (3, 5) |
| SBP *, median (IQR), mmHg | 138.5 (125.5, 153.5) | 142 (130, 156) | 135 (122, 150.5) |
| DBP *, mean (SD), mmHg | 84.2 (11.2) | 86.5 (11) | 82.2 (11) |
| Stage 1 Hypertension †, N (%) | 68,284 (23.1) | 31,492 (23.5) | 36,792 (22.8) |
| Stage 2 Hypertension ‡, N (%) | 153,474 (52) | 79,680 (59.4) | 73,794 (45.8) |
| Townsend Deprivation Index, median (IQR) | −2.4 (−3.8, −0.06) | −2.4 (−3.8; −0.02) | −2.4 (−3.8; −0.1) |
| Urinary sodium, median (IQR) § | 68 (42.7, 102.8) | 81.9 (53.4, 117.5) | 57.6 (36.5, 88.3) |
| Composite cardiovascular disease, N (%) | 8688 (2.9) | 5808 (4.3) | 2880 (1.8) |
| Stroke, N (%) | 1857 (0.6) | 1048 (0.8) | 809 (0.5) |
| Regression Model | N Non-Cases/ N Cases | Effect Estimates * | 95% CI | p Value for GRS † | p Value for Interaction Term | PH ‡ | |
|---|---|---|---|---|---|---|---|
| SBP | Linear | 286,806 | 0.25 | 0.18, 0.32 | 3.12 × 10−12 | 0.052 | NR |
| DBP | Linear | 286,806 | 0.14 | 0.10, 0.19 | 3.50 × 10−12 | 0.33 | NR |
| Stage 1 Hypertension | Logistic | 71,322/66,395 | 1.01 | 1.00, 1.02 | 0.10 | 0.32 | NR |
| Stage 2 Hypertension | Logistic | 137,717/149,089 | 1.02 | 1.02, 1.03 | 9.78 × 10−9 | 0.03 | NR |
| Stroke | Cox PH | 285,012/1794 | 1.06 | 1.01, 1.11 | 0.02 | 0.72 | 0.23 |
| Composite CVD | Cox PH | 278,418/8388 | 1.01 | 0.99, 1.03 | 0.37 | 0.03 | 0.45 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jiang, X.; Anasanti, M.D.; Drenos, F.; Blakemore, A.I.; Pazoki, R. Urinary Sodium Excretion Enhances the Effect of Alcohol on Blood Pressure. Healthcare 2022, 10, 1296. https://doi.org/10.3390/healthcare10071296
Jiang X, Anasanti MD, Drenos F, Blakemore AI, Pazoki R. Urinary Sodium Excretion Enhances the Effect of Alcohol on Blood Pressure. Healthcare. 2022; 10(7):1296. https://doi.org/10.3390/healthcare10071296
Chicago/Turabian StyleJiang, Xiyun, Mila D. Anasanti, Fotios Drenos, Alexandra I. Blakemore, and Raha Pazoki. 2022. "Urinary Sodium Excretion Enhances the Effect of Alcohol on Blood Pressure" Healthcare 10, no. 7: 1296. https://doi.org/10.3390/healthcare10071296
APA StyleJiang, X., Anasanti, M. D., Drenos, F., Blakemore, A. I., & Pazoki, R. (2022). Urinary Sodium Excretion Enhances the Effect of Alcohol on Blood Pressure. Healthcare, 10(7), 1296. https://doi.org/10.3390/healthcare10071296






