Abstract
The solution of the so-called mixed-integer optimization problem is an important challenge for modern life sciences. A wide range of methods has been developed for its solution, including metaheuristics approaches. Here, a modification is proposed of the differential evolution entirely parallel (DEEP) method introduced recently that was successfully applied to mixed-integer optimization problems. The triangulation recombination rule was implemented and the recombination coefficients were included in the evolution process in order to increase the robustness of the optimization. The deduplication step included in the procedure ensures the uniqueness of individual integer-valued parameters in the solution vectors. The developed algorithms were implemented in the DEEP software package and applied to three bioinformatic problems. The application of the method to the optimization of predictors set in the genomic selection model in wheat resulted in dimensionality reduction such that the phenotype can be predicted with acceptable accuracy using a selected subset of SNP markers. The method was also successfully used to optimize the training set of samples for such a genomic selection model. According to the obtained results, the developed algorithm was capable of constructing a non-linear phenomenological regression model of gene expression in developing a Drosophila eye with almost the same average accuracy but significantly less standard deviation than the linear models obtained earlier.
1. Introduction
Modern life sciences, such as bioinformatics, heavily rely on numerical methods and computer algorithms. Optimization problems in which some unknown parameters of mathematical models are to be determined from data arise in many practical and theoretical studies, such as the identification of genes, proteins and regulatory networks, association between genotype and phenotype, prediction of agronomically important traits, and others. Unknown parameters often represent not only coefficients of floating point type, but also indices of some kind and thus are integer numbers. Consequently, the solution of so-called mixed-integer optimization problems is an important challenge for modern bioinformatics.
A wide range of methods has been developed for solution of such problems. Particularly, stochastic optimizers can be employed to obtain a global minimum of objective function given diverse biological data of a big volume. Different types of simulated annealing (SA) and genetic algorithms (GA) were successfully applied to biological problems [1]. Modern evolutionary algorithms, such as evolution strategies (ESs) or differential evolution (DE) [2], can outperform other methods in the estimation of parameters of several biological models [3,4,5]. In a recent study [6], it was shown that DE is robust against structural bias that may cause the optimization procedure to favor certain regions of the search space over others. The development of new methods for solution of mixed-integer optimization problem continues. In [7], mixed-integer evolution strategies (MIES) were presented, which are natural extensions of ES for mixed-integer optimization problems. A two-timescale duplex neurodynamic approach to mixed-integer optimization, based on a biconvex optimization problem reformulation with additional bilinear equality or inequality constraints was proposed in [8]. A genetic algorithm for model-based mixed-integer optimization (GAMBIT) was developed and extended with a parameterless scheme in [9]. Several approaches to mixed-integer optimization can be found within a commercial general algebraic modeling system (GAMS) framework [10]. Other alternatives include the covariance matrix adaptation evolutionary strategy CMA-ES [11], mixed-integer sequential quadratic programming (MISQP) [12,13] and a Bayesian network-based approach with the mixed-integer Bayesian optimization algorithm (MIBOA) by [14]. A method for the solution of very large mixed-integer dynamic optimization (MIDO) based on an parallel cooperative scatter search metaheuristic with new mechanisms of self-adaptation and specific extensions to handle large mixed-integer problems was introduced in [15].
The global optimization methods that may depend on problem-specific assumptions introduce difficulties in adaptation to other types of problems together with challenges in efficient implementation suitable in different areas. The suboptimal algorithm of temperature reduction, termed the cooling schedule, in SA can increase the computational time and lower the accuracy of the final solution. The correct choice of mutation, recombination and selection operators is crucial for success of GA application. The evolutionary algorithms are highly dependent on the parameters of the evolution model. A wide range of metaheuristic approaches have been developed for parameter estimation in biology. For example, enhanced scatter search, proposed in [16], is computationally expensive but is capable of obtaining integer and real parameters with high accuracy and outperforming the state-of-the-art methods [17].
Differential evolution (DE) [2,18] is a metaheuristic algorithm proposed by Rainer Storn and Kenneth Price. In the standard DE, population is randomly initialized and evolved using mutation and crossover operators to produce trial offspring together with a selection operator that accepts or rejects those trials into the population, comparing their values of the objective function.
The standard DE works with floating point parameters; therefore, DE algorithms for discrete parameters [19] were proposed. A modified binary DE (MBDE) algorithm and logical mutation operator was proposed in [20] based on the structure of binary bit strings. In a binary-adapted DE (BADE), developed in [21], the scaling factor is considered the probability that the scaled difference bit becomes “1”. In a discrete DE algorithm, proposed in [22], the real value obtained from the DE operators is converted to binary values using the sigmoid constraint function. The works [23,24] used the adaptive DE algorithm for feature selection, where parameters in DE are automatically adapted depending on the task. A hybrid approach, proposed in [25], combined strong exploratory properties of differential evolution with a modified artificial bee colony observation process for trait selection. In [26] the authors developed differential evolution with a binary mutation scheme for feature selection. A novel adapted mixed-integer differential evolution algorithm was designed in [27]. In [28], an improved binary difference evolution algorithm was developed for the feature selection problem for general classification problems in data mining.
In this work, we propose a modification of the differential evolution entirely parallel (DEEP) method introduced recently [29,30]. DEEP was successfully applied in mixed-integer optimization problems, such as the identification of the biogeographic origins of highly mixed individuals implemented in the online ancestry prediction tool [31], regression and dynamical models of flowering time in chickpea [32,33,34] and mungbean [35]. The DEEP method operates on real-valued parameters that can be converted to integers, using either rounding operation or array index transformation in which the resulting integers are the indices in the sorted array of real parameters. The rounding is better suited for many practical tasks, as a drawback of another transformation is that a relatively small change in real values can lead to a big perturbation of the solution. DEEP implements a parameter adaptation scheme based on preserving the diversity of solutions [36] that was developed for classic DE strategies. The local search capabilities of DE can be enhanced by including additional differences in the mutation. Consequently, in order to increase the robustness of the procedure, the triangulation recombination rule was implemented based on [37]. To control the diversity of the integer-valued parameters in intermediate and final solution vectors, the deduplication step was included based on [38]. To implement a new adaptation scheme, the control parameters of the resulting algorithm are made separate for each vector in the population and included in the evolution process.
Thus the contribution of the paper is three fold: (i) DEEP was enhanced with the triangulation recombination rule and deduplication step; (ii) the control parameters were included in the evolution process for self-adaptation; (iii) the resulting method implemented in the open-source software [39] was applied to three bioinformatic problems. The first two of them, namely the optimization of the set predictors and the compilation of the training sample, are important to reduce the dimensionality of the genomic selection model. The third example illustrates the construction of the regression model of gene expression in analytic form.
We compared the results for the genomic selection model with those for the DEEP method without the proposed enhancements and the genetic algorithm and the results for the third example were compared with a state-of-the-art package gramEvol specifically designed for building models in analytic form using the genetic algorithm for optimization [40].
The next sections of the paper give detailed description of the developed methods, test problems and results. The papers on modifications of the DE are surveyed in Section 2. The formal problem is stated in Section 3.1, and the differential evolution entirely parallel method and proposed enhancements are presented in Section 3.2–Section 3.4. The bioinformatic problems considered in this research are stated in Section 3.5–Section 3.8. The results are presented in Section 4 followed by their discussion in Section 5, and Section 6 provides the conclusions.
2. Related Work
Numerous variants of DE have been developed to improve the reliability, scalability, accuracy, convergence speed and other aspects of the method. Parameter adaptation methods were introduced that resulted in self-adaptive DE variants, such as SaDE [41] and ensemble-based SAEDE [42]. The mutation rule was improved by using the best member from the selected fraction of individuals and an external archive containing suboptimal individuals JADE [43]. Success history-based parameter adaptation SHADE was introduced in [44]. The classic mutation operator DE/rand/1 was enhanced by attaching a benchmark factor to the basis vector in IMMSADE [45]. A crossover probability sorting mechanism and dynamic population reduction strategy was proposed in EJADE [46]. Scale factor and crossover probability obeyed the Cauchy distribution and uniform distribution, respectively, and were adaptively adjusted, according to their respective weighted Lehmer means in ACDE [47]. Several ways for handling constraints imposed for parameters were introduced and investigated [48].
Different variants of improved parameter control were introduced, such as a multiple exponential recombination [49] and encoding of control parameters in individuals [50]. Improved individual-based parameter setting and selection strategy was developed in IDEI [51]. A DE variant with novel control parameter adaptation that includes a grouping strategy for adjusting the scaling factor and crossover probability and a parabolic reduction rule for changing NP was suggested in PaDE [52]. Mechanisms of population size adaption were also developed. In [53], the algorithm was proposed that evaluated the current population diversity based on the European distance and adjusted the NP size according to the evaluation results. The size of the population was also dynamically adjusted in APTS [54]. The population was divided into subpopulations in mDE-bES [55]. The population was clustered in multiple tribes in sTDE-dR [56].
