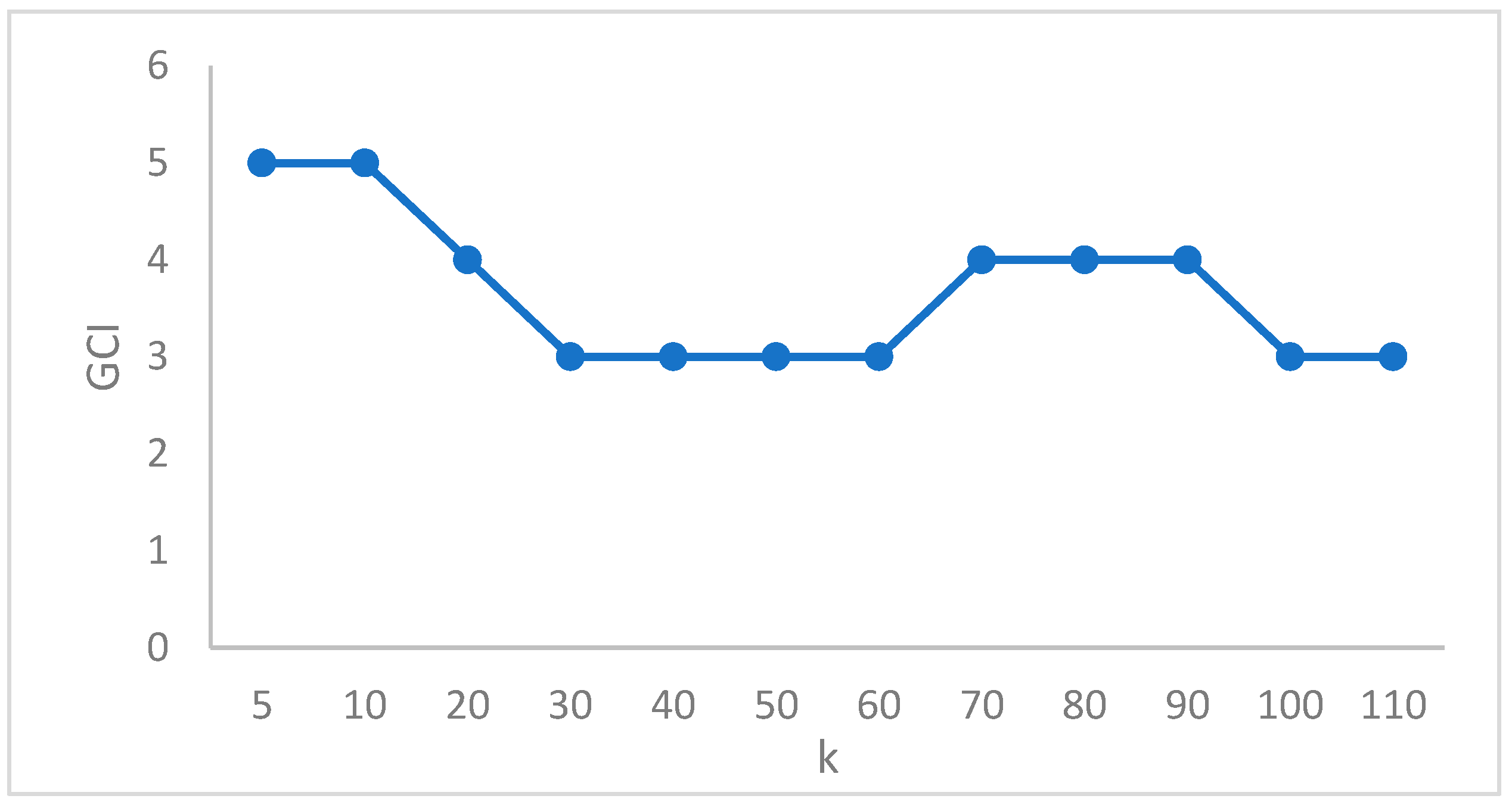
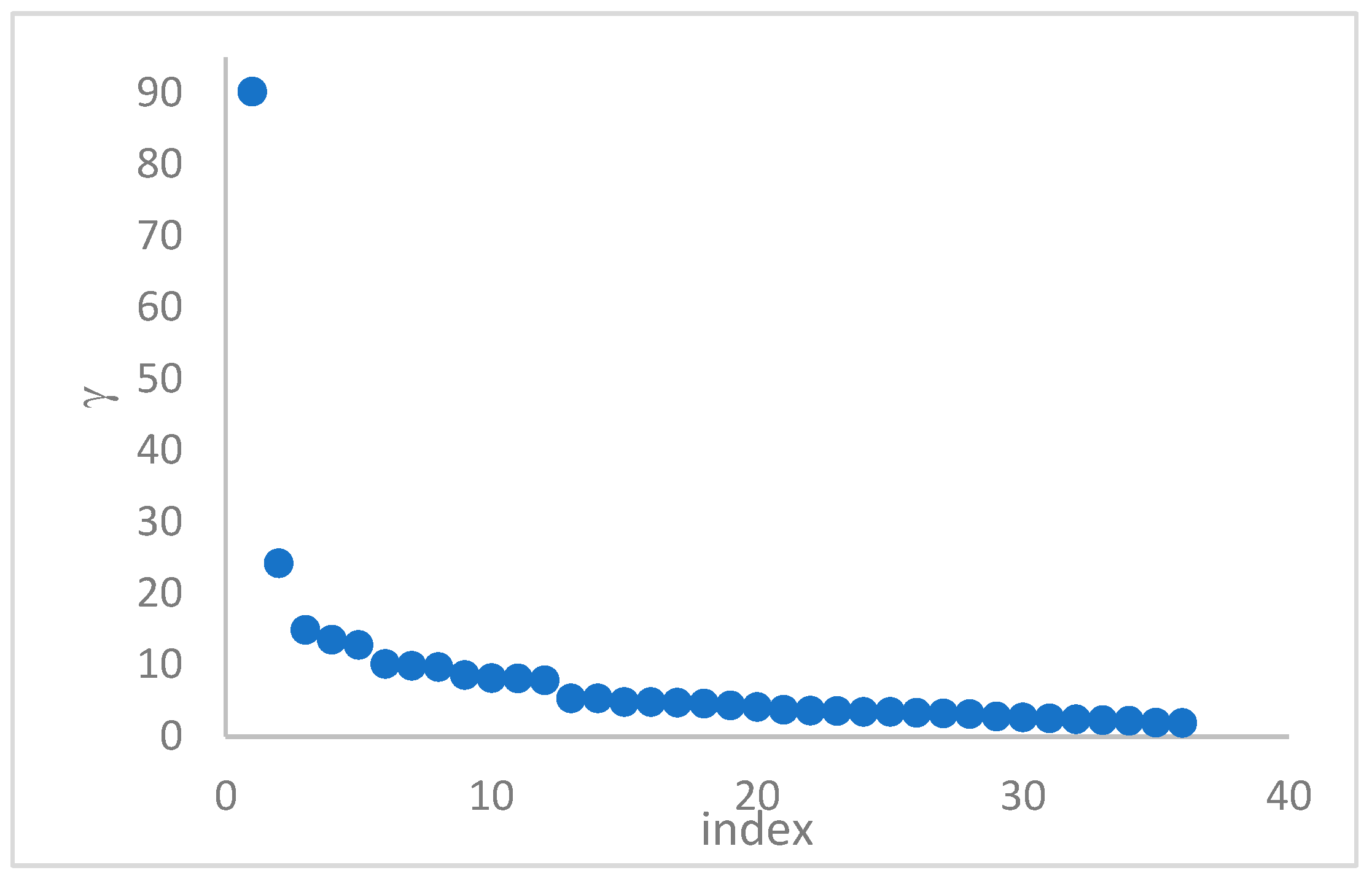
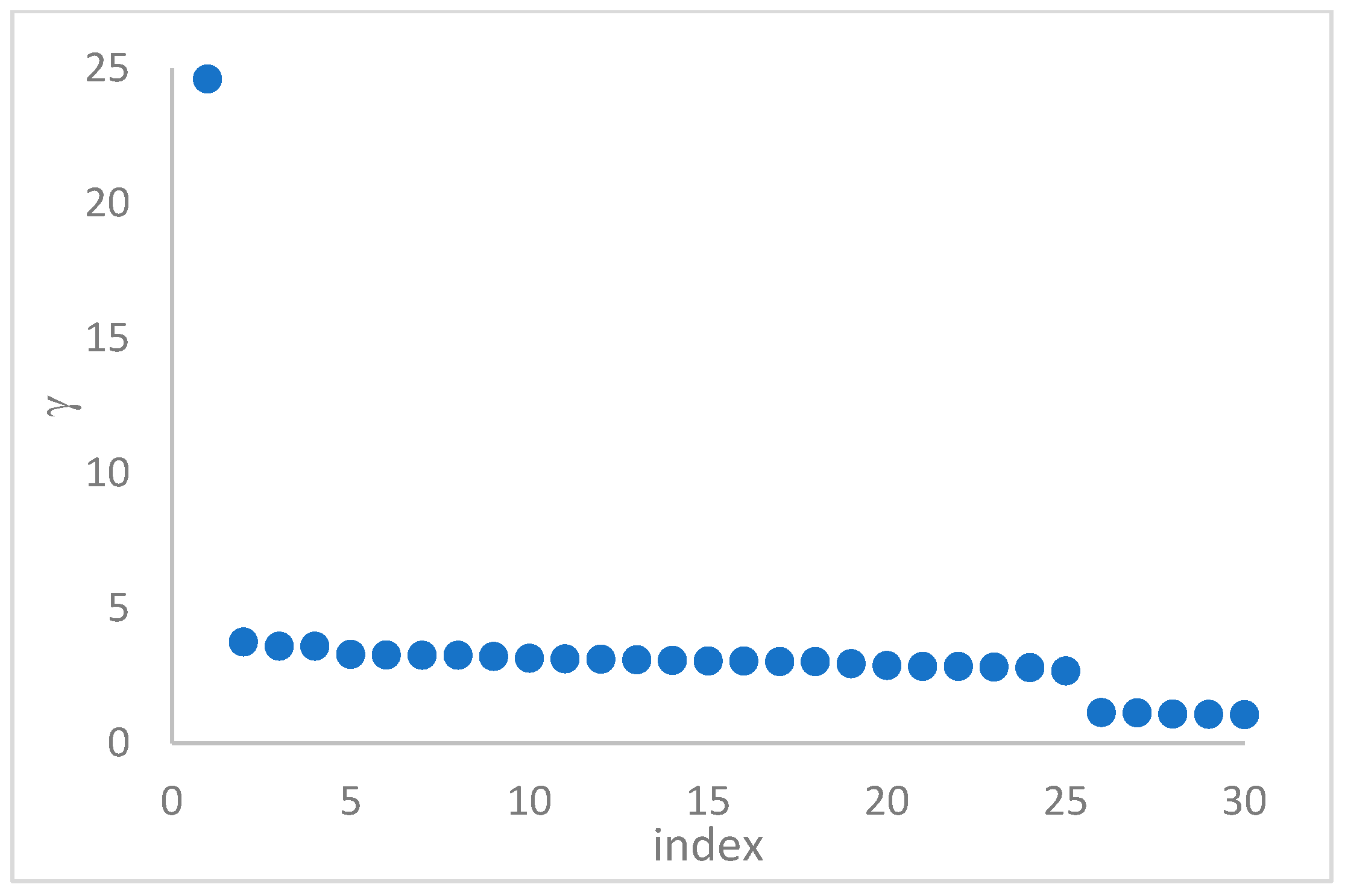
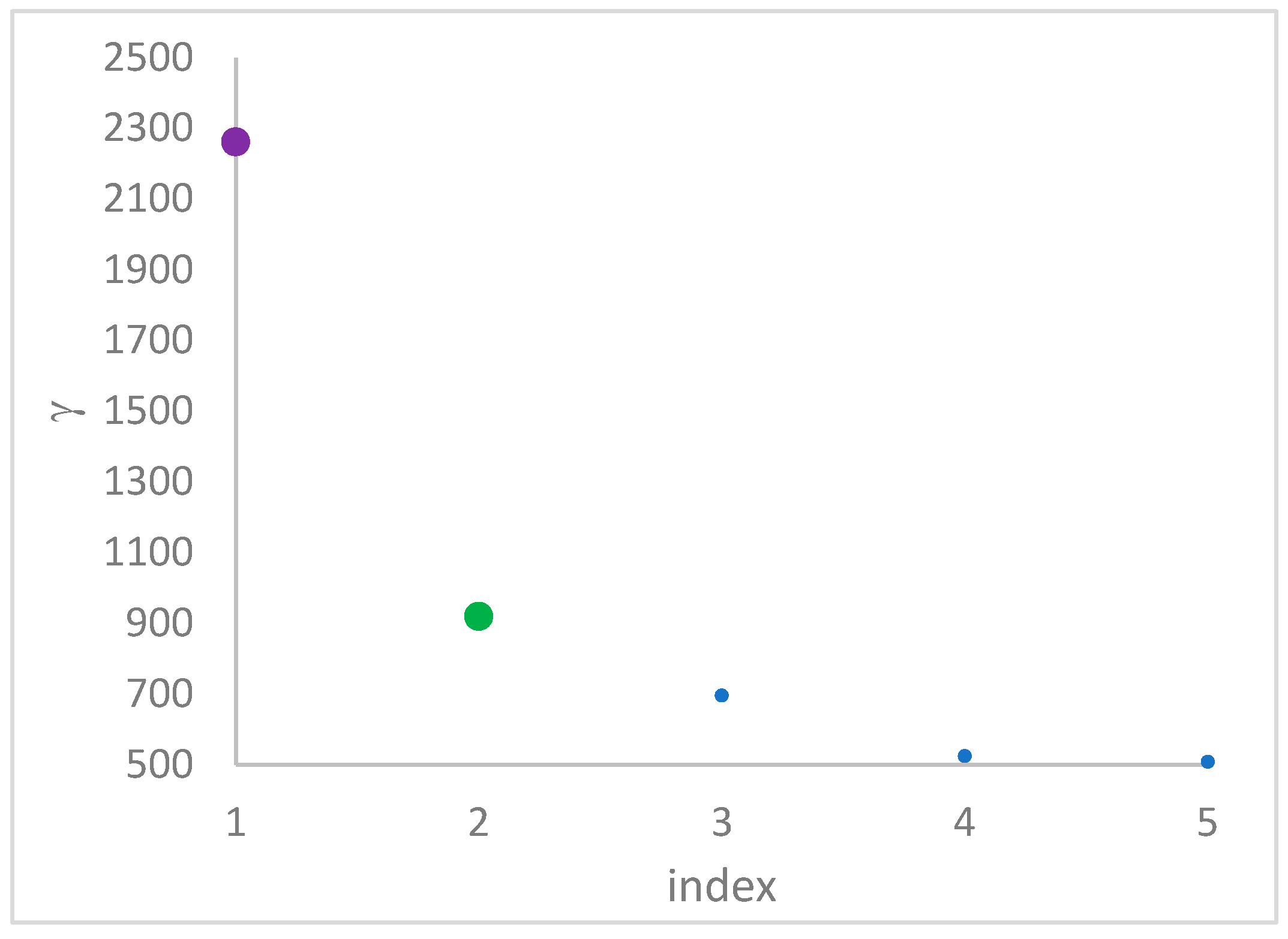
1. Introduction
Unsupervised clustering [
1] is a common technique for mining information from the sample data points of a dataset. Application domains include image segmentation, machine learning, artificial intelligence, medicine, biology, etc. The goal is to partition the points of a dataset,
, into groups (said clusters) in such a way that the points of the same cluster are similar, but dissimilar to the points assigned to other clusters. Each cluster is characterized by a representative point called its
centroid . The similarity function often coincides with the Euclidean function when the dataset points are vectors with
numerical coordinates (dimensions):
.
An often-used centroids-based clustering algorithm is K-Means [
2,
3], where the number
of clusters/centroids is an input parameter. K-Means’s recurrent use owes to its simplicity and efficiency [
4]. K-Means, though, is more suited to datasets with spherical clusters. In addition, it strongly depends on the initialization of centroids (
seeding) [
5,
6]. A bad initialization quickly causes K-Means to become stuck in a local sub-optimal solution. K-Means acts as a
local refiner of centroids and cannot search for centroids globally in the dataset. To control these limitations, careful seeding methods can be used [
7,
8,
9] together with the concept of repeating it many times (Repeated K-Means), each time with a different initialization, toward the obtainment of an acceptable solution, as mirrored by some clustering accuracy measures. Random Swap [
10,
11] integrates K-Means in its behavior and bases its operation on a
global search for centroids. At each iteration, a swap is accomplished, which replaces a randomly chosen centroid with a randomly selected dataset point. Provided a suited number of swap iterations are executed, Random Swap has been proven to be able, in many cases, to find a solution close to the optimal one.
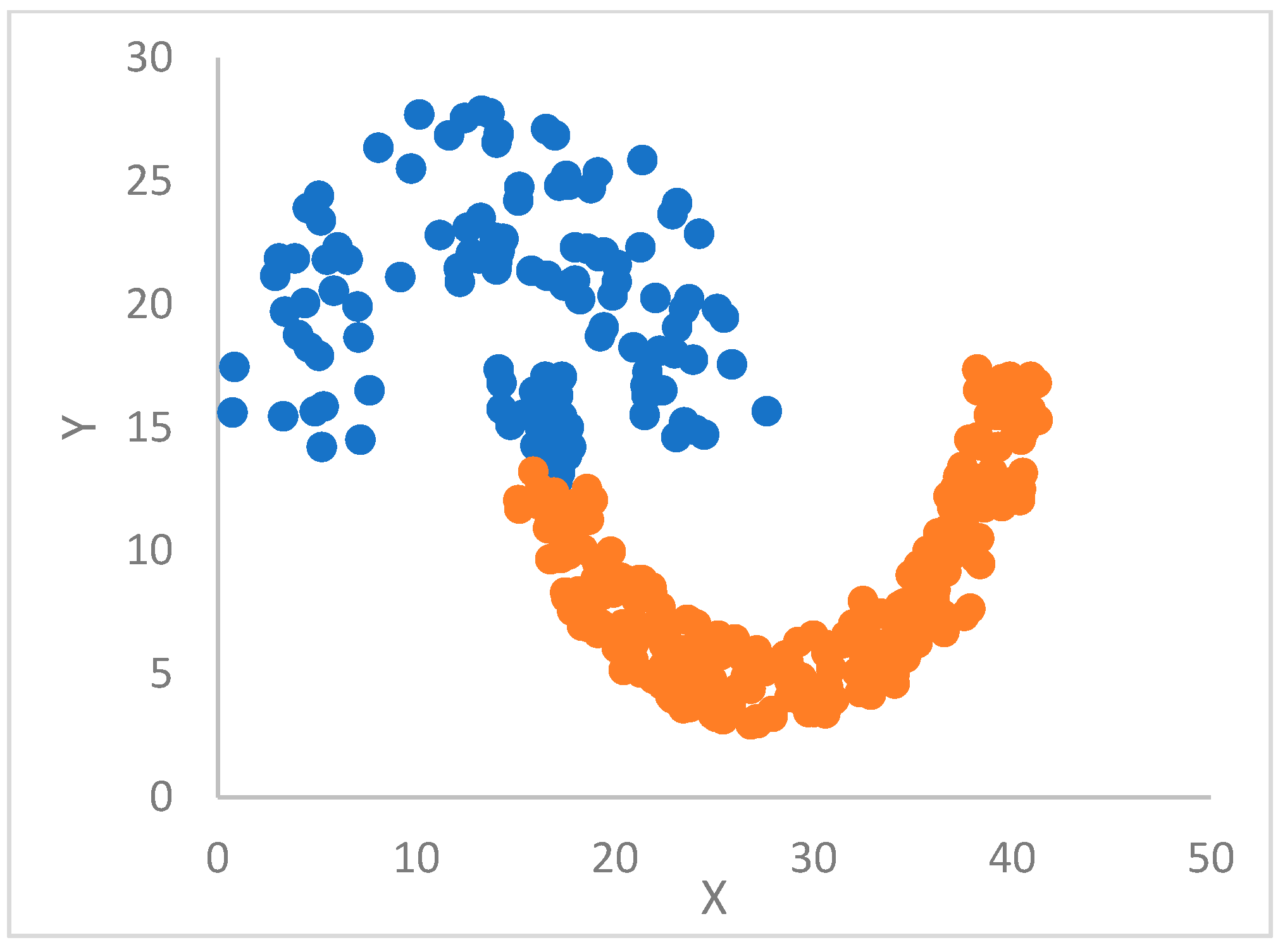
A different yet popular clustering algorithm, designed for non-spherical clusters, is based on the concept of
density peaks (DP) [
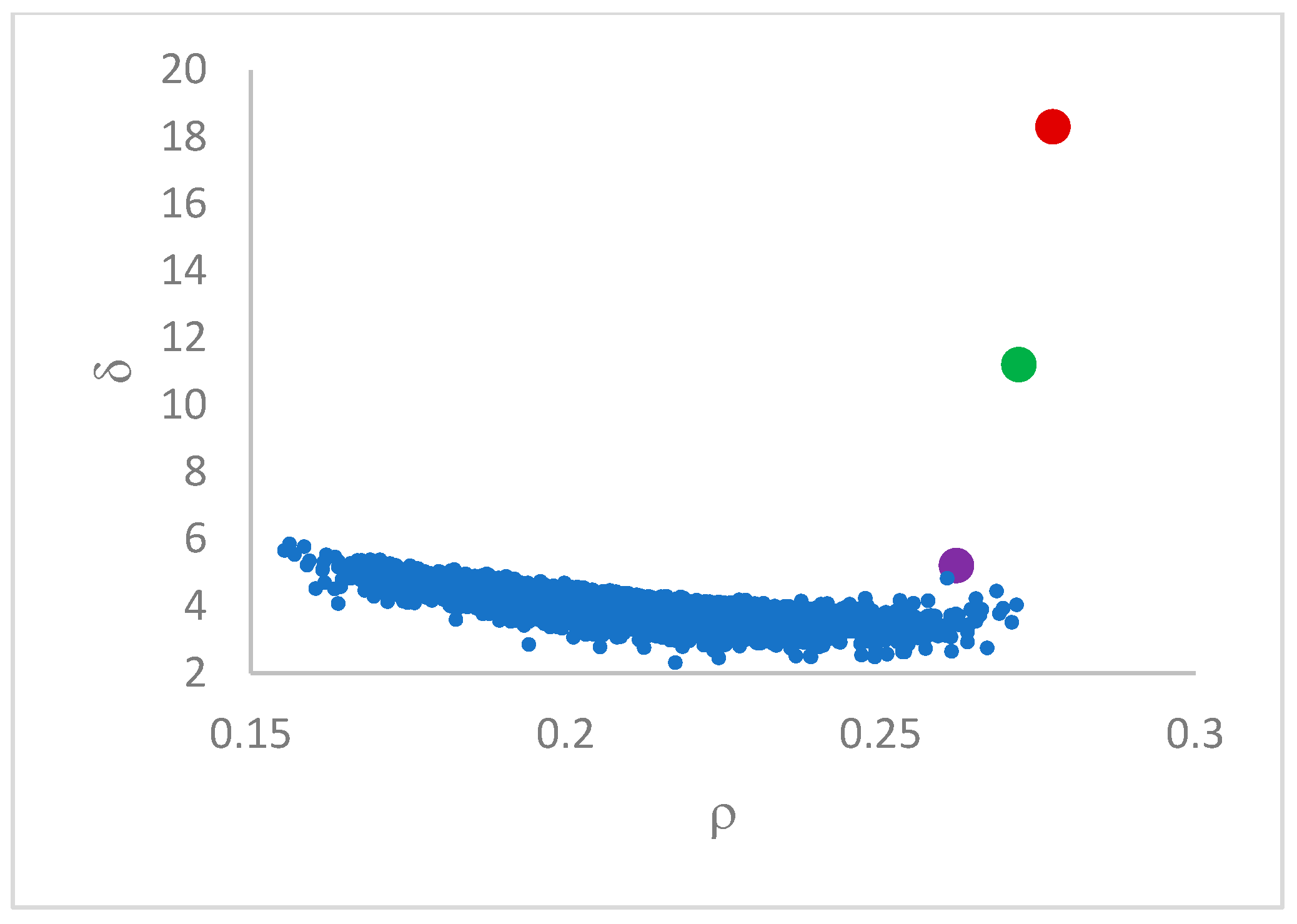
12]. DP relies on the fact that, naturally, centroids are points of higher local density, whose neighborhood is made up of points with lesser density. In addition, a candidate centroid is a point of higher density relatively far from points with a higher density. Two data point quantities regulate the application of DP: density (
rho) and minimal distance to the nearest point with a higher density (
delta). A fundamental problem is the estimation of the point density. In the original proposal [
12], the concept of a
cutoff distance (distance threshold) is introduced. Such distance is the radius of a hyperball in the D-dimensional space. The density is established by (logically) counting points that fall in the ball. The authors in [
12] adopted the “thumb rule” of choosing a value for
, which can ensure that from 1% to 2% of the dataset, the number of points falls in the hyperball. DP clustering is finalized by considering both the rho and delta values of points. Centroids are chosen as points with high values of both rho and delta. The remaining points are assigned to clusters/centroids according to the delta attribute. However, it has been pointed out that the DP basic approach cannot recognize clusters with an arbitrary geometry.
In the last few years, several proposals appeared in the literature to improve specifically the estimation of density and thus handling clusters of an arbitrary shape. A notable class of works is based on the
k-Nearest Neighbors (kNN) technique, with
as an input parameter. In [
13], from the
-th Nearest Neighbor distance of a data point, p, indicated as
, the kNN neighborhood of a data point, p (
), is first defined:
, where
is the distance between p and q. Then, the local density of p is inferred through a Gaussian exponential kernel. The proposal in [
13] adds to the clustering method the
principal component analysis (PCA) to deal with the “curse of dimensionality”, that is, the problem of a high number of point coordinates (dimensions) that can cause the Euclidean distance to lose its meaning. A (hopefully) reduced number of coordinates is preliminarily detected to guarantee data variations and the consistency of the Euclidean distance.
The kNN approach was further specialized with the concepts of
reverse and
natural Nearest Neighbors [
14,
15]. From the k-th nearest point of a point, p,
, the
Reverse Nearest Neighbors (RNN) of p,
, is defined as the set of points, q, which have p within the distance
, that is:
. Two nearest neighbor points, p and q, are said natural neighbors when, as in the friendship relationship of human societies, each one belongs to the RNN of the partner:
. A further concept concerns the definition of the
Mutual Nearest Neighbors (NMM) set of a point, p, defined as:
. A benefit of the RNN-based approaches in [
14,
15] is the possibility of inferring a suitable value of
in the algorithm, by automatically checking different radius values, starting from 2, until a stable search state is reached, where the friendship relation among points stabilizes.
A common problem of density estimation by kNN-based approaches is the high computational cost,
, due to the need to compute all the pairwise distances of points. Such a cost can limit the applicability of kNN to large datasets. Fränti et al. [
16,
17] developed a fast-density peak algorithm (FastDP) based on the approximate construction of the
graph through an efficient divisive technique. The divisive technique randomly selects two points,
and
, in the dataset, and then the points of the dataset are partitioned into two groups according to the nearer distance from
or
. The two sub-sets are recursively split until their size appears under a given threshold, in which case a corresponding kNN subgraph is directly built (brute force). The divisive technique is repeated many times in the algorithm’s first part until the edge changes in the last iteration becomes smaller than a given tolerance. In the second part, the divisive technique is continued by combining sub-graphs by
neighborhood propagation (NN-Descent method [
16,
18]), that is by evaluating neighbors of neighbors. Instead of creating the graph from scratch, the previous graph is updated by reviewing the k-Nearest Neighbors of each point by considering the information that emerged in the latest solution. The approximate kNN graph is finally used to estimate each point’s density and delta values. Density is evaluated as the inverse of the mean distance to the k-Nearest Neighbors. As demonstrated in [
16], FastDP can handle different data types, provided a distance function is available.
This paper proposes ParDP, a density peaks-based algorithm and tool that depends on kNN to estimate density. ParDP does not build an approximate KNN graph as in [
16]. Anyway, the
cost is smoothed by exploiting Java parallelism [
4,
9,
11,
19].
A preliminary version of ParDP, where a suitable global value,
, was initially tentatively tuned, was presented as a conference paper [
20]. Concerning the conference paper, the following new contributions are added.
Using the kNN approach for inferring points’ density and delta values. In particular, ParDP can exploit kNN for inferring a global value of from which a Gaussian kernel evaluates densities. Alternatively, densities can directly be estimated from the kNN distances.
Supporting the principal component analysis (PCA) for (hopefully) reducing the high number of dimensions of some datasets while preserving the meaning of the Euclidean distance.
Optimizing the use of Java parallel streams [
19] in all the basic steps of ParDP, ranging from evaluating the density and delta values to final clustering to implementing various clustering measures.
Applying ParDP to several benchmark and real-world datasets.
Adapting the Java implementation of ParDP for an achievement, PCA included, of the Du et al. [
13] density peaks algorithm (here referred to as ParDPC-KNN) as well as a density peaks algorithm based on the Mutual Nearest Neighbors (MNN) approach [
19] (here referred to as ParDPC-MNN).
Verifying ParDP correctness and performance by comparing it to ParDPC-KNN and ParDPC-MNN.
This paper is structured as follows.
Section 2 reviews the fundamental concepts of the basic density peaks clustering algorithm (DP).
Section 3 discusses the design and implementation aspects of the proposed ParDP algorithm.
Section 4 provides a summary of the clustering accuracy indexes used in the practical experiments.