Improved mutation and crossover strategies were proposed, such as a novel framework based on the proximity characteristics among the individual solutions in Pro-DE [57], a variant of the classical DE/current-to-best/1 scheme that uses the best of a selected group individual in MDE-pBX [58], a collective information-powered DE proposed in CIPDE [59], an improved DE algorithm with dual mutation strategies collaboration termed DMCDE [60], DE with a fitness and diversity ranking-based mutation operator called FDDE [61], ReDE [62] for which a revised mutation strategy for DE was introduced, RNDE [63] which used a new mutation operator, and DE/neighbor/1 which balanced the exploration and exploitation ability of the DE process. Other examples include DE with eigenvector-based crossover [64], DE with multiple exponential crossover [65], a switched parameter DE [66], a classification-based mutation strategy DE [67], and an orthogonal crossover (OX) operator DE [68].
Various offspring generation strategies were combined to produce an algorithm with enhanced properties, such as DE with composite strategies and parameters CoDE [65], DE that selects mutation strategies and control parameters from the strategy and parameter pools, respectively, to produce successful offspring population EPSDE [69], the framework with two mechanisms, namely, the multiple variants adaptive selecting mechanism and the multiple variants adaptive solutions preserving mechanisms [70], and improved ensemble of differential evolution variants (IEDEV) [71]. A method with multiple variant coordination was proposed in [72], the archiving of promising directions was suggested [73], and IMO-CADE was introduced in [74], where mutation operators were selected adaptively.
Hybrid algorithms were introduced with intent to integrate the advantages of DE and other meta-heuristic algorithms, such as a hybrid DE with the gaining–sharing knowledge algorithm (GSK) and Harris hawks optimization (HHO), DEGH [75], a hybrid artificial bee colony with DE (HABCDE) [76], a semi-parametric adaptation method in the LSHADE hybridized with covariance matrix adaptation evolution strategy (LSHADE-SPACMA) [77], a hybrid algorithm based on self-adaptive gravitational search algorithm (SGSADE) [78], a hybrid algorithm for DE and particle swarm optimization (DEPSO) [79], mixed DE with whale optimization algorithm (MDE-WOA) [80], a hybrid adaptive teaching–learning-based optimization algorithm with DE (ATLDE) [81], a hybrid symbiotic DE moth flame optimization algorithm (HSDE-MFO) [82], a modified Boltzmann annealing [83] differential evolution algorithm (BADE) [84], an adaptive DE with PSO (A-DEPSO) [85], a hybrid differential symbiotic organism search (HDSOS) algorithm [86], a new local search scheme based on the Hadamard matrix (HLS) [87], an opposition-based learning DE (ODE) [88], DE/EDA [89], which combined DE with the estimation of distribution algorithm, a DE variant with commensal learning and uniform local search, named CUDE [90], and ESADE [91], which combines simulated annealing in the selection stage.
3. Materials and Methods
3.1. Problem Statement
Let us consider mixed-integer optimization problem (1) that combines unknowns that take real and integer values.
where is the vector of parameters of the model of the length K that is to be found to give the minimal value to the objective function Q and satisfy the constraints with upper and lower limits and , respectively. Q computes the deviation of the model with parameters from experimental data . The subset I holds the indices of parameters that are integer numbers.
3.2. Differential Evolution Entirely Parallel Method
Differential evolution (DE) was proposed in [18] as an effective stochastic method for function minimization. DE operates on a set called population of parameter vectors called individuals that are initially generated randomly. The size of population is constant, and on each iteration, the set of parameter vectors may be referred to as generation. The differential evolution entirely parallel (DEEP) method [29,30,92] combines enhancements proposed in the literature as well as some specifically developed modifications. These enhancements include the two-step scheme for adaptation of algorithmic parameters based on the control of the population diversity developed in [36], an optional scatter search step [16] that is performed each iteration, where is a control parameter, and a selection rule [93] for consideration of several different objective functions. To avoid premature convergence, the predefined number of individuals that were unable to update the parameters after the predefined number of iterations are substituted with the same number of ones with the lowest values of the objective function. To satisfy the constraints in the form of inequalities imposed for the model parameters, the infeasible solutions that might be generated by the method are substituted with the values uniformly sampled from the constrained interval.
DE operates on floating point parameters. The conversion from real to integer values can be performed using one of the two approaches implemented in the DEEP method. The first option employs the rounding operation that substitutes a real value with the nearest integer number. The second approach involves the following operations:
- The values are sorted in ascending order.
- The index of the parameter in the floating point array becomes the value of the parameter in the integer array.
The rounding method is more convenient for the solution of practical problems because in the second procedure, a relatively small change in the real value of the parameter can lead to a relatively important perturbation of the solution. The local search capabilities of classic DE strategies can be improved by including additional differences in the mutation formula. This was done using the triangulation recombination rule based on [37].
As the parameter adaptation scheme based on preserving the population diversity [36] was developed for classic DE strategies, another approach is introduced in this work. To avoid equally valued integer parameters in the trial and final solution vectors, the deduplication procedure is performed on each iteration based on [38]. The control parameters of the triangulation recombination rule are made different for each individual vector in the population. They are included in the evolution process and updated on each iteration with the same formulae.
The operation of the method, including the two enhancements proposed in this work, is described in the Algorithm 1 insertion. The input to the algorithm includes the following:
- The specification of the problem to be solved:
- -
- The objective function;
- -
- The data with which the solution is to be compared;
- -
- Constraints for the parameters;
- Control parameters of the algorithm.
The execution starts with the initialization step that randomly assigns the values satisfying the given constraints , to a set of parameter vectors for starting generation .
The main loop of the algorithm consists of recombination, evaluation and selection steps. Calculations are terminated when the stopping criterion is satisfied and can be defined by the maximal allowed number of generations or the number of consecutive iterations during which the objective function variation is less than a predefined small value. Objective function Q for several vectors is calculated in parallel.
The number of individuals in population , the frequencies and , the number of substituted individuals and stopping criterion parameter are the main control parameters of the method that are to be set by the user.
The output of the algorithm contains the values of unknowns that provides the minimal value of the objective function.
| Algorithm 1: Differential evolution entirely parallel method |
| INITIALIZATION: |
| The individuals of the population are initialized randomly. |
| Generation |
| repeat |
| RECOMBINATION: |
| if The predefined number of iterations passed. then |
| Make Scatter Search Step |
| else |
| for all individuals in population do |
| Triangulation Recombination of model parameters (2)-(6) |
| Update scaling coefficients and weights (7), (8) |
| end for |
| end if |
| for all offsprings do |
| Make Deduplication of integer-valued parameters |
| end for |
| EVALUATION: |
| for all offsprings do |
| Calculate objective function Q |
| end for |
| SELECTION: |
| for all offsprings do |
| Accept or Reject an offspring to the next generation |
| end for |
| if The predefined number of generations passed then |
| Substitute oldest individuals with the best ones |
| end if |
| until Stopping criterion is met |
3.3. Triangulation Recombination Rule
In order to increase the robustness of the procedure, the triangulation recombination rule was implemented based on [37]. The trial vector for each i-th vector from population for current generation G is obtained according to formulae (2)–(6):
where b, t, and r are random indices sampled from the uniform distribution so that , , and are the best, better and the worst vectors with respect to objective function Q; and are the normalized and nonnormalized weights, respectively; and are the scaling coefficients.
To adaptively change the weights and scaling coefficients, they are evolved using the same formulae (4)–(6):
where and are trial vectors of scaling coefficients and nonnormalized weights, respectively, and and are the sets of all such vectors for the current generation G.
3.4. Deduplication
To avoid equal integer-valued parameters in intermediate and final solution vectors, the deduplication step for each offspring was included based on [38]. The algorithm performs the following operations for each trial vector :
- -
- Sample another index h uniformly, different from b, t and r used in recombination.
- -
- Create a helper array: assign 1 if the value firstly occurs in , assign 2 if the value firstly occurs in and 3—if the value is duplicated.
- -
- Sort the helper array in ascending order.
- -
- Fill with the values that corresponds to the first K elements of the helper array.
The execution of the algorithm is illustrated in Table 1. Here, the values 8 (in the second column), 22 and 15 are firstly met in ; 8 in the fourth column is duplicated; and values 84, 47 and 105 are firstly met in . The helper array is filled accordingly and then sorted. The values that correspond to the first K indices in the sorted helper array are included in the updated .

Table 1.
Deduplication algorithm. The first row is the concatenation of two individual vectors of parameters, and . The next two rows illustrate original and sorted helper arrays. The last row shows the result of the procedure.
The time need to complete the deduplication procedure depends on the number of parameters K and, with the fast sorting algorithm, is estimated as .
3.5. Genomic Selection
More recently, it has become clear that improved crop production can only be achieved if the potential of varieties can be assessed at different locations and seasons and in combination with a range of management options that cannot be tested in vivo [94,95,96]. Thus, world-class breeding programs have reformed their yield improvement strategy by integrating traditional yield estimates with gene–phenotype–system modeling tools [97,98,99,100]. This integration allows in silico identification of genotypes associated with phenotypes that are likely to improve crop production [101,102,103]. Such approaches led to a 3–4% increase in yields per year under the Australian sorghum program and ≈1% growth under the U.S. Northern Production Zone corn program. These techniques have not only proven to be effective, but are rapidly improving [100].
Breeding value assessment models suffer from the so-called curse of dimensionality—a scenario in which the number of possible SNP markers is significantly larger compared to the available observations [104].
The standard approach to calculating genomic breeding value is to use the best linear unbiased predictor (BLUP), using a linear model and ridge regression [105]. Bayesian methods have also found widespread use [104]. In this work, an implementation provided in rrBLUP package [106] for R software system [107] is utilized.
Here, we apply the developed algorithm to two independent problems with two different datasets.
3.6. Optimization of the Set of Predictors
Ridge regression helps to overcome the dimensionality problem, but the set of predictors can be optimized to obtain the same accuracy with less SNPs used [28]. Thus the optimization problem is formulated to select the subset of predictors that maximize the correlation between the estimated and measured phenotype. To reduce overfitting, 4-fold cross-validation is utilized, and the objective function equals the mean Pearson’s correlation coefficient for the model built by rrBLUP [106].
The algorithm was applied to a wheat dataset [108] phenotyped for baking quality traits that was split into training and test samples of 1516 and 300 elements, respectively, and genotyped for 5021 SNP markers.
3.7. Compilation of Training Sample
An important problem in the wide adoption of genomic selection is that the accuracy of the prediction of the phenotype for new genotypes is hard to estimate beforehand. It was shown that it is possible to achieve a substantial gain in accuracy by careful compilation of the sample on which the genomic selection model is trained. As a result, EthAcc (estimated theoretical accuracy) was proposed in [109] as an objective function for the training sample optimization. EthAcc is based on causal quantitative trait loci assessed from a genome-wide association study. EthAcc does not evaluate the training set but the genomic prediction accuracy based on the training set. The choice of the best training set from the historical panel of genotyped and phenotyped samples is a key task in the evaluation of newly created breeding resources.
The developed algorithm was applied to a wheat dataset provided in [109] containing information on 1368 SNPs and 237 samples that were phenotyped for the heading date. The algorithm and data for EthAcc computation were taken from the original paper [109].
3.8. Regression Model of Gene Expression
Gene expression controls all biological processes and gene regulation can be modeled in several ways: differential equations for dynamical models and linear or non-linear regression in statistical models [110]. The fruit fly Drosophila is an ideal model organism for different biological studies. In [111], the robustness in development was investigated using quantitative spatial gene expression in the morphogenetic furrow (MF), a wave of differentiation that patterns the developing eye disc. The MF passes from the posterior to the anterior of the disc over a period of two days (90 min per adjacent row of cells). The method of hybridization chain reaction (HCR) was used to quantitatively measure spatial gene expression, co-staining for several genes simultaneously. Three dimensional images of mounted, HCR stained eye discs were acquired on a Zeiss LSM 780 laser scanning microscope. Gene transcripts were detected within image stacks after processing.
The gene atonal plays a central role in establishing the correct eye structure. It is known that its expression is under the control of hedgehog and Delta genes. In [111], a regulation was modeled by linear regression function. A more general approach called grammatical evolution (GE) [112] allows the user to determine the analytic form of functional dependence by minimizing the deviation of model solution from data. In GE, as in a genetic programming method, the function is constructed according to the rules of the given context-free grammar from “words” that are integer-valued vectors of parameters.
The data represent a quantitative description of gene expression, presented in the form of bursts of intensities obtained by the processing of images of the eye discs of fruit fly Drosophila larvae. The information is provided for 85 discs in the form of the table for each processed sample with one row for each cell in a disc, approximately 1000 cells per disc. The rows contain values of measured gene expression.
3.9. Experimental Setup
The described algorithm was implemented in C programming language in the framework of the open-source software DEEP [39]. For each case study, the experimental setup contained the formulation of the objective function Q to be minimized, the upper and lower limits for unknown model parameters and selected control parameters for the algorithm.
The control parameters , , , and were chosen to be the same as those used in the successful previous applications of the method [32,35]. The objective function Q used in the evaluation step in Algorithm 1 measures the deviation of the model solution from provided experimental data. In practice, any executable program written in any programming language can be utilized; two R-scripts and a program written in C++ were used in the case studies described in Section 3.6–Section 3.8, respectively.
The specification of the problem is stored in the INI-file that is loaded by the DEEP software.
4. Results
4.1. Optimization of the Set of Predictors
The developed algorithm was used to optimize the set of SNPs that serve as predictors in a linear regression model of genomic selection. The integer-valued parameters represent the indices of SNPs that are included in the model that was built using the rrBLUP package for R statistical software. The Pearson’s correlation coefficient between predicted and measured phenotypes was used as an objective function. Several runs with different seeds for a random number generator were performed that took, on average, hours on a cluster node equipped with 2xIntel Xeon CPU E5-2697 v3, 2.60GHz.
Each resulting model reduced the number of needed SNPs from 5021 to 1800–3800, with the correlation coefficient ranging from to and from to for training and test data, respectively. The accuracy is not dependent on the number of predictors in these results (see Figure 1). The average number of selected SNPs was 2939, and the average correlation coefficient is for the test data and for the training data. The smallest model contains 1456 SNPs, and the correlation coefficient equals for the test set.
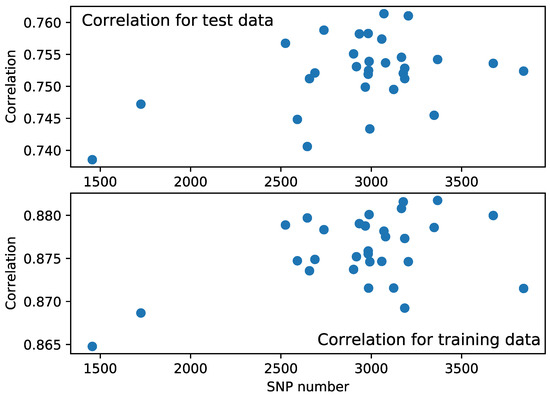
Figure 1.
Pearson correlation coefficient between data and model prediction with respect to the number of SNPs used in the model for test and training samples.
Different combination of SNPs were obtained in different runs, and each SNP was selected in aa different number of combinations (see Figure 2). The average frequency of the SNP selection was 65%, and 264 SNPs were selected in more than 80% combinations.
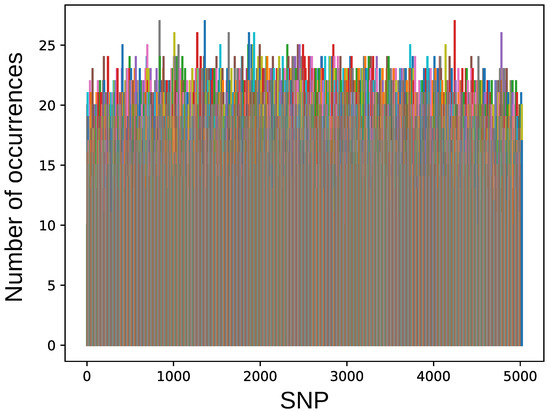
Figure 2.
The number of occurrences for each SNP in obtained solutions. The colors are made random for better visual appearance.
Three SNPs named IWA837, IWA1360, and IWA4243 were selected in 90% solutions and six SNPs named IWA1008, IWA1273, IWA1637, IWA1870, IWA1929, and IWA4784 were selected in 87% solutions.
The results obtained with the developed method were compared with those for the base DEEP method without proposed enhancements. The new method outperforms the base one in terms of the smallest model, as the base method needs 2400 SNPs instead of 1456 with almost the same correlation coefficient, equal to , for the test set.