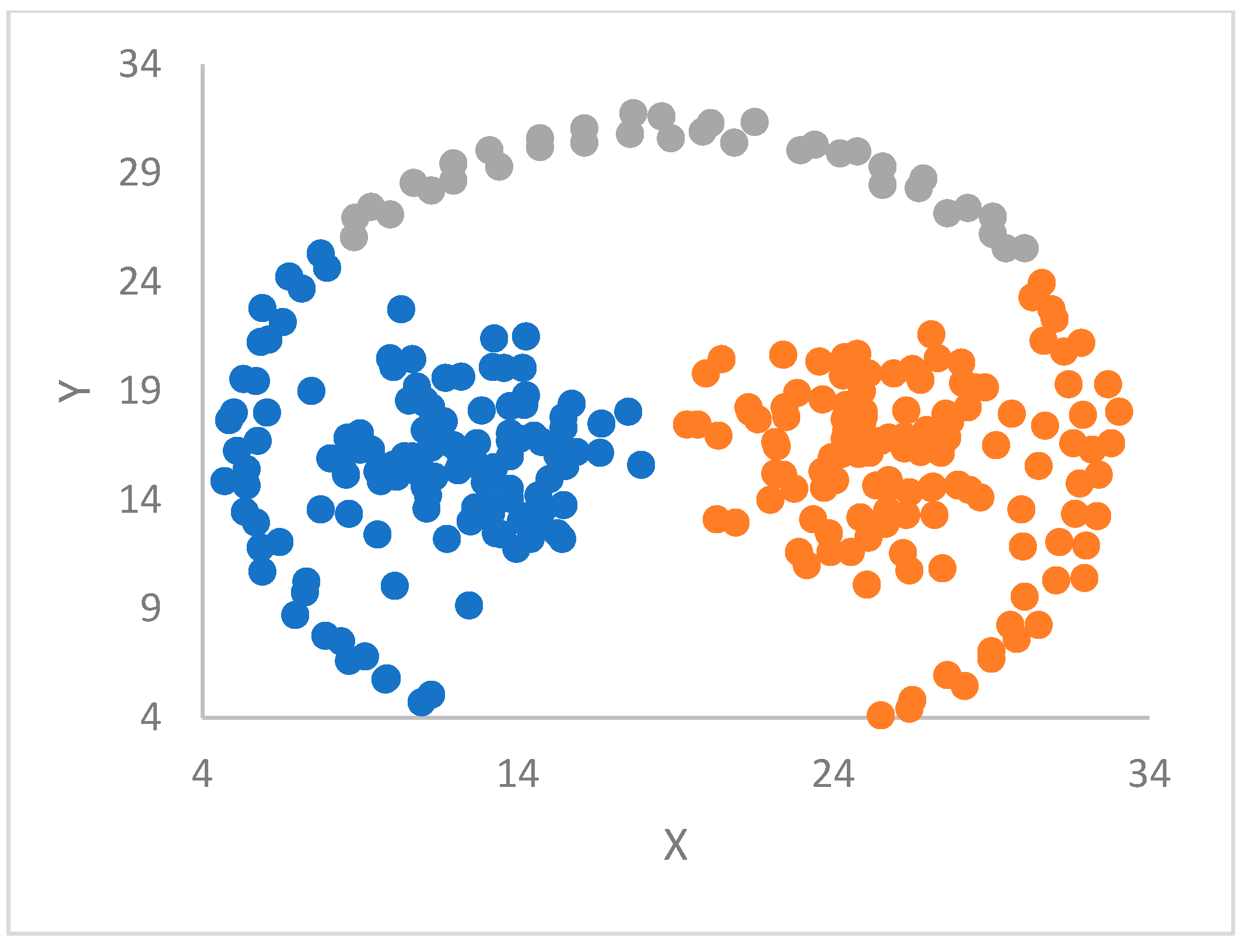
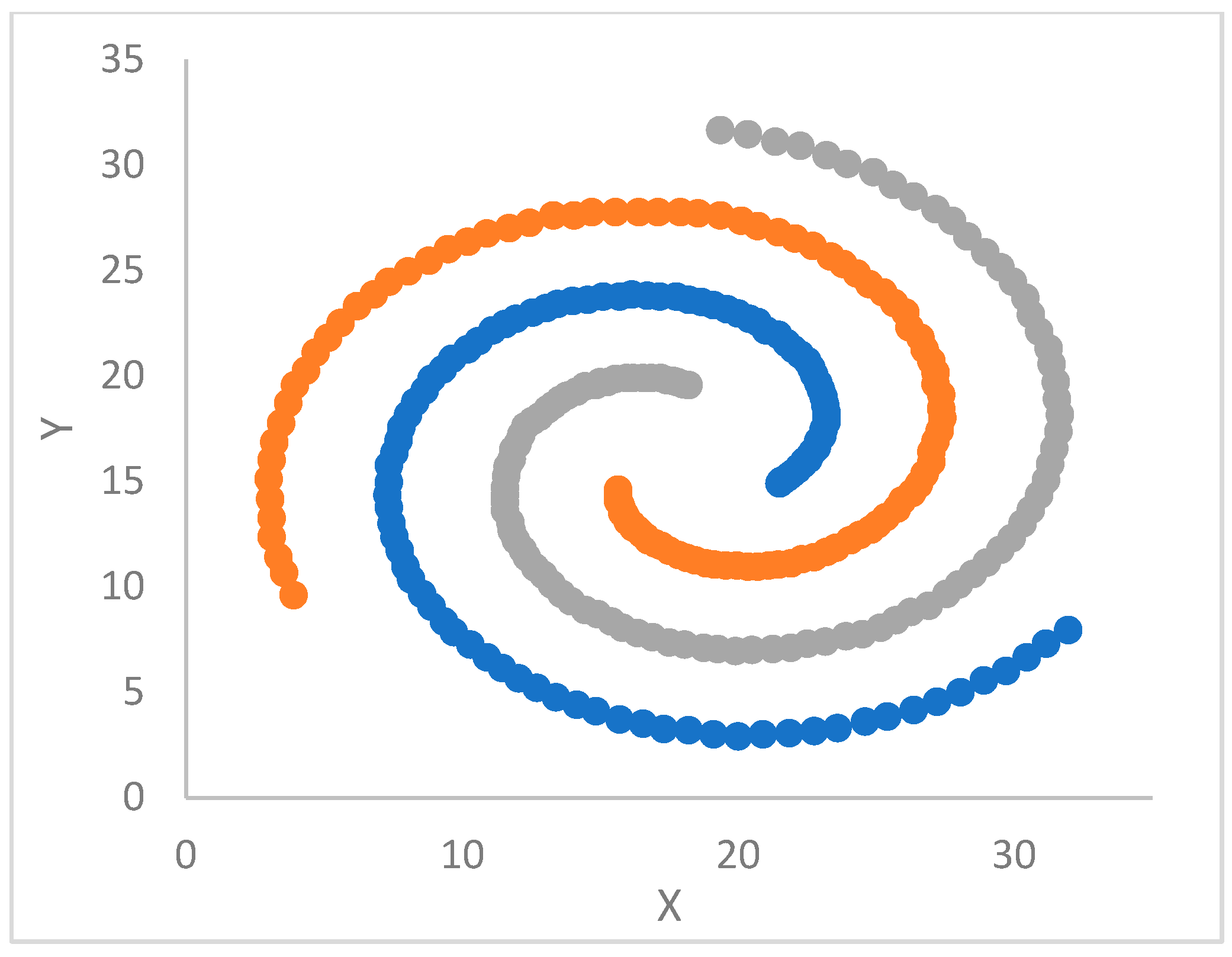
Section 5 is devoted to the experimental work. ParDP is applied to 23 challenging synthetic and real-world datasets, split into three groups. The clustering results are compared with those achieved by competing tools. Finally,
Section 6 draws some conclusions about the developed work and indicates directions for future work.
2. Density Peaks-Basic Algorithm
The density peaks basic algorithm (DP) [