4.2. Compilation of Training Sample
The DEEP method with proposed modifications was applied to a problem of compilation of the set of samples from the given dataset to be used for training of regression model for genomic selection using EthAcc as an objective function. The implementation of the EthAcc computation algorithm and additional data were taken from the original paper [109]. The integer-valued parameters of the model represented the indices of the observations to be included in training. Several runs were performed with different initial values for the random number generator, with the mean runtime of 42 minutes on a cluster node equipped with 2xIntel Xeon CPU E5-2697 v3, 2.60 GHz.
The value of the objective function was reduced almost twice for a typical run according to the convergence curve (see Figure 3). The value of the objective function EthAcc was significantly positively correlated (, p-value ) with the accuracy of the final model expressed as the correlation between the predicted and measured phenotypes (see Figure 4), as was expected in [109].
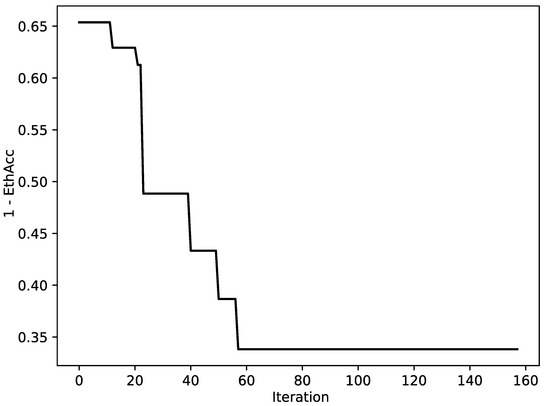
Figure 3.
The convergence curve for a typical optimization run. The value (1—EthAcc) is plotted as a function of iteration number.
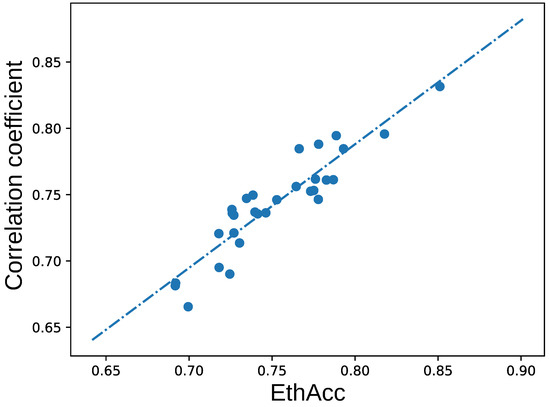
Figure 4.
Pearson correlation coefficient between predicted and measured values for models with different EthAcc values. The strait line illustrates the linear dependence.
The frequency of inclusion of individual samples into training set varies greatly between obtained solutions (see Figure 5). The mean number of samples in the resulting training set was 70 out of 237 records, with the mean accuracy of ; 28 samples were included in 70% of solutions.
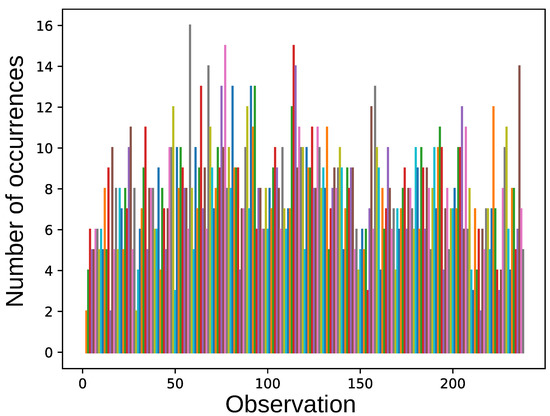
Figure 5.
The number of occurrences of each observation in the optimized training set. The colors are made random for better visual appearance.
The results obtained with the developed method were compared with those for the base DEEP method without proposed enhancements and the state-of-the-art genetic algorithm implemented in package GA [113] for R statistical software. According to the Mann–Whitney criterion, the comparison was statistically significant (p-value ); consequently, the developed method outperformed the competitors in all cases (see Table 2).

Table 2.
Comparison of results for the problem of compilation of training sample for the method developed in this work (new), for DEEP without enhancements with DE/rand/1/bin, DE/rand/1/bin, DE/best/1/bin and DE/best/1/exp strategies and for GA with real-valued (GA/rvd) and binary encoding (GA/bin), also with a additional local search step (GA/rvd/loc and GA/bin/loc).
4.3. Regression Model of Gene Expression
The developed algorithm was used to optimize the analytic form of the phenomenological function that describes how the expression of the atonal gene depends on the expression of hedgehog and Delta genes in cells of morphogenetic furrow of developing eye disc. The data on gene expression were split into folds for cross validation. The model was trained on parts of the sample and tested on a remaining part.
Parameters of the mixed-integer optimization problem include integer indices interpreted according to the grammatical evolution rules and real-valued coefficients. The best solution (9) with respect to the root mean squared () difference between predicted and measured gene expression suggests the non-linear dependence between gene expression levels (see Figure 6).
where is an adjusted coefficient of determination, , and are expressions of genes hedgehog, Delta and atonal, respectively. The average accuracy of the obtained non-linear solutions () is slightly better than that of linear regressions () obtained in [111], but the deviation of accuracy is statistically significantly smaller () according to the F-test (p-value ).
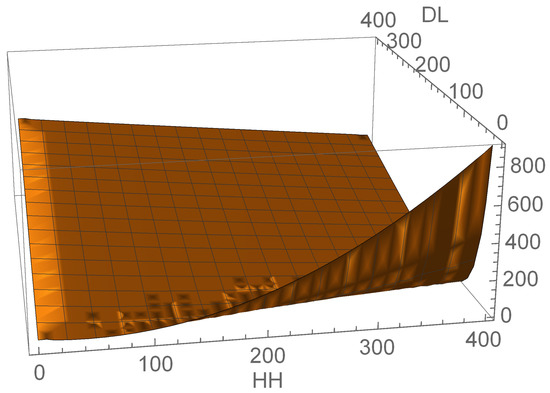
Figure 6.
The non-linear dependence of atonal expression on that of hedgehog and Delta genes in the cells of developing Drosophila eye.
We compared the obtained results with a state-of-the-art method for grammatical evolution implemented in the gramEvol [40] package for R statistical software [107]. We used default recommended settings 40, 200 and 300 for chromosome size, population size and iterations, respectively. The default mutation probability was set to . The mean adjusted coefficient of determination for the developed method was statistically significantly better than that of the competitor, , according to the Mann–Whitney criterion (, ).
5. Discussion
Feature selection methods are often subdivided in two main groups [114,115]: filtering and wrapper methods. The filtering methods select features based on information content or statistics and do not depend on any learning algorithm. Thus, the filtering techniques are generally less computationally expensive, and are fast, and suitable for large datasets. Moreover, they are easily applicable to various learning algorithms. For ranking features, filtering methods are commonly used, such as Fisher’s score, T statistic, entropy-based feature ranking, Bhattacharya distance, etc. [116,117]. However, the filtering methods ignore the interaction of features and the performance of the classification algorithm with the selected features, while the wrapper methods evaluate subsets of features based on the classification efficiency, which is highly dependent on the specific algorithm used. It is known that wrapper methods, such as, for example, SVM (SVM-R), cooperative index (CI), K-top scoring pair (k-TSP), microarray significance analysis (SAM), positive synergy index (PSI) and greedy forward selection (GFS) [118,119] are more accurate than filtering methods, but require more computation and are more likely to lead to overfitting. In [28], an improved binary differential evolution (BDE) algorithm was developed for the feature selection problem for general classification problems in data mining.
The differential evolution entirely parallel (DEEP) method introduced recently [29,30] is capable of finding integer-valued parameters that can represent indices of selected features. In this work, the method was enhanced by the deduplication step based on [38] to avoid equal integer-valued parameters in the intermediate and final solution vectors and by the triangulation recombination rule based on [37] to increase the robustness of the procedure.
The implementation of DEEP [39] was applied to mixed-integer problems, such as the optimization of the predictors set and training set for genomic selection and the gene regulation regression model in symbolic form.
The accuracy of regression models in genomic selection is suppressed by the fact that the number of SNP markers is usually much larger than the number of observations—the so-called curse of dimensionality [104]. Consequently, the dimensionality reduction and training set optimization are important challenges.
The ability of the developed algorithm to optimize the set of SNP predictors for genomic selection was evaluated on the wheat dataset presented in [108]. The developed algorithm successfully constructed a model with fewer predictors and acceptable accuracy, thereby reducing the dimensionality of the problem. The average number of selected SNPs was 2939, and the average correlation coefficient was for the test data and for the training data. The small set of 9 markers—IWA1008, IWA1273, IWA1637, IWA1870, IWA1929, IWA4784, IWA837, IWA1360, and IWA4243—was selected in more than 87% of solutions, which suggests their high impact on the modeled phenotype.
The developed algorithm was successfully used to determine the optimal training set for genomic selection for the heading date in wheat, using the maximization of EthAcc (estimated theoretical accuracy) as an objective function [109]. The mean number of samples in the resulting training set was 70 out of 237 records with the mean accuracy of ; 28 samples were included in 70% of solutions.
The DEEP method with proposed modifications was used to obtain a non-linear phenomenological model of the expression of atonal gene expression under the control of hedgehog and Delta genes in the developing Drosophila eye, using the grammatical evolution (GE) approach [112]. The quantitative data on gene expression obtained by processing of images of the eye discs were used for model construction [111]. The obtained solutions are highly non-linear, and the adjusted coefficient of determination showed almost the same average accuracy but significantly less deviation than the linear models obtained earlier.
These problems represent important areas of application of mathematical modeling in biology. The prediction of important phenotypic characteristics based on the combination of genetic markers can be used in selection, as in the tested case study, and also in biomedicine for early screening of diseases. The models of the regulation of gene expression help to decipher the mechanism of development. The successful application of the method to these diverse problems supports its applicability in other biological tasks.
6. Conclusions
The differential evolution entirely parallel method was enhanced by the deduplication step and by the triangulation recombination rule for solving mixed-integer optimization problems.
The application of the method resulted in dimensionality reduction in the genomic selection model in wheat such that the phenotype can be predicted with acceptable accuracy, using a selected subset of SNP markers. The method was also successfully used to optimize the training set of samples for such a genomic selection model.
According to the obtained results, the developed algorithm is capable of constructing a non-linear phenomenological model of gene expression in the developing Drosophila eye with almost the same average accuracy but significantly less deviation than the linear models obtained earlier.
Author Contributions
Conceptualization, M.S. and K.K.; methodology, M.S. and K.K.; software, S.S., D.M. and K.K.; validation, K.K.; formal analysis, investigation, S.S., D.M. and K.K.; resources, data curation, M.S.; writing—original draft preparation, review and editing, S.S., D.M., M.S. and K.K.; visualization, S.S. and D.M.; supervision, project administration, funding acquisition, M.S. All authors have read and agreed to the published version of the manuscript.
Funding
The research was funded by the Ministry of Science and Higher Education of the Russian Federation as part of World-class Research Center program: Advanced Digital Technologies (contract No. 075-15-2020-934 dated 17 November 2020).
Data Availability Statement
The data analyzed in this study are available in Zenodo at 10.5281/zenodo.5792935.
Acknowledgments
Calculations were performed in Supercomputer center of Peter the Great St.Petersburg Polytechnic University.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Jaeger, J.; Surkova, S.; Blagov, M.; Janssens, H.; Kosman, D.; Kozlov, K.N.; Myasnikova, E.; Vanario-Alonso, C.E.; Samsonova, M.; Sharp, D.H.; et al. Dynamic control of positional information in the early Drosophila embryo. Nature 2004, 430, 368–371. [Google Scholar] [CrossRef]
- Storn, R.; Price, K. Differential Evolution—A Simple and Efficient Heuristic for Global Optimization over Continuous Spaces; Technical Report Technical Report TR-95-012; International Computer Science Institute: Berkeley, CA, USA, 1995. [Google Scholar]
- Fomekong-Nanfack, Y.; Kaandorp, J.A.; Blom, J. Efficient parameter estimation for spatio-temporal models of pattern formation: Case study of Drosophila melanogaster. Bioinformatics 2007, 23, 3356–3363. [Google Scholar] [CrossRef]
- Fomekong-Nanfack, Y.; Postma, M.; Kaandorp, J.A. Inferring Drosophila gap gene regulatory network: A parameter sensitivity and perturbation analysis. BMC Syst. Biol. 2009, 3, 94. [Google Scholar] [CrossRef] [PubMed]
- Suleimenov, Y.; Ay, A.; Samee, M.A.H.; Dresch, J.M.; Sinha, S.; Arnosti, D.N. Global parameter estimation for thermodynamic models of transcriptional regulation. Methods 2013, 62, 99–108. [Google Scholar] [CrossRef]
- Caraffini, F.; Kononova, A.V.; Corne, D. Infeasibility and structural bias in differential evolution. Inf. Sci. 2019, 496, 161–179. [Google Scholar] [CrossRef]
- Li, R.; Emmerich, M.T.; Eggermont, J.; Bäck, T.; Schütz, M.; Dijkstra, J.; Reiber, J. Mixed Integer Evolution Strategies for Parameter Optimization. Evol. Comput. 2013, 21, 29–64. [Google Scholar] [CrossRef] [PubMed]
- Che, H.; Wang, J. A Two-Timescale Duplex Neurodynamic Approach to Mixed-Integer Optimization. IEEE Trans. Neural Networks Learn. Syst. 2021, 32, 36–48. [Google Scholar] [CrossRef]
- Sadowski, K.L.; Thierens, D.; Bosman, P.A. GAMBIT: A Parameterless Model-Based Evolutionary Algorithm for Mixed-Integer Problems. Evol. Comput. 2018, 26, 117–143. [Google Scholar] [CrossRef] [PubMed]
- Bussieck, M.R.; Meeraus, A. General Algebraic Modeling System (GAMS). In Modeling Languages in Mathematical Optimization; Series Title: Applied, Optimization; Pardalos, P.M., Hearn, D.W., Kallrath, J., Eds.; Springer: Boston, MA, USA, 2004; Volume 88, pp. 137–157. [Google Scholar] [CrossRef]
- Hansen, N. ACMA-ES for Mixed-Integer Nonlinear Optimization; Technical Report 7751; Institut National De Recherche En Informatique Et En Automatique: Le Chesnay-Rocquencourt, France, 2011. [Google Scholar]
- Exler, O.; Schittkowski, K. A trust region SQP algorithm for mixed-integer nonlinear programming. Optim. Lett. 2007, 1, 269–280. [Google Scholar] [CrossRef]
- Exler, O.; Lehmann, T.; Schittkowski, K. A comparative study of SQP-type algorithms for nonlinear and nonconvex mixed-integer optimization. Math. Program. Comput. 2012, 4, 383–412. [Google Scholar] [CrossRef]
- Emmerich, M.T.M.; Li, R.; Zhang, A.; Lucas, P.; Flesch, I. Mixed-Integer Bayesian Optimization Utilizing A-Priori Knowledge on Parameter Dependences. In Proceedings of the 20th Belgium-Netherlands Conference on Artificial Intelligence, BNAIC, Leiden, The Netherlands, 19–20 November 2008; pp. 65–72. [Google Scholar]
- Penas, D.R.; Henriques, D.; González, P.; Doallo, R.; Saez-Rodriguez, J.; Banga, J.R. A parallel metaheuristic for large mixed-integer dynamic optimization problems, with applications in computational biology. PLoS ONE 2017, 12, e0182186. [Google Scholar] [CrossRef]
- Egea, J.A.; Martí, R.; Banga, J.R. An evolutionary method for complex-process optimization. Comput. Oper. Res. 2010, 37, 315–324. [Google Scholar] [CrossRef]
- Egea, J.A.; Henriques, D.; Cokelaer, T.; Villaverde, A.F.; MacNamara, A.; Danciu, D.P.; Banga, J.R.; Saez-Rodriguez, J. MEIGO: An open-source software suite based on metaheuristics for global optimization in systems biology and bioinformatics. BMC Bioinform. 2014, 15, 136. [Google Scholar] [CrossRef]