12] rests on the intuition that centroids are points of higher local density, surrounded by points of lesser density. As a consequence, a centroid is relatively distant from other points of a higher density. To formalize this intuition, two quantities are introduced for each point,
: its density (
) and its nearest distance (
) to a point with a higher density. Such a nearest point is called, in [
16,
17], the
big brother of
. It is evident that for a point to be a candidate centroid, it should have a high value of both
and
. Equivalently, a candidate centroid should have a high value of a third quantity, (
gamma)
. In this paper, as in [
16], the
gamma strategy is adopted for choosing the centroids.
Unlike, e.g., K-Means [
2,
3], the number
of centroids is not necessarily an input parameter for DP. The value of
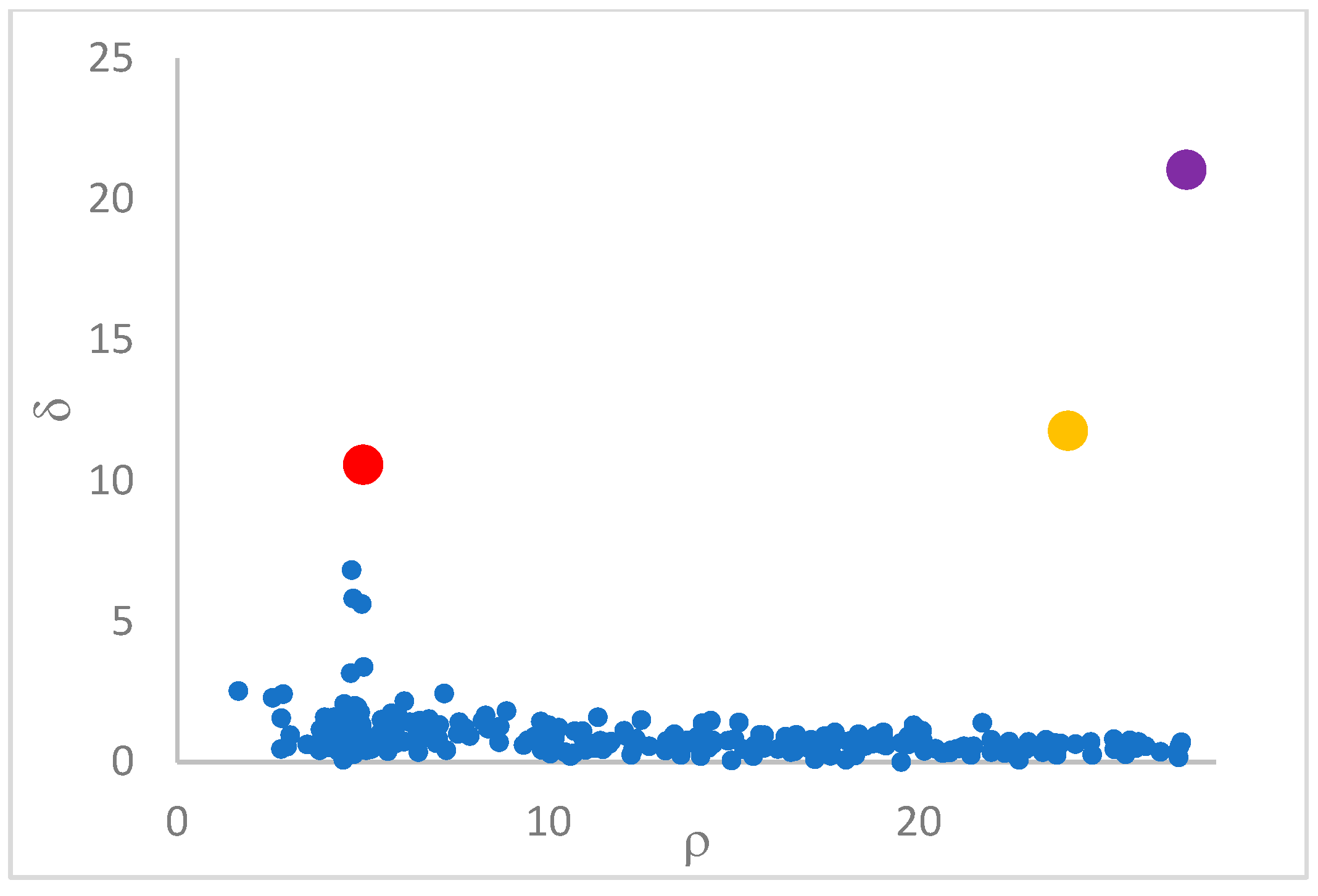
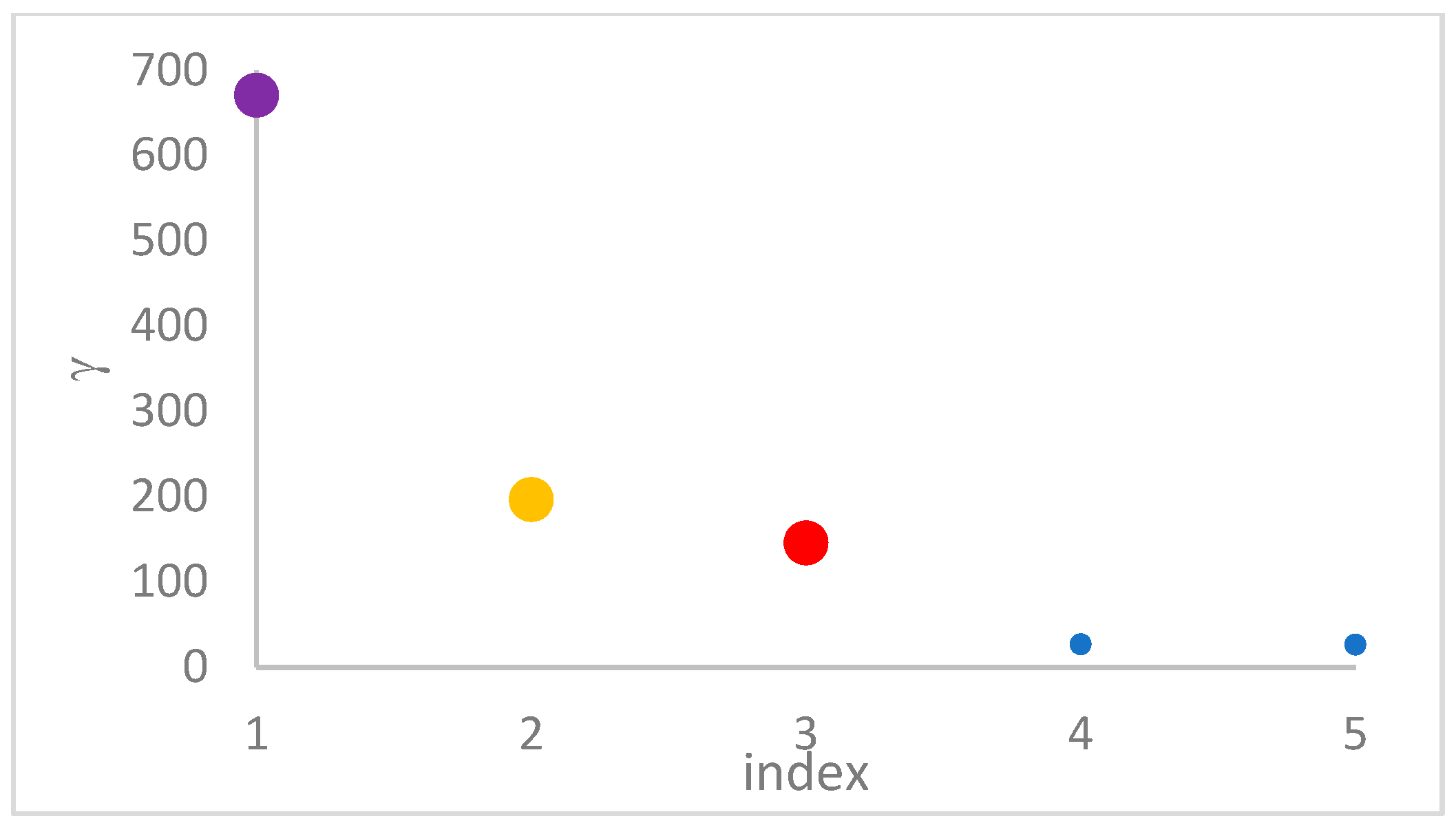
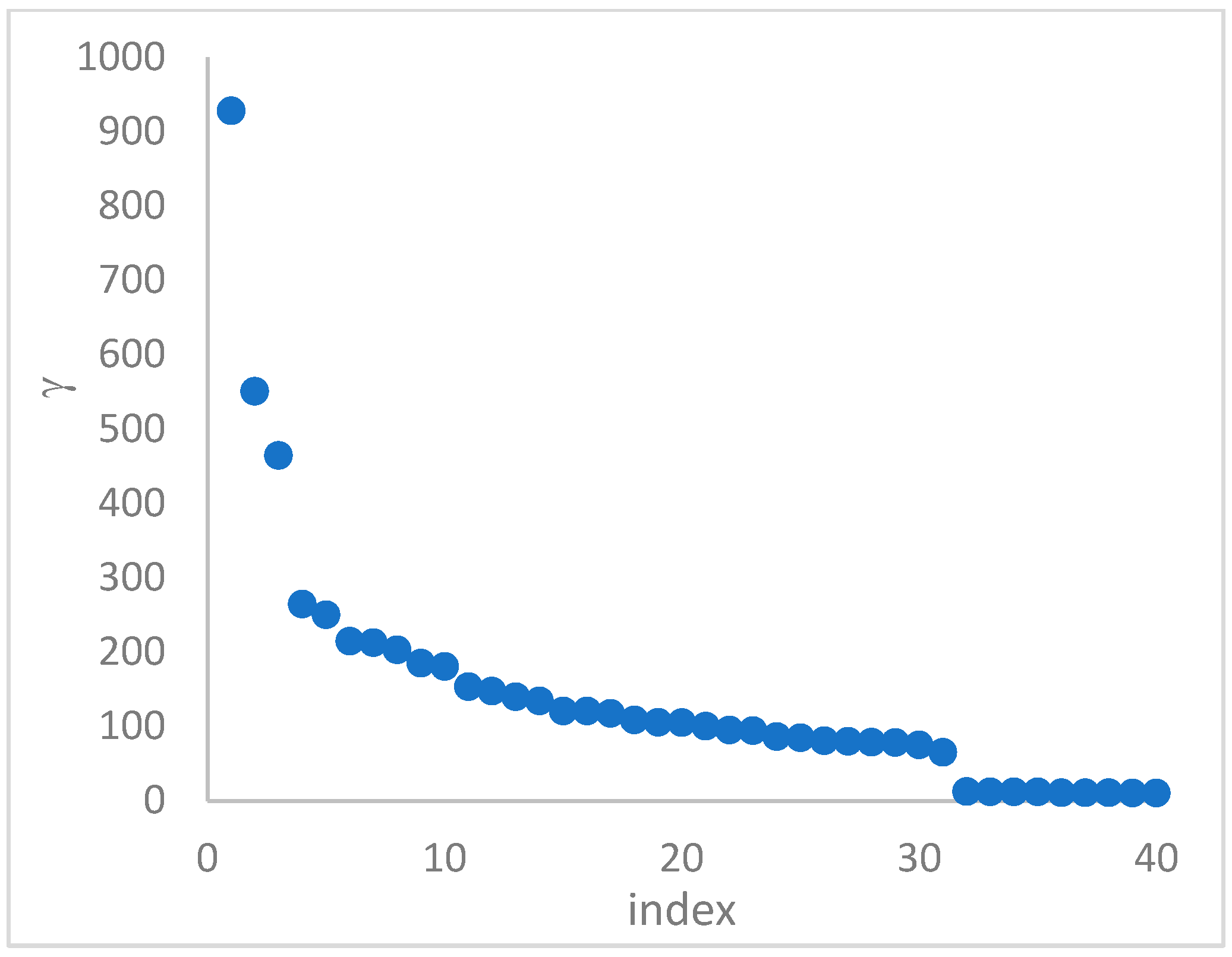
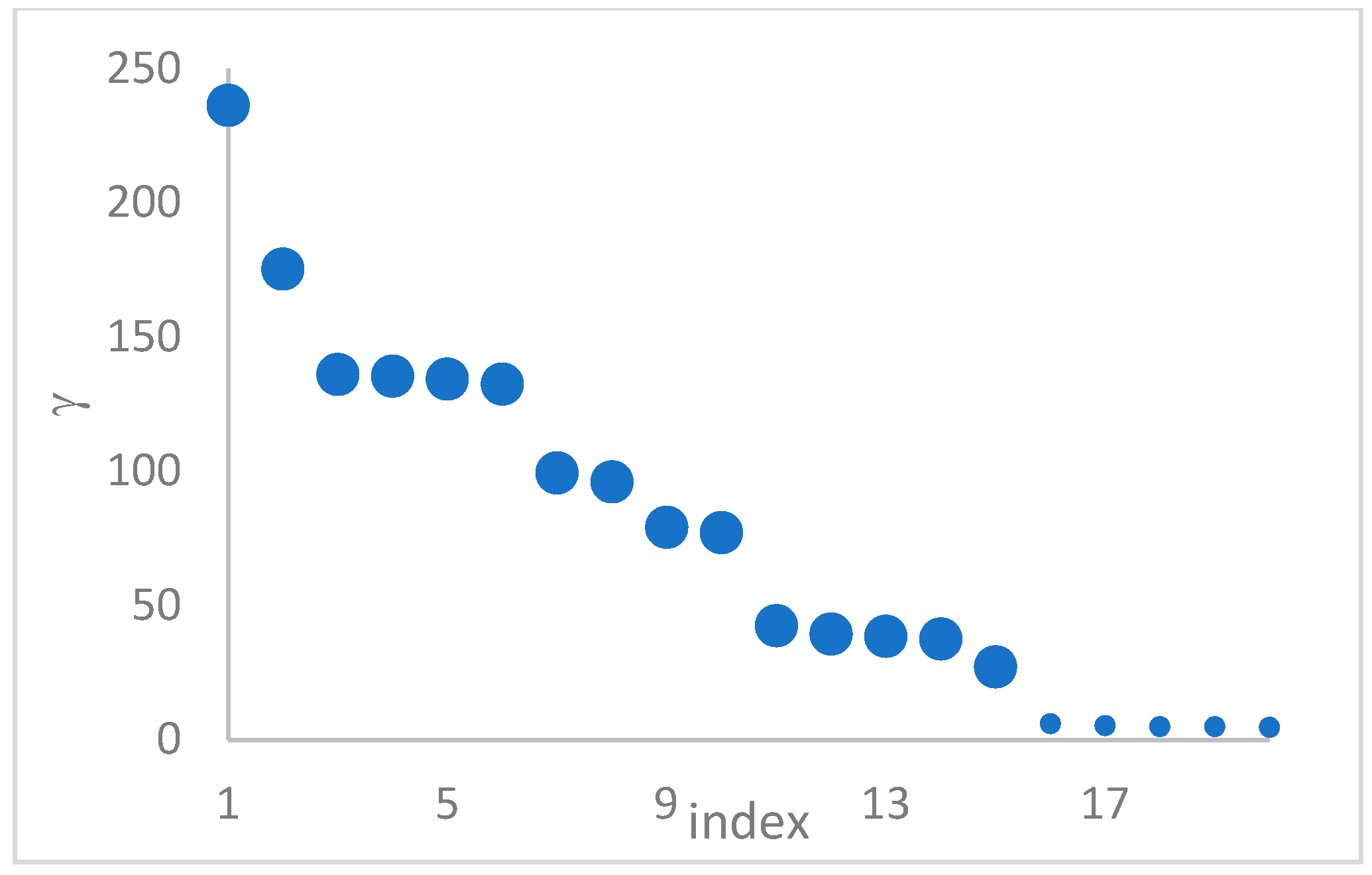