- Storn, R. Differential Evolution—A Simple and Efficient Heuristic for Global Optimization over Continuous Spaces. J. Glob. Optim. 1997, 11, 341–359. [Google Scholar] [CrossRef]
- Khushaba, R.N.; Al-Jumaily, A.; Al-Ani, A. Evolutionary fuzzy discriminant analysis feature projection technique in myoelectric control. Pattern Recognit. Lett. 2009, 30, 699–707. [Google Scholar] [CrossRef]
- Wu, C.Y.; Tseng, K.Y. Truss structure optimization using adaptive multi-population differential evolution. Struct. Multidiscip. Optim. 2010, 42, 575–590. [Google Scholar] [CrossRef]
- Gong, T.; Tuson, A.L. Differential Evolution for Binary Encoding. In Soft Computing in Industrial Applications; Saad, A., Dahal, K., Sarfraz, M., Roy, R., Eds.; Springer: Berlin/Heidelberg, Germany, 2007; Volume 39, pp. 251–262. [Google Scholar] [CrossRef]
- He, X.; Zhang, Q.; Sun, N.; Dong, Y. Feature Selection with Discrete Binary Differential Evolution. In Proceedings of the 2009 International Conference on Artificial Intelligence and Computational Intelligence, Shanghai, China, 7–8 November 2009; pp. 327–330. [Google Scholar] [CrossRef]
- Ghosh, A.; Datta, A.; Ghosh, S. Self-adaptive differential evolution for feature selection in hyperspectral image data. Appl. Soft Comput. 2013, 13, 1969–1977. [Google Scholar] [CrossRef]
- Akbar, A.; Kuanar, A.; Joshi, R.K.; Sandeep, I.S.; Mohanty, S.; Naik, P.K.; Mishra, A.; Nayak, S. Development of Prediction Model and Experimental Validation in Predicting the Curcumin Content of Turmeric (Curcuma longa L.). Front. Plant Sci. 2016, 7, 1507. [Google Scholar] [CrossRef] [PubMed]
- Zorarpacı, E.; Özel, S.A. A hybrid approach of differential evolution and artificial bee colony for feature selection. Expert Syst. Appl. 2016, 62, 91–103. [Google Scholar] [CrossRef]
- Chattopadhyay, S.; Mishra, S.; Goswami, S. Feature selection using differential evolution with binary mutation scheme. In Proceedings of the 2016 International Conference on Microelectronics, Computing and Communications (MicroCom), Durgapur, India, 23–25 January 2016; pp. 1–6. [Google Scholar] [CrossRef]
- Zhao, W.; Beach, T.H.; Rezgui, Y. A systematic mixed-integer differential evolution approach for water network operational optimization. Proc. R. Soc. A 2018, 474, 20170879. [Google Scholar] [CrossRef]
- Zhao, X.S.; Bao, L.L.; Ning, Q.; Ji, J.C.; Zhao, X.W. An Improved Binary Differential Evolution Algorithm for Feature Selection in Molecular Signatures. Mol. Inform. 2018, 37, 1700081. [Google Scholar] [CrossRef] [PubMed]
- Kozlov, K.; Samsonov, A. DEEP—differential evolution entirely parallel method for gene regulatory networks. J. Supercomput. 2011, 57, 172–178. [Google Scholar] [CrossRef] [PubMed]
- Kozlov, K.; Samsonov, A.M.; Samsonova, M. A software for parameter optimization with Differential Evolution Entirely Parallel method. PeerJ Comput. Sci. 2016, 2, e74. [Google Scholar] [CrossRef][Green Version]
- Kozlov, K.; Chebotarev, D.; Hassan, M.; Triska, M.; Triska, P.; Flegontov, P.; Tatarinova, T. Differential Evolution approach to detect recent admixture. BMC Genom. 2015, 16, S9. [Google Scholar] [CrossRef]
- Ageev, A.; Aydogan, A.; Bishop-von Wettberg, E.; Nuzhdin, S.V.; Samsonova, M.; Kozlov, K. Simulation Model for Time to Flowering with Climatic and Genetic Inputs for Wild Chickpea. Agronomy 2021, 11, 1389. [Google Scholar] [CrossRef]
- Ageev, A.Y.; Bishop-von Wettberg, E.J.; Nuzhdin, S.V.; Samsonova, M.G.; Kozlov, K.N. Forecasting the Timing of Floral Initiation in Wild Chickpeas under Climate Change. Biophysics 2021, 66, 107–116. [Google Scholar] [CrossRef]
- Kozlov, K.; Singh, A.; Berger, J.; Wettberg, E.B.V.; Kahraman, A.; Aydogan, A.; Cook, D.; Nuzhdin, S.; Samsonova, M. Non-linear regression models for time to flowering in wild chickpea combine genetic and climatic factors. BMC Plant Biol. 2019, 19, 94. [Google Scholar] [CrossRef] [PubMed]
- Kozlov, K.; Sokolkova, A.; Lee, C.R.; Ting, C.T.; Schafleitner, R.; Bishop-von Wettberg, E.; Nuzhdin, S.; Samsonova, M. Dynamical climatic model for time to flowering in Vigna radiata. BMC Plant Biol. 2020, 20, 202. [Google Scholar] [CrossRef]
- Zaharie, D. Parameter Adaptation in Differential Evolution by Controlling the Population Diversity. In Seria Matematica-Informatica, Proceedings of the 4th InternationalWorkshop on Symbolic and Numeric Algorithms for Scientific Computing, Timişoara, Romania, 9–12 October 2002; Petcu, D., Ed.; Analele Universitatii Timisoara: Timisoara, Romania, 2002; Volume XL, pp. 385–397. [Google Scholar]
- Mohamed, A.W.; Almazyad, A.S. Differential Evolution with Novel Mutation and Adaptive Crossover Strategies for Solving Large Scale Global Optimization Problems. Appl. Comput. Intell. Soft Comput. 2017, 2017, 7974218. [Google Scholar] [CrossRef]
- Uher, V.; Gajdoš, P.; Radecký, M.; Snášel, V. Utilization of the Discrete Differential Evolution for Optimization in Multidimensional Point Clouds. Comput. Intell. Neurosci. 2016, 2016, 6329530. [Google Scholar] [CrossRef]
- Kozlov, K. DEEP. Available online: https://gitlab.com/mackoel/deepmethod (accessed on 29 April 2021).
- Noorian, F.; de Silva, A.M.; Leong, P.H.W. gramEvol: Grammatical Evolution in R. J. Stat. Softw. 2016, 71, 1–26. [Google Scholar] [CrossRef]
- Qin, A.; Huang, V.; Suganthan, P. Differential Evolution Algorithm with Strategy Adaptation for Global Numerical Optimization. IEEE Trans. Evol. Comput. 2009, 13, 398–417. [Google Scholar] [CrossRef]
- Wang, S.L.; Morsidi, F.; Ng, T.F.; Budiman, H.; Neoh, S.C. Insights into the effects of control parameters and mutation strategy on self-adaptive ensemble-based differential evolution. Inf. Sci. 2020, 514, 203–233. [Google Scholar] [CrossRef]
- Zhang, J.; Sanderson, A. JADE: Adaptive Differential Evolution with Optional External Archive. IEEE Trans. Evol. Comput. 2009, 13, 945–958. [Google Scholar] [CrossRef]
- Tanabe, R.; Fukunaga, A. Success-history based parameter adaptation for Differential Evolution. In Proceedings of the 2013 IEEE Congress on Evolutionary Computation, Cancun, Mexico, 20–23 June 2013; pp. 71–78. [Google Scholar] [CrossRef]
- Wang, S.; Li, Y.; Yang, H. Self-adaptive differential evolution algorithm with improved mutation mode. Appl. Intell. 2017, 47, 644–658. [Google Scholar] [CrossRef]
- Li, S.; Gu, Q.; Gong, W.; Ning, B. An enhanced adaptive differential evolution algorithm for parameter extraction of photovoltaic models. Energy Convers. Manag. 2020, 205, 112443. [Google Scholar] [CrossRef]
- Xue, X.; Chen, J. Matching biomedical ontologies through compact differential evolution algorithm. Syst. Sci. Control. Eng. 2019, 7, 85–89. [Google Scholar] [CrossRef]
- Kononova, A.V.; Caraffini, F.; Bäck, T. Differential evolution outside the box. Inf. Sci. 2021, 581, 587–604. [Google Scholar] [CrossRef]
- Qiu, X.; Tan, K.C.; Xu, J.X. Multiple Exponential Recombination for Differential Evolution. IEEE Trans. Cybern. 2017, 47, 995–1006. [Google Scholar] [CrossRef]
- Brest, J.; Zumer, V.; Maucec, M. Self-Adaptive Differential Evolution Algorithm in Constrained Real-Parameter Optimization. In Proceedings of the 2006 IEEE International Conference on Evolutionary Computation, Vancouver, BC, Canada, 16–21 July 2006; pp. 215–222. [Google Scholar] [CrossRef]