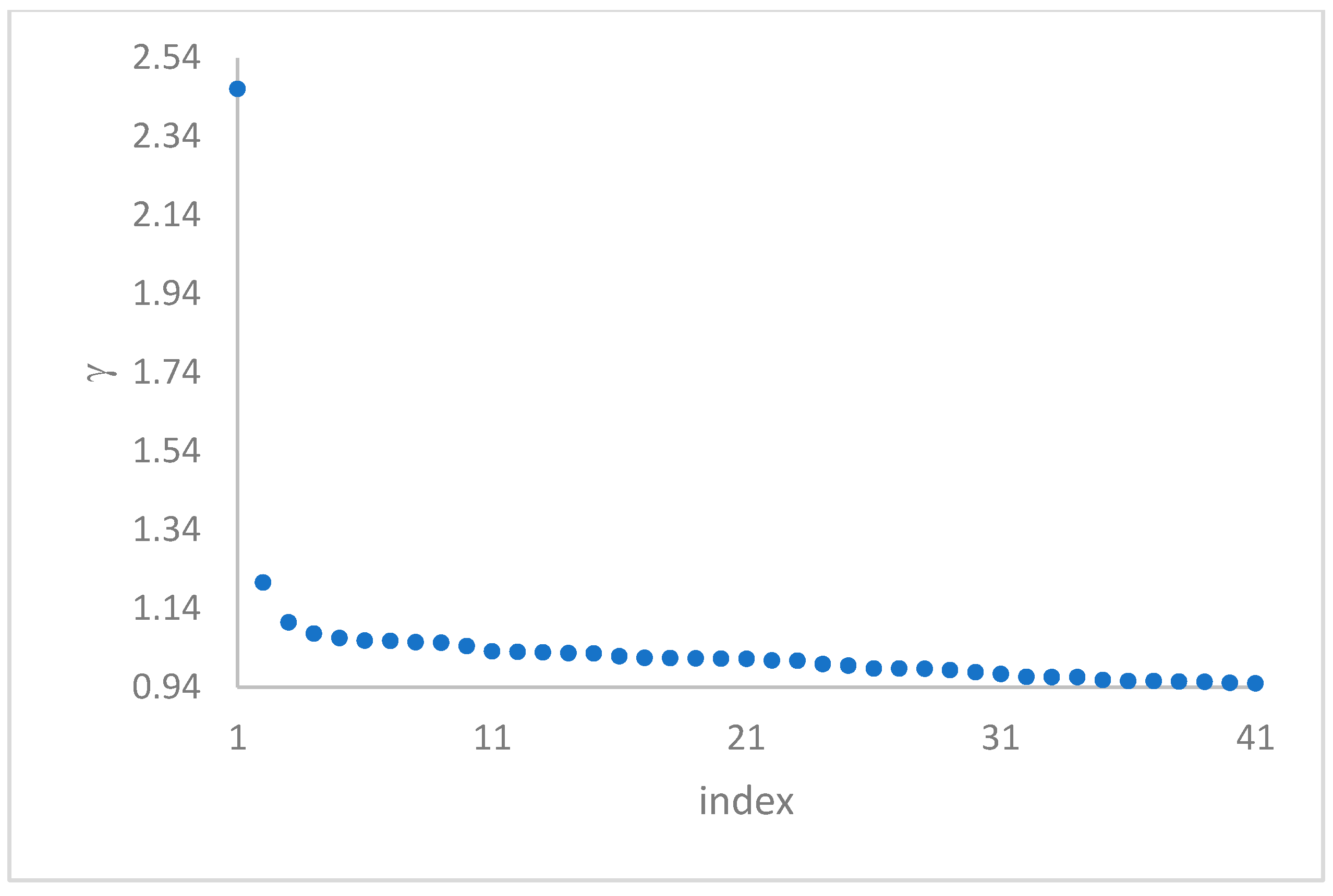
can, in many cases, be deducted by using the so-called
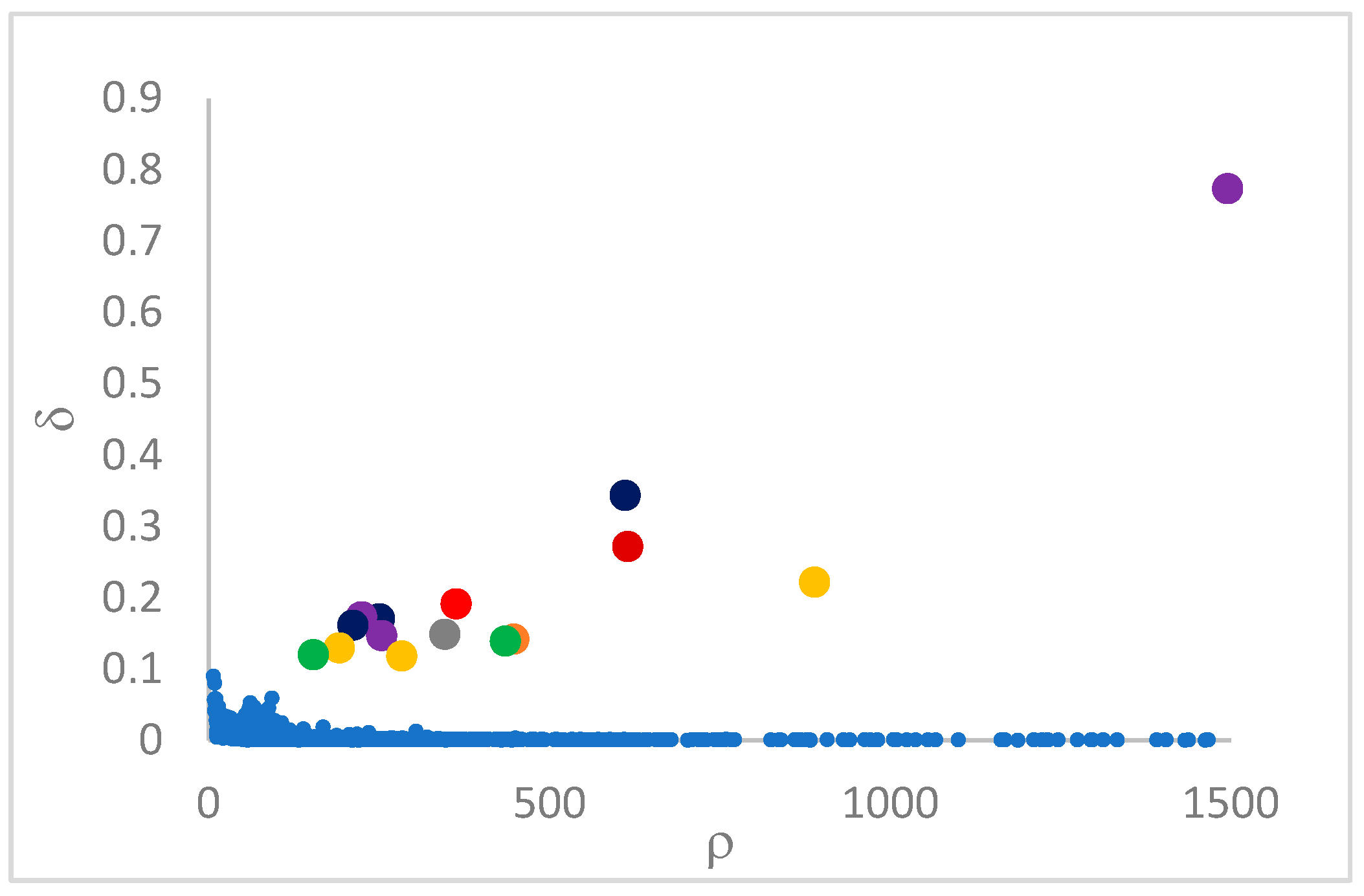
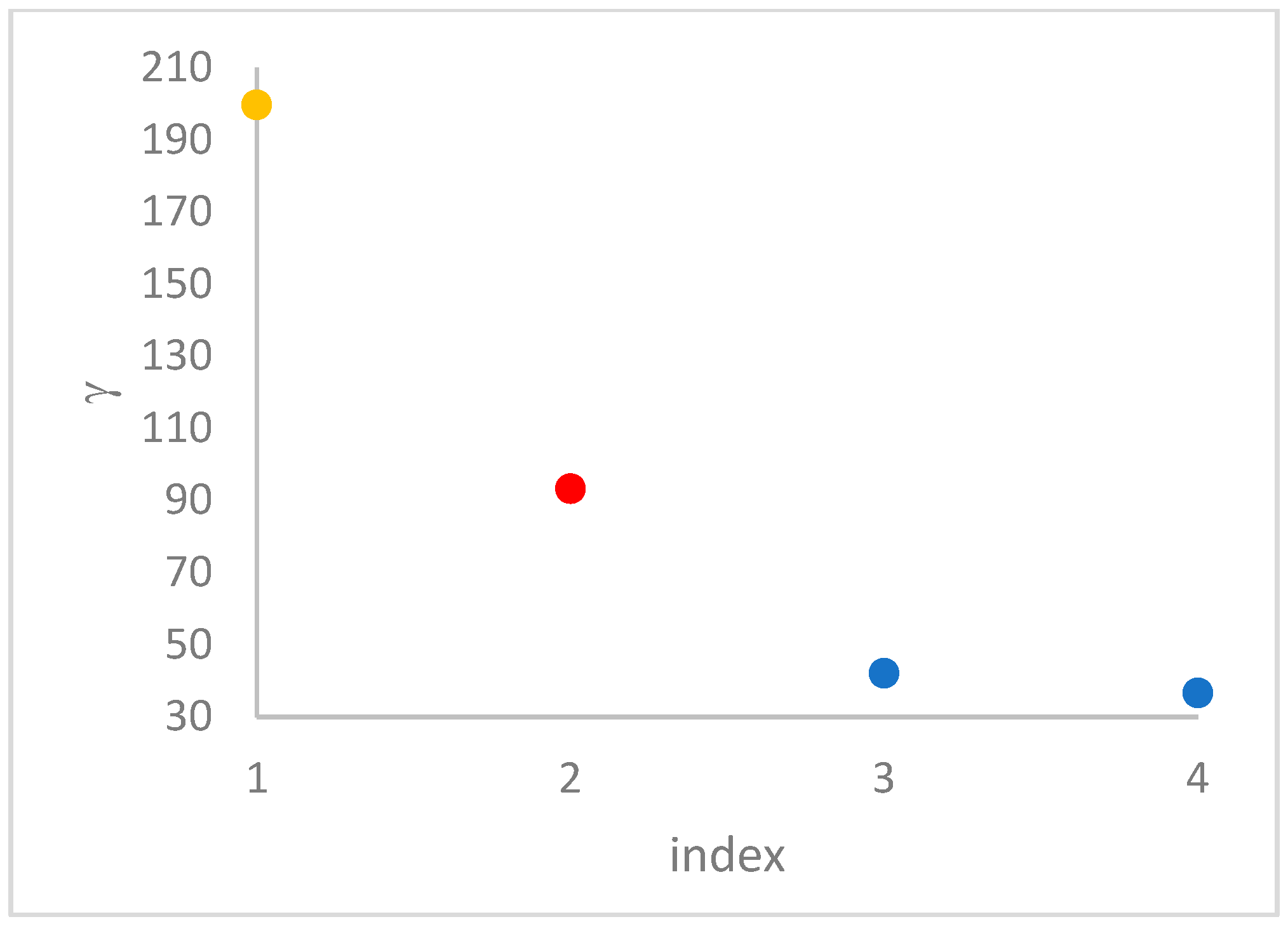
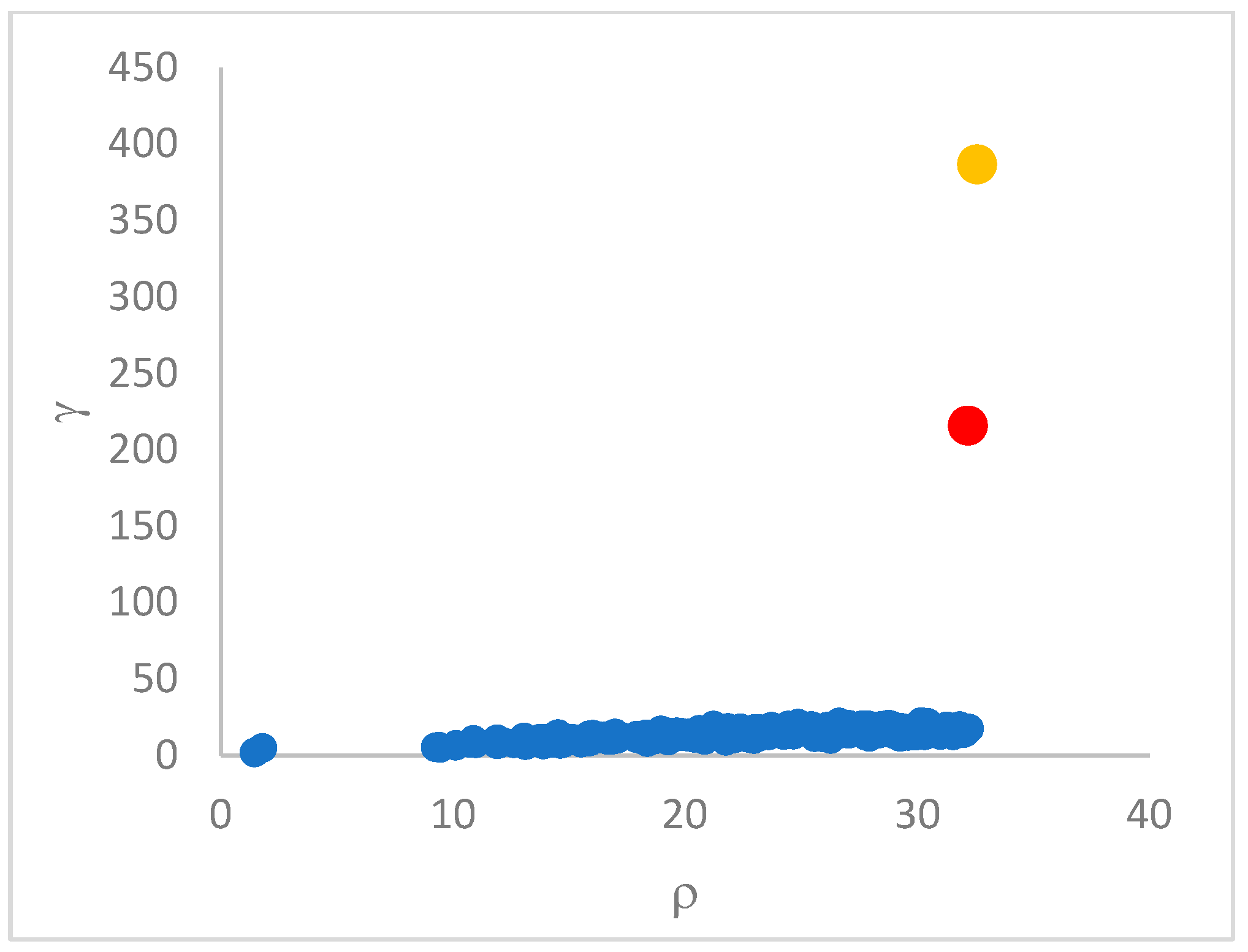
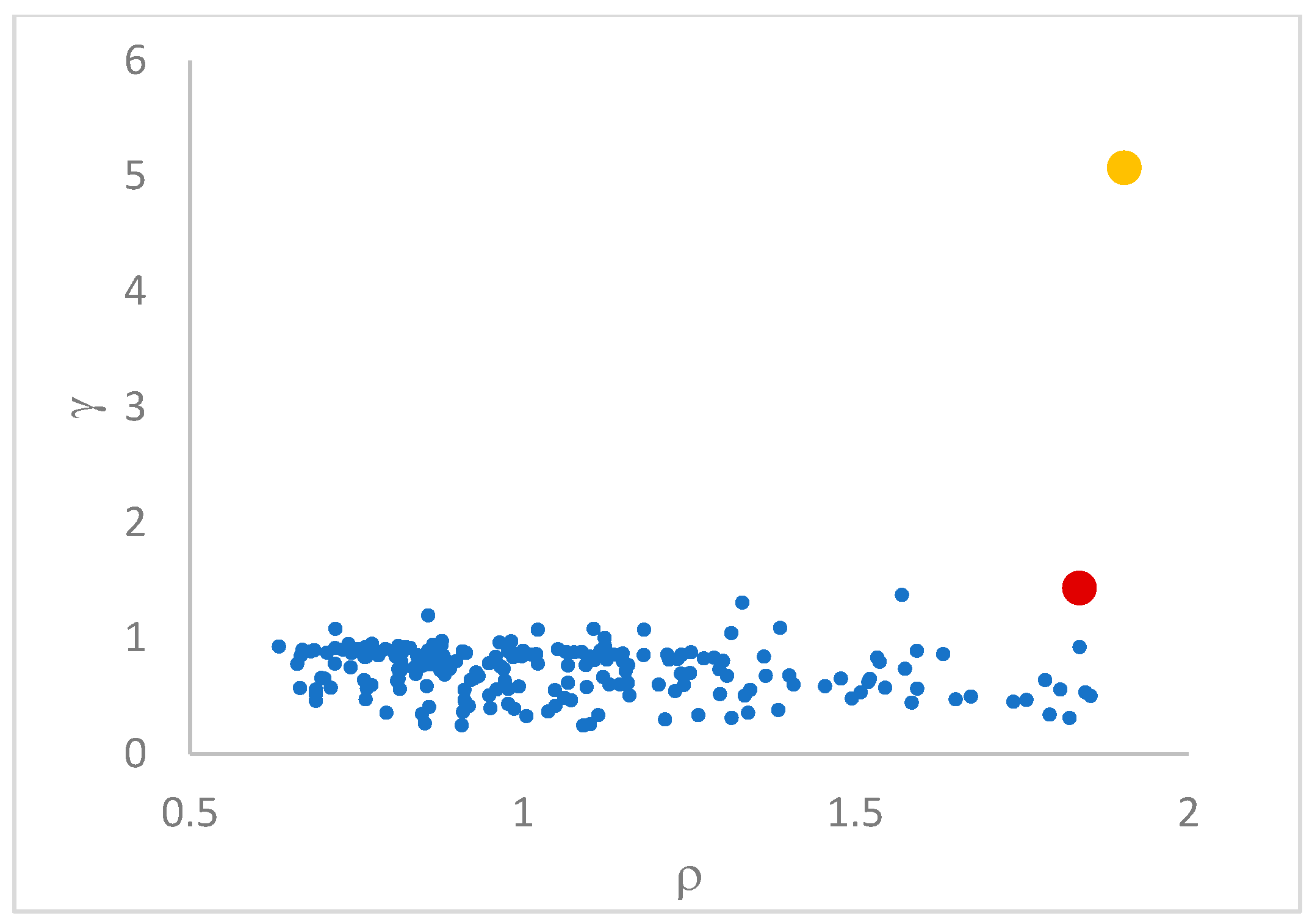
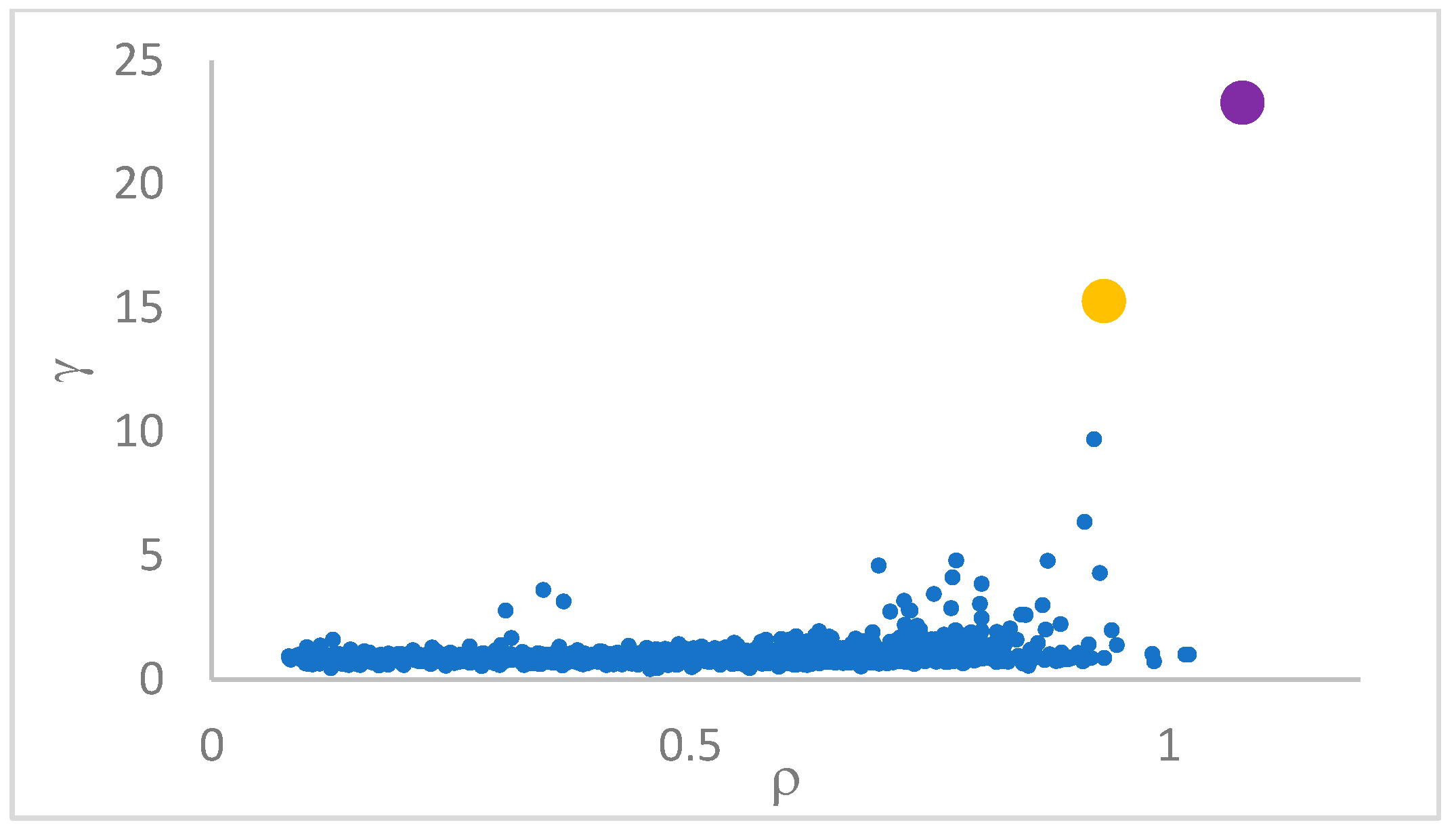
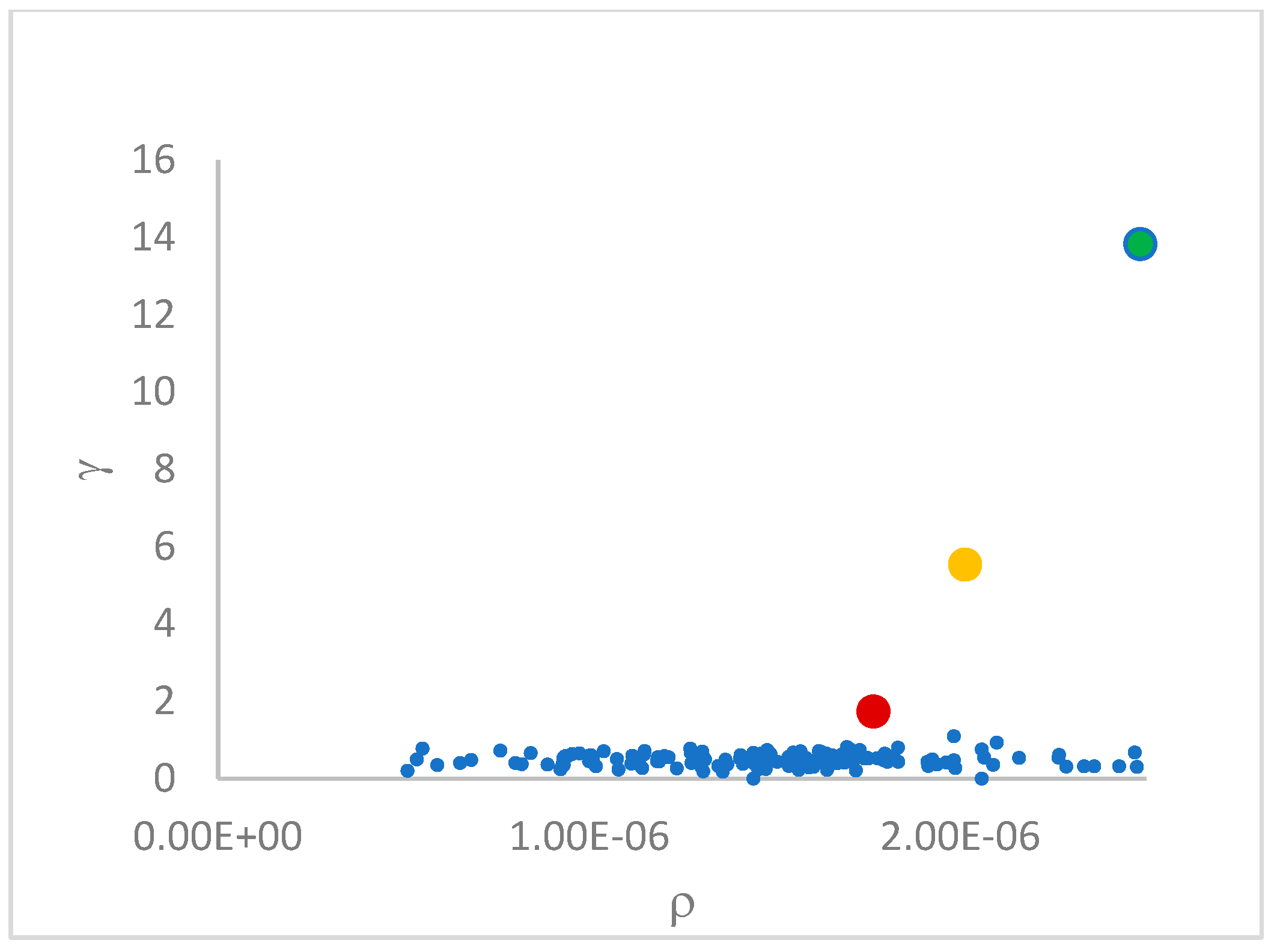
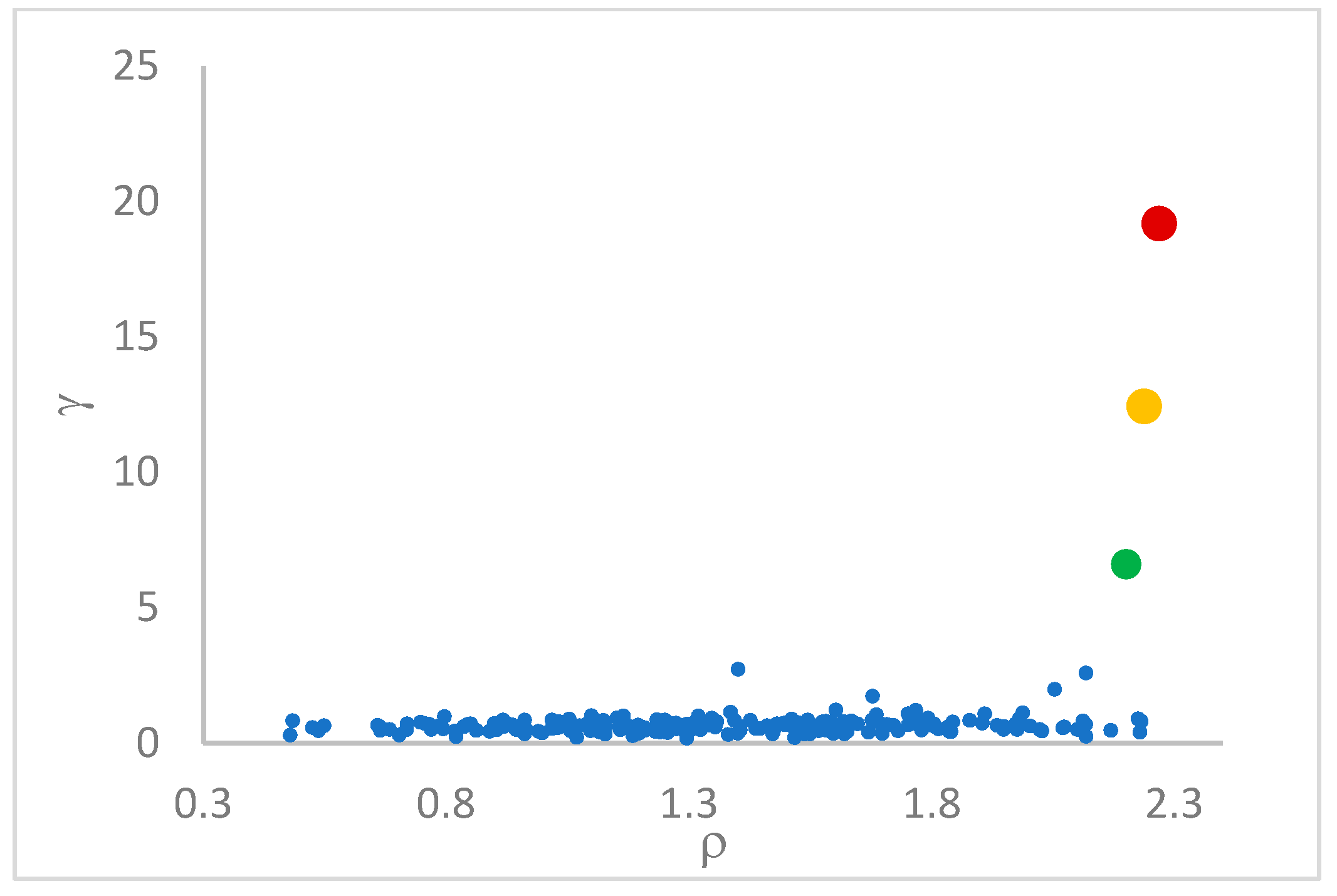
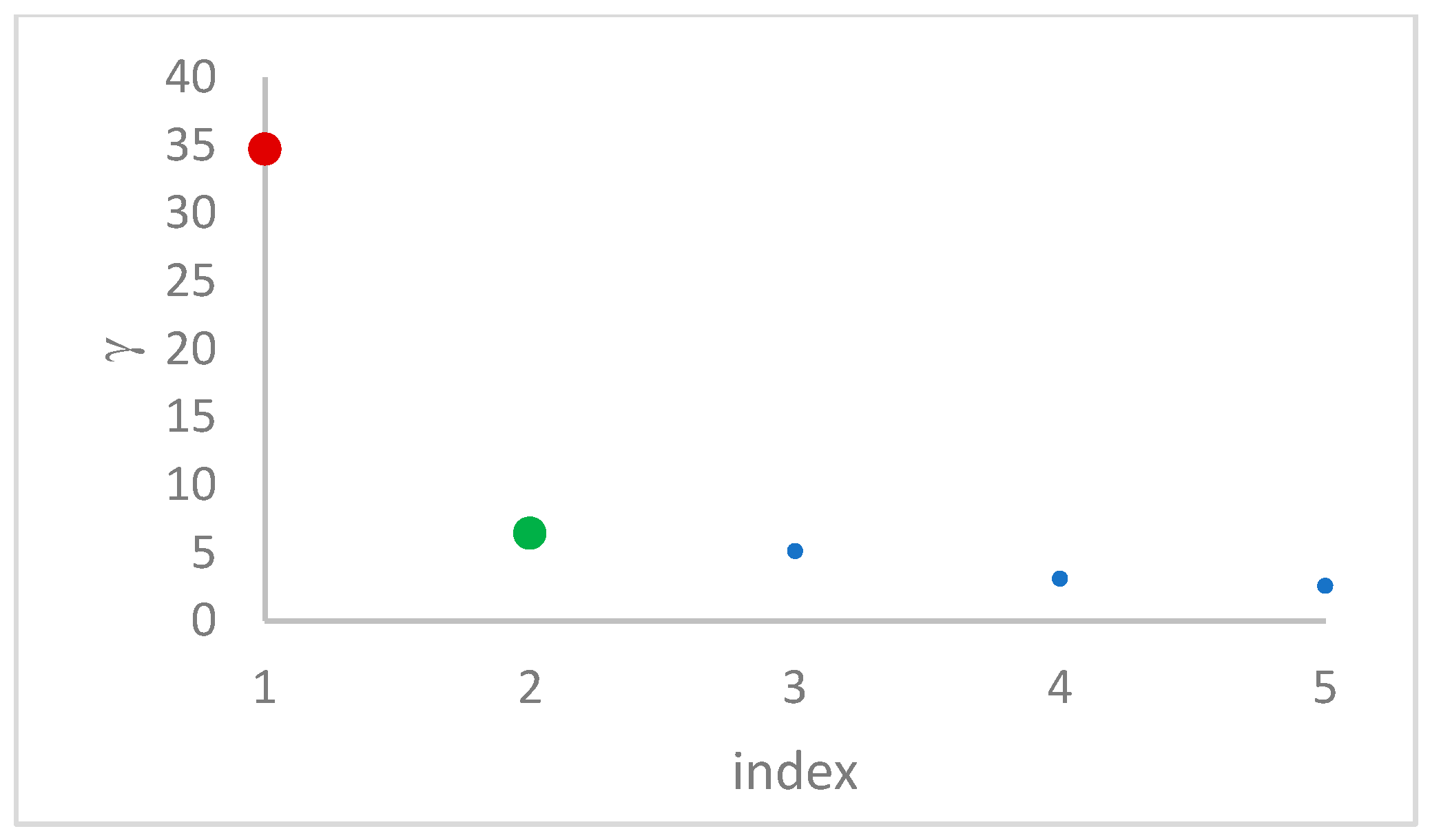
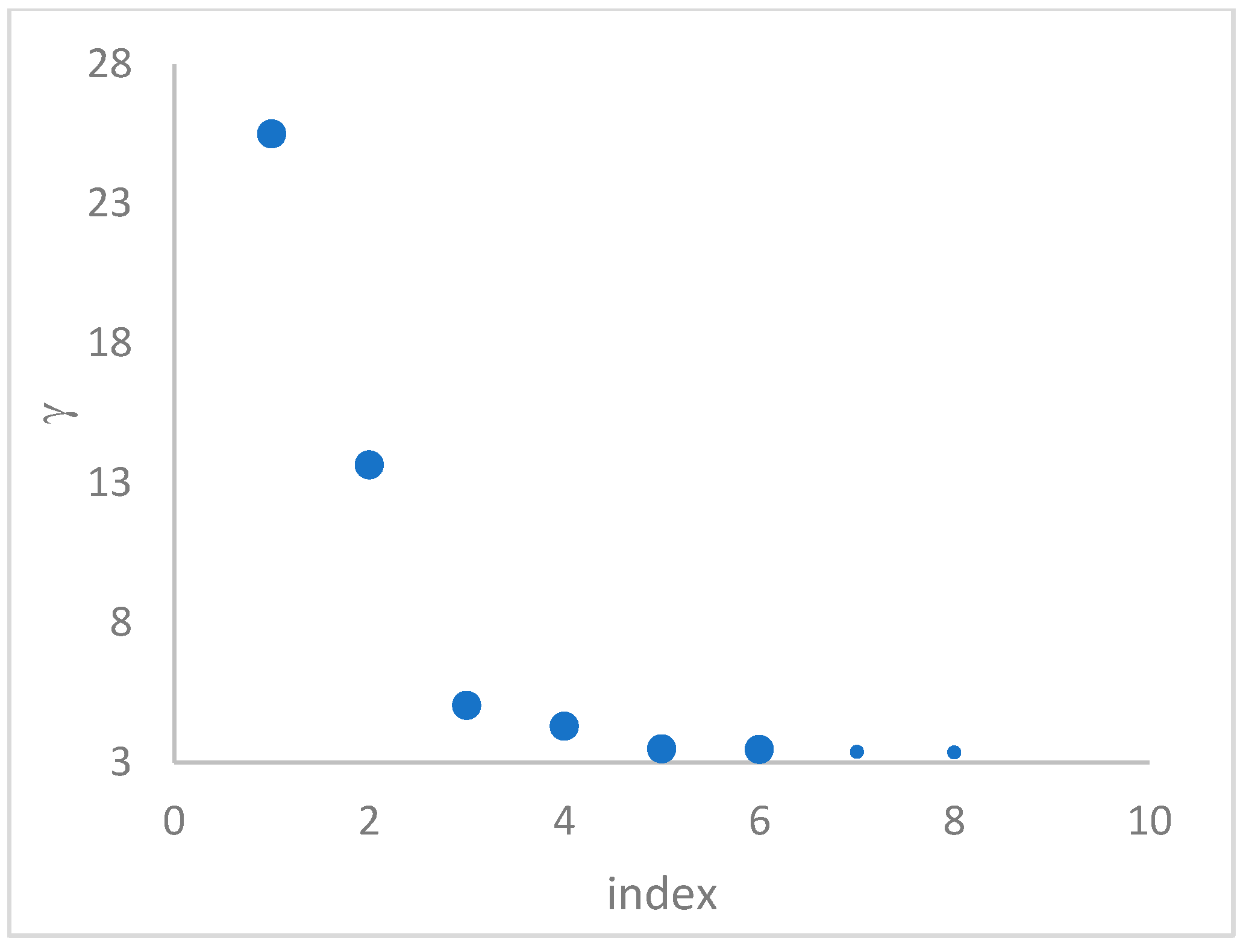
decision graph, which depicts
vs.
or, better,
vs.