- Tian, M.; Gao, X.; Dai, C. Differential evolution with improved individual-based parameter setting and selection strategy. Appl. Soft Comput. 2017, 56, 286–297. [Google Scholar] [CrossRef]
- Meng, Z.; Pan, J.S.; Tseng, K.K. PaDE: An enhanced Differential Evolution algorithm with novel control parameter adaptation schemes for numerical optimization. Knowl.-Based Syst. 2019, 168, 80–99. [Google Scholar] [CrossRef]
- Poláková, R.; Tvrdík, J.; Bujok, P. Differential evolution with adaptive mechanism of population size according to current population diversity. Swarm Evol. Comput. 2019, 50, 100519. [Google Scholar] [CrossRef]
- Zhu, W.; Tang, Y.; Fang, J.A.; Zhang, W. Adaptive population tuning scheme for differential evolution. Inf. Sci. 2013, 223, 164–191. [Google Scholar] [CrossRef]
- Ali, M.Z.; Awad, N.H.; Suganthan, P.N. Multi-population differential evolution with balanced ensemble of mutation strategies for large-scale global optimization. Appl. Soft Comput. 2015, 33, 304–327. [Google Scholar] [CrossRef]
- Ali, M.Z.; Awad, N.H.; Suganthan, P.N.; Reynolds, R.G. An Adaptive Multipopulation Differential Evolution with Dynamic Population Reduction. IEEE Trans. Cybern. 2017, 47, 2768–2779. [Google Scholar] [CrossRef]
- Epitropakis, M.G.; Tasoulis, D.K.; Pavlidis, N.G.; Plagianakos, V.P.; Vrahatis, M.N. Enhancing Differential Evolution Utilizing Proximity-Based Mutation Operators. IEEE Trans. Evol. Comput. 2011, 15, 99–119. [Google Scholar] [CrossRef]
- Islam, S.M.; Das, S.; Ghosh, S.; Roy, S.; Suganthan, P.N. An Adaptive Differential Evolution Algorithm with Novel Mutation and Crossover Strategies for Global Numerical Optimization. IEEE Trans. Syst. Man Cybern. Part B (Cybern.) 2012, 42, 482–500. [Google Scholar] [CrossRef]
- Zheng, L.M.; Zhang, S.X.; Tang, K.S.; Zheng, S.Y. Differential evolution powered by collective information. Inf. Sci. 2017, 399, 13–29. [Google Scholar] [CrossRef]
- Li, Y.; Wang, S.; Yang, B. An improved differential evolution algorithm with dual mutation strategies collaboration. Expert Syst. Appl. 2020, 153, 113451. [Google Scholar] [CrossRef]
- Cheng, J.; Pan, Z.; Liang, H.; Gao, Z.; Gao, J. Differential evolution algorithm with fitness and diversity ranking-based mutation operator. Swarm Evol. Comput. 2021, 61, 100816. [Google Scholar] [CrossRef]
- Ramadas, M.; Abraham, A.; Kumar, S. ReDE-A Revised mutation strategy for Differential Evolution Algorithm. Int. J. Intell. Eng. Syst. 2016, 9, 51–58. [Google Scholar] [CrossRef]
- Peng, H.; Guo, Z.; Deng, C.; Wu, Z. Enhancing differential evolution with random neighbors based strategy. J. Comput. Sci. 2018, 26, 501–511. [Google Scholar] [CrossRef]
- Guo, S.M.; Yang, C.C. Enhancing Differential Evolution Utilizing Eigenvector-Based Crossover Operator. IEEE Trans. Evol. Comput. 2015, 19, 31–49. [Google Scholar] [CrossRef]
- Wang, Y.; Cai, Z.; Zhang, Q. Differential Evolution with Composite Trial Vector Generation Strategies and Control Parameters. IEEE Trans. Evol. Comput. 2011, 15, 55–66. [Google Scholar] [CrossRef]
- Ghosh, A.; Das, S.; Mullick, S.S.; Mallipeddi, R.; Das, A.K. A switched parameter differential evolution with optional blending crossover for scalable numerical optimization. Appl. Soft Comput. 2017, 57, 329–352. [Google Scholar] [CrossRef]
- Gong, W.; Cai, Z. Differential Evolution with Ranking-Based Mutation Operators. IEEE Trans. Cybern. 2013, 43, 2066–2081. [Google Scholar] [CrossRef]
- Wang, Y.; Cai, Z.; Zhang, Q. Enhancing the search ability of differential evolution through orthogonal crossover. Inf. Sci. 2012, 185, 153–177. [Google Scholar] [CrossRef]
- Mallipeddi, R.; Suganthan, P.N. Differential Evolution Algorithm with Ensemble of Parameters and Mutation and Crossover Strategies. In Swarm, Evolutionary, and Memetic Computing; Lecture Notes in Computer, Science; Panigrahi, B.K., Das, S., Suganthan, P.N., Dash, S.S., Eds.; Springer: Berlin/Heidelberg, Germany, 2010; Volume 6466, pp. 71–78. [Google Scholar] [CrossRef]
- Zhang, J.; Naik, H.S.; Assefa, T.; Sarkar, S.; Reddy, R.V.C.; Singh, A.; Ganapathysubramanian, B.; Singh, A.K. Computer vision and machine learning for robust phenotyping in genome-wide studies. Sci. Rep. 2017, 7, 44048. [Google Scholar] [CrossRef] [PubMed]
- Yao, J.; Chen, Z.; Liu, Z. Improved ensemble of differential evolution variants. PLoS ONE 2021, 16, e0256206. [Google Scholar] [CrossRef]
- Zhou, X.G.; Zhang, G.J. Abstract Convex Underestimation Assisted Multistage Differential Evolution. IEEE Trans. Cybern. 2017, 47, 2730–2741. [Google Scholar] [CrossRef]
- Ghosh, A.; Das, S.; Das, A.K.; Gao, L. Reusing the Past Difference Vectors in Differential Evolution—A Simple But Significant Improvement. IEEE Trans. Cybern. 2020, 50, 4821–4834. [Google Scholar] [CrossRef]
- Li, W.; Gong, W. An Improved Multioperator-Based Constrained Differential Evolution for Optimal Power Allocation in WSNs. Sensors 2021, 21, 6271. [Google Scholar] [CrossRef] [PubMed]
- Zhong, X.; Duan, M.; Zhang, X.; Cheng, P. A hybrid differential evolution based on gaining-sharing knowledge algorithm and harris hawks optimization. PLoS ONE 2021, 16, e0250951. [Google Scholar] [CrossRef]
- Jadon, S.S.; Tiwari, R.; Sharma, H.; Bansal, J.C. Hybrid Artificial Bee Colony algorithm with Differential Evolution. Appl. Soft Comput. 2017, 58, 11–24. [Google Scholar] [CrossRef]
- Mohamed, A.W.; Hadi, A.A.; Fattouh, A.M.; Jambi, K.M. LSHADE with semi-parameter adaptation hybrid with CMA-ES for solving CEC 2017 benchmark problems. In Proceedings of the 2017 IEEE Congress on Evolutionary Computation (CEC), Donostia-San Sebastián, Spain, 5–8 June 2017; pp. 145–152. [Google Scholar] [CrossRef]
- Zhao, F.; Xue, F.; Zhang, Y.; Ma, W.; Zhang, C.; Song, H. A hybrid algorithm based on self-adaptive gravitational search algorithm and differential evolution. Expert Syst. Appl. 2018, 113, 515–530. [Google Scholar] [CrossRef]
- Wang, S.; Li, Y.; Yang, H. Self-adaptive mutation differential evolution algorithm based on particle swarm optimization. Appl. Soft Comput. 2019, 81, 105496. [Google Scholar] [CrossRef]
- Luo, J.; Shi, B. A hybrid whale optimization algorithm based on modified differential evolution for global optimization problems. Appl. Intell. 2019, 49, 1982–2000. [Google Scholar] [CrossRef]
- Li, S.; Gong, W.; Wang, L.; Yan, X.; Hu, C. A hybrid adaptive teaching-learning-based optimization and differential evolution for parameter identification of photovoltaic models. Energy Convers. Manag. 2020, 225, 113474. [Google Scholar] [CrossRef]
- Wu, Y.; Chen, R.; Li, C.; Zhang, L.; Cui, Z. Hybrid Symbiotic Differential Evolution Moth-Flame Optimization Algorithm for Estimating Parameters of Photovoltaic Models. IEEE Access 2020, 8, 156328–156346. [Google Scholar] [CrossRef]
- Aarts, E.H.L.; Korst, J.H.M. Boltzmann machines as a model for parallel annealing. Algorithmica 1991, 6, 437–465. [Google Scholar] [CrossRef]
- Li, H.; Wang, H.; Wang, L.; Zhou, X. A modified Boltzmann Annealing Differential Evolution algorithm for inversion of directional resistivity logging-while-drilling measurements. J. Pet. Sci. Eng. 2020, 188, 106916. [Google Scholar] [CrossRef]