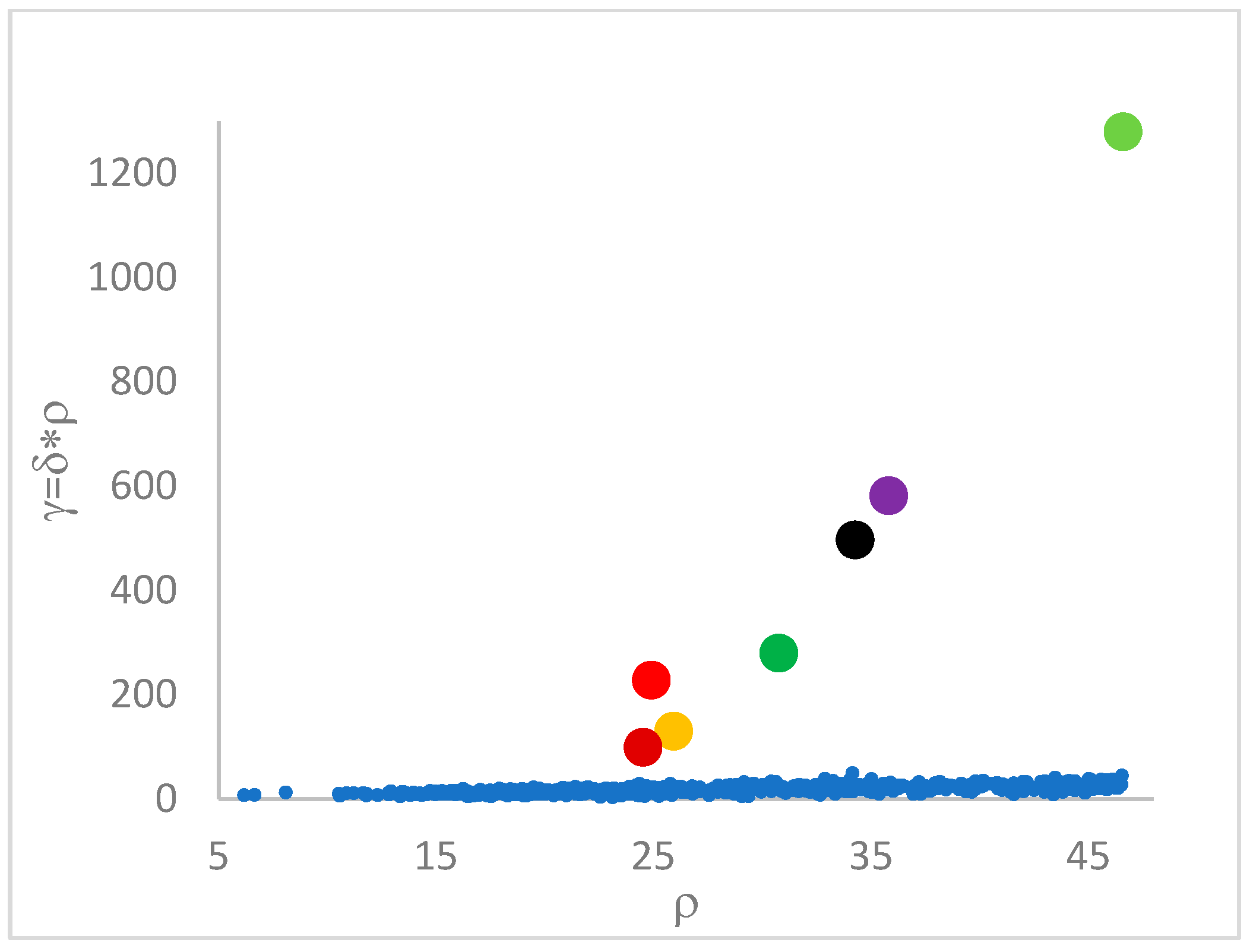
. Candidate centroids are expected to naturally emerge as points with the highest values of
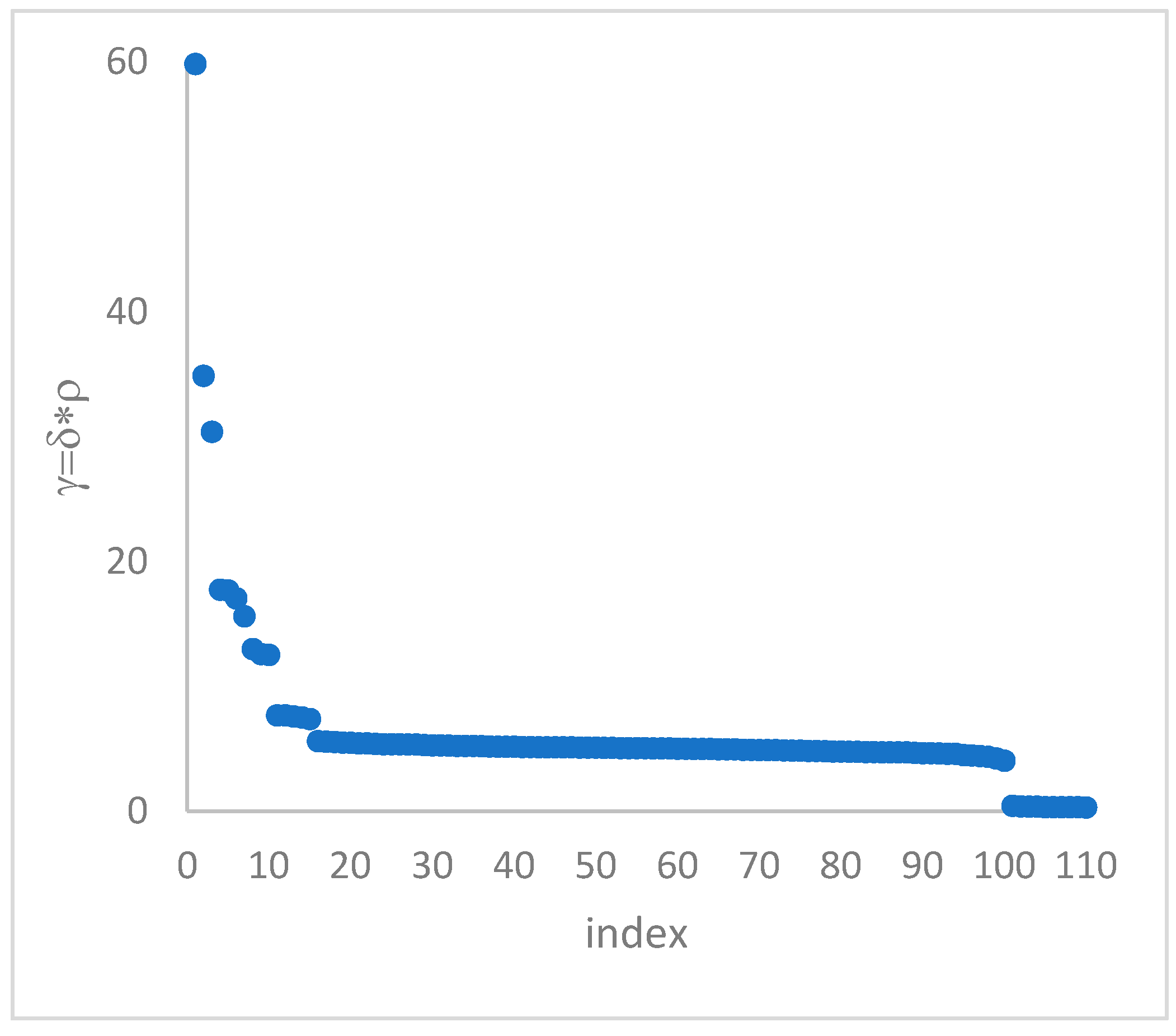
in the decision graph. In the case the dataset is (logically) sorted by decreasing
values, as it is presented in this work, candidate centroids are the “
first” points after which a sudden decrease (knee point) occurs for the
vs. dataset point indexes. However, the reading of centroids from the decision graph shows it is not always clear. In addition, some datasets come with a desired value of
, which suggests picking up the first
points in the sorted dataset by decreasing
.
A fundamental problem in DP is the definition of points’ density. In [
12], the
cutoff distance,
, is assumed, that is the common radius of a hyperball in the D-dimensional space, centered on any point,
. All the points that fall within
establish the density
:
where
is the distance between
and
, and
if
; otherwise,
. In reality, in the implementation of DP, the authors of [
12] used a Gaussian kernel instead for estimating the density:
Such a choice [
21] can be preferable because it avoids points having the same density as it may occur by counting with the cutoff distance. The following defines the
delta feature of a point,
:
This definition ensures that the point with the maximum density has its set to the maximum distance between and any other point in the dataset. Otherwise, is set to the distance of the nearest point with a higher density.
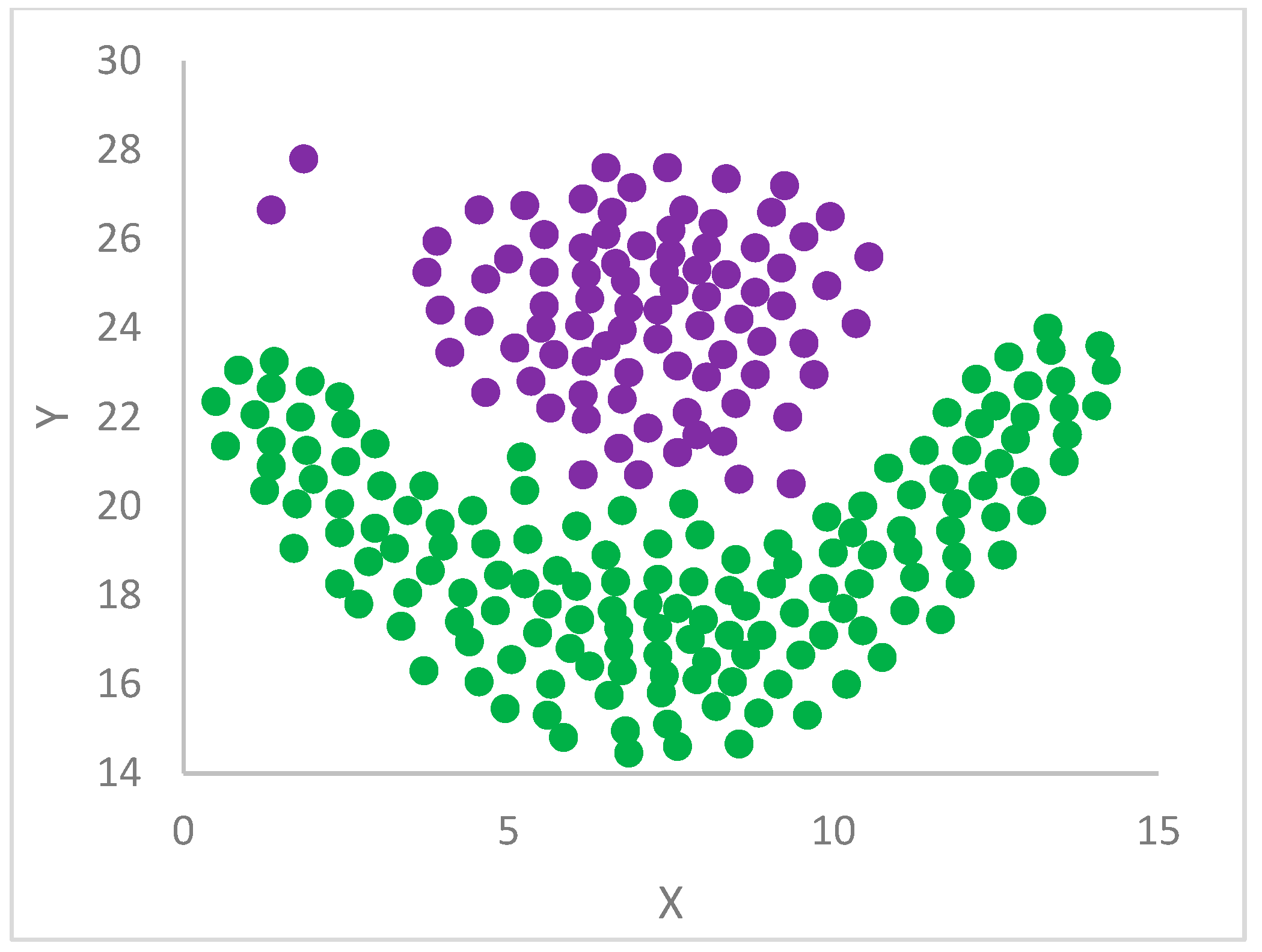
Following the evaluation of , , and then of , DP finalizes the clustering in a standard way. First, centroids are selected, according to the values, and then they are assigned a label. After that, the labels of centroids are propagated to all the remaining points through the rule that a point acquires the label of its big brother.