- Ahmadianfar, I.; Kheyrandish, A.; Jamei, M.; Gharabaghi, B. Optimizing operating rules for multi-reservoir hydropower generation systems: An adaptive hybrid differential evolution algorithm. Renew. Energy 2021, 167, 774–790. [Google Scholar] [CrossRef]
- Huo, L.; Zhu, J.; Li, Z.; Ma, M. A Hybrid Differential Symbiotic Organisms Search Algorithm for UAV Path Planning. Sensors 2021, 21, 3037. [Google Scholar] [CrossRef]
- Deng, C.; Dong, X.; Tan, Y.; Peng, H. Enhanced Differential Evolution Algorithm with Local Search Based on Hadamard Matrix. Comput. Intell. Neurosci. 2021, 2021, 8930980. [Google Scholar] [CrossRef]
- Rahnamayan, S.; Tizhoosh, H.; Salama, M. Opposition-Based Differential Evolution. IEEE Trans. Evol. Comput. 2008, 12, 64–79. [Google Scholar] [CrossRef]
- Sun, J.; Zhang, Q.; Tsang, E. DE/EDA: A new evolutionary algorithm for global optimization. Inf. Sci. 2005, 169, 249–262. [Google Scholar] [CrossRef]
- Peng, H.; Wu, Z.; Deng, C. Enhancing Differential Evolution with Commensal Learning and Uniform Local Search. Chin. J. Electron. 2017, 26, 725–733. [Google Scholar] [CrossRef]
- Guo, H.; Li, Y.; Li, J.; Sun, H.; Wang, D.; Chen, X. Differential evolution improved with self-adaptive control parameters based on simulated annealing. Swarm Evol. Comput. 2014, 19, 52–67. [Google Scholar] [CrossRef]
- Kozlov, K.; Ivanisenko, N.; Ivanisenko, V.; Kolchanov, N.; Samsonova, M.; Samsonov, A.M. Enhanced Differential Evolution Entirely Parallel Method for Biomedical Applications. In Parallel Computing Technologies; Hutchison, D., Kanade, T., Kittler, J., Kleinberg, J.M., Mattern, F., Mitchell, J.C., Naor, M., Nierstrasz, O., Pandu Rangan, C., Steffen, B., et al., Eds.; Springer: Berlin/Heidelberg, Germany, 2013; Volume 7979, pp. 409–416. [Google Scholar] [CrossRef]
- Kozlov, K.N.; Baumann, P.; Waldmann, J.; Samsonova, M.G. TeraPro, a system for processing large biomedical images. Pattern Recognit. Image Anal. 2013, 23, 488–497. [Google Scholar] [CrossRef]
- Hammer, G.L.; Vaderlip, R.; Gibson, G.; Wade, L.; Henzell, R.; Younger, D.; Warren, J.; Dale, A. Genotype-by-environment interaction in grain sorghum. II. Effects of temperature and photoperiod on ontogeny. Crop Sci. 1989, 29, 376–384. [Google Scholar] [CrossRef]
- Van Oosterom, E.; Bidinger, F.; Weltzien, E. A yield architecture framework to explain adaptation of pearl millet to environmental stress. Field Crop. Res. 2003, 80, 33–56. [Google Scholar] [CrossRef]
- Whish, J.; Butler, G.; Castor, M.; Cawthray, S.; Broad, I.; Carberry, P.; Hammer, G.; McLean, G.; Routley, R.; Yeates, S. Modelling the effects of row configuration on sorghum yield reliability in north-eastern Australia. Aust. J. Agric. Res. 2005, 56, 11. [Google Scholar] [CrossRef]
- Jordan, D.R.; Tao, Y.Z.; Godwin, I.D.; Henzell, R.G.; Cooper, M.; McIntyre, C.L. Comparison of identity by descent and identity by state for detecting genetic regions under selection in a sorghum pedigree breeding program. Mol. Breed. 2005, 14, 441–454. [Google Scholar] [CrossRef]
- Cooper, M.; van Eeuwijk, F.A.; Hammer, G.L.; Podlich, D.W.; Messina, C. Modeling QTL for complex traits: Detection and context for plant breeding. Curr. Opin. Plant Biol. 2009, 12, 231–240. [Google Scholar] [CrossRef] [PubMed]
- Sinclair, T.R.; Messina, C.D.; Beatty, A.; Samples, M. Assessment across the United States of the Benefits of Altered Soybean Drought Traits. Agron. J. 2010, 102, 475–482. [Google Scholar] [CrossRef]
- Technow, F.; Messina, C.D.; Totir, L.R.; Cooper, M. Integrating Crop Growth Models with Whole Genome Prediction through Approximate Bayesian Computation. PLoS ONE 2015, 10, e0130855. [Google Scholar] [CrossRef] [PubMed]
- Boote, K.J.; Jones, J.W.; Hoogenboom, G. Incorporating realistic trait physiology into crop growth models to support genetic improvement. Silico Plants 2021, 3, diab002. [Google Scholar] [CrossRef]
- Asseng, S.; van Herwaarden, A.F. Analysis of the benefits to wheat yield from assimilates stored prior to grain filling in a range of environments. Plant Soil 2003, 256, 217–229. [Google Scholar] [CrossRef]
- Hammer, G.L.; Chapman, S.; van Oosterom, E.; Podlich, D.W. Trait physiology and crop modelling as a framework to link phenotypic complexity to underlying genetic systems. Aust. J. Agric. Res. 2005, 56, 947. [Google Scholar] [CrossRef]
- Meuwissen, T.H.; Solberg, T.R.; Shepherd, R.; Woolliams, J.A. A fast algorithm for BayesB type of prediction of genome-wide estimates of genetic value. Genet. Sel. Evol. 2009, 41, 2. [Google Scholar] [CrossRef]
- De los Campos, G.; Hickey, J.M.; Pong-Wong, R.; Daetwyler, H.D.; Calus, M.P.L. Whole-Genome Regression and Prediction Methods Applied to Plant and Animal Breeding. Genetics 2013, 193, 327–345. [Google Scholar] [CrossRef]
- Endelman, J.B. Ridge Regression and Other Kernels for Genomic Selection with R Package rrBLUP. Plant Genome J. 2011, 4, 250. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2018. [Google Scholar]
- Lado, B.; Vázquez, D.; Quincke, M.; Silva, P.; Aguilar, I.; Gutiérrez, L. Resource allocation optimization with multi-trait genomic prediction for bread wheat (Triticum aestivum L.) baking quality. Theor. Appl. Genet. 2018, 131, 2719–2731. [Google Scholar] [CrossRef] [PubMed]
- Mangin, B.; Rincent, R.; Rabier, C.E.; Moreau, L.; Goudemand-Dugue, E. Training set optimization of genomic prediction by means of EthAcc. PLoS ONE 2019, 14, e0205629. [Google Scholar] [CrossRef] [PubMed]
- Kozlov, K.; Gursky, V.; Kulakovskiy, I.; Samsonova, M. Sequence-based model of gap gene regulatory network. BMC Genom. 2014, 15, S6. [Google Scholar] [CrossRef]
- Ali, S.; Signor, S.A.; Kozlov, K.; Nuzhdin, S.V. Novel approach to quantitative spatial gene expression uncovers genetic stochasticity in the developing Drosophila eye. Evol. Dev. 2019, 21, 157–171. [Google Scholar] [CrossRef] [PubMed]
- O’Neill, M. Grammatical Evolution. IEEE Trans. Evol. Comput. 2001, 5, 349–358. [Google Scholar] [CrossRef]
- Scrucca, L. On Some Extensions to GA Package: Hybrid Optimisation, Parallelisation and Islands Evolution. R J. 2017, 9, 20. [Google Scholar] [CrossRef]
- Guyon, I.; Elisseeff, A. An Introduction to Variable and Feature Selection. J. Mach. Learn. Res. 2003, 3, 1157–1182. [Google Scholar]
- Kohavi, R.; John, G.H. Wrappers for feature subset selection. Artif. Intell. 1997, 97, 273–324. [Google Scholar] [CrossRef]
- Furey, T.S.; Cristianini, N.; Duffy, N.; Bednarski, D.W.; Schummer, M.; Haussler, D. Support vector machine classification and validation of cancer tissue samples using microarray expression data. Bioinformatics 2000, 16, 906–914. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y. A Comparative Study on Feature Selection Methods for Drug Discovery. J. Chem. Inf. Comput. Sci. 2004, 44, 1823–1828. [Google Scholar] [CrossRef] [PubMed]
- Meinshausen, N.; Bühlmann, P. Stability selection: Stability Selection. J. R. Stat. Soc. Ser. B (Stat. Methodol.) 2010, 72, 417–473. [Google Scholar] [CrossRef]
- Li, X.J.; Hayward, C.; Fong, P.Y.; Dominguez, M.; Hunsucker, S.W.; Lee, L.W.; McLean, M.; Law, S.; Butler, H.; Schirm, M.; et al. A Blood-Based Proteomic Classifier for the Molecular Characterization of Pulmonary Nodules. Sci. Transl. Med. 2013, 5, 207ra142. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).