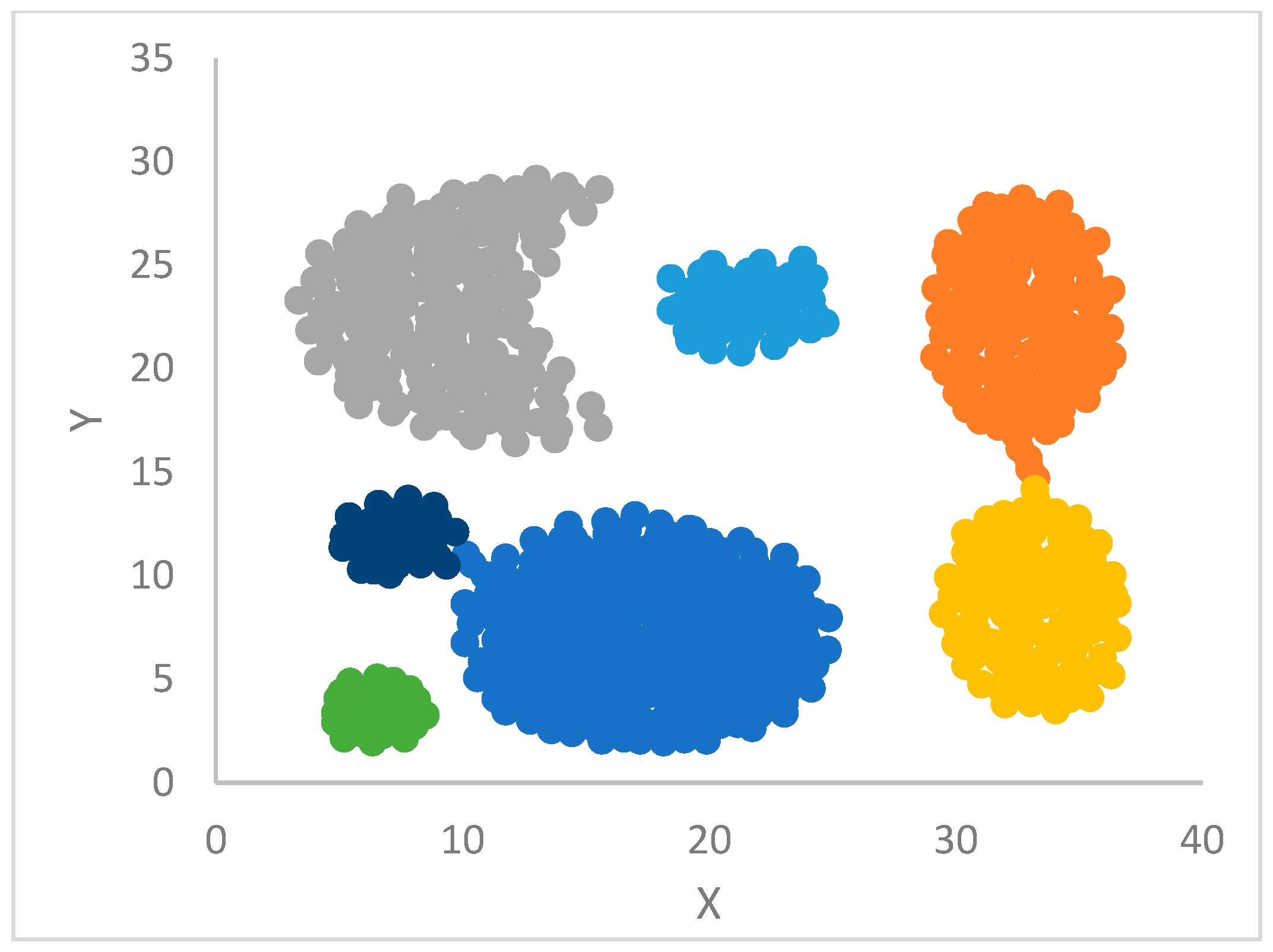
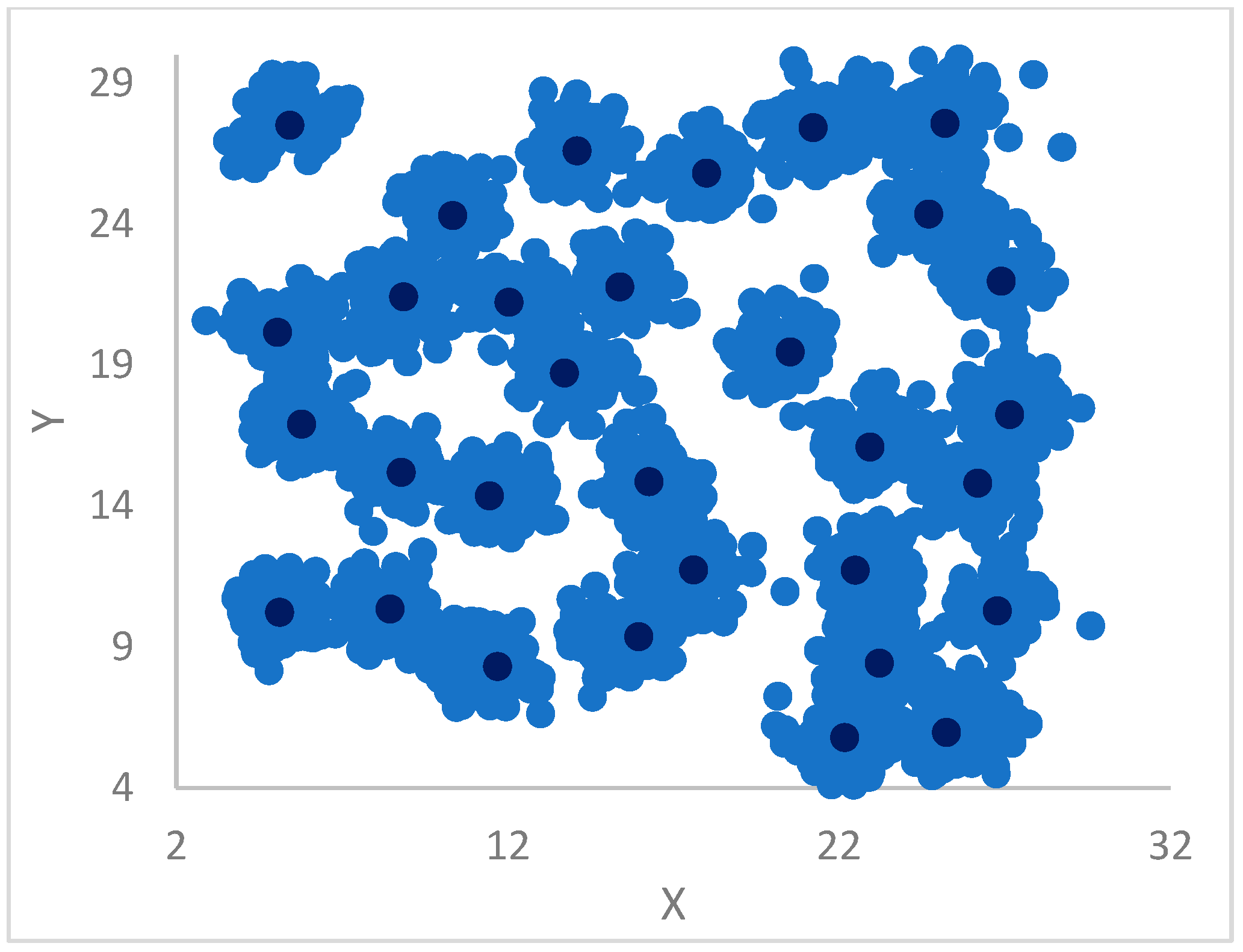
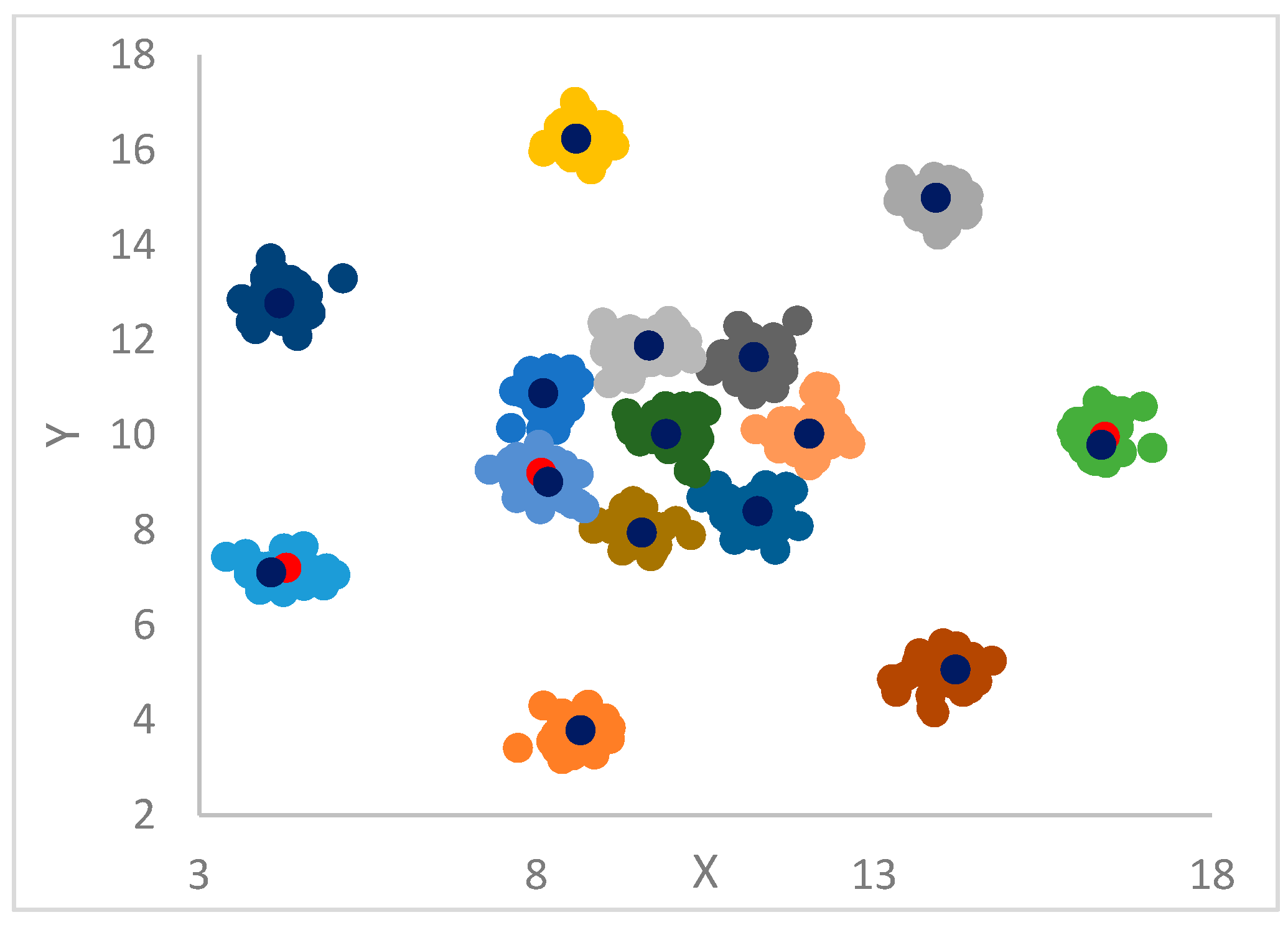
DP can identify outliers by introducing a threshold, , so that points that have a density less than are potential outliers or noise points. qualifies as the maximum density of points in the halo region of a cluster, which has points assigned to this cluster, but that are within the radius of points belonging to other clusters.
3. Development of the ParDP Algorithm
The design of ParDP was influenced by the basic DP work [
12] and by work on kNN clustering [
13,
14,
15,
16,
22]. The concepts of DP are inherited as ρ, δ, γ features of dataset points and the standard recursive clustering algorithm. Differences from DP are introduced in the way points’ density is evaluated.
Two kNN-based approaches are supported. As a preliminary task, the first
distinct distances to the nearest neighbors are determined for each data point. Then the average of these
distances is computed. In the first approach, similar to the work described in [
16], the point density (mass divided by volume) is immediately estimated as the inverse of the average distance. In the second approach, the average distance is maintained in the data point as a
local . When all the dataset points have their local
in place, a
global is defined by sorting in ascending order all the local
values and by extracting the
median value as the common radius [
12,
22] for scanning the points’ density through a Gaussian kernel.
Both approaches naturally tend to limit the influence of outliers on the clustering process. For an outlier, the average of the first k nearest distances tends to be higher than the mean distance of non-outlier points. As a consequence, in the first approach, the density of the outlier is minimal, and the point has a lower probability of being chosen as a centroid. In the second approach, the choice of the median value of local (s), paired with the use of a Gaussian kernel, again ensures that the density measure of the outlier point is very small.
The validity and efficacy of each supported kNN approach depend ultimately on the cluster shapes of the handled dataset, as confirmed experimentally (see later in this paper).
Algorithm 1 provides a pseudo-code of ParDP’s behavior. For some datasets, it can be helpful to preliminarily scale the dataset point coordinates, e.g., by a
normalization, or dividing each coordinate by the maximum value of all features, and so forth. Steps such as pca(), kNN(), delta(), gamma, and clustering(), directly correspond to Java methods in the ParDP implementation.
| Algorithm 1. Pseudo-code of basic ParDP operations. |
Input: dataset X and (if there are any) ground truth centroids or partitions
Scaling data if required
pca()
kNN()
delta()
gamma()
if(decision graph is required)
Output: persist rho, delta and gamma values of the data points
else
clustering()
Output: some accuracy measures of the achieved clustering solution |
3.1. Step pca()
The following steps of
principal component analysis [
13,
23] detect the most important coordinates of data points.
Make each coordinate dimension (feature) have a 0 mean.
Define the covariance matrix of the matrix, , associated with the dataset.
Compute the eigenvalues
that is the diagonal elements,
, of matrix
by the internal products:
Extract the vector of eigenvalues from the matrix, .
Sort by decreasing values the eigenvalues vector.
Select the first
sorted eigenvalues that would ensure, e.g.,
Practically, only the indexes of the most important coordinates (those having higher eigenvalues) are retained during the sorting of the eigenvalues. Such indexes are then used when computing the Euclidean distance between two data points.
3.2. Step kNN()
This step realizes the two approaches to the density evaluation of ParDP. A global boolean
parameter permits kNN() to adapt its behavior to one of the two approaches. The value of the parameter
is important. Algorithm 1 is supposed to be repeated for different values of
, whose “best” value must be tuned to the dataset. For example, several datasets used in the experiments reported in this paper were investigated by using the range [5…200] for
. The effects of a particular value of
on the clustering can be checked on the decision graph or by observing some clustering measures (see later in this paper). The execution of the kNN() step (see also
Section 2) assigns to each point
its estimated density but also the maximal distance of
from any other point in the dataset. Such information is used by the subsequent delta() step. For each value of
, the kNN() step costs
to compute all the pairwise distances of points. To this cost, the heap sorting cost,
, of the dataset by ascending local cutoff distances of data points is added if the cutoff approach of ParDP is chosen. The dominant
cost is smoothed by using Java parallel streams (see
Section 3.6).
3.3. Step delta()
This step is responsible for assigning the information about big brother into each data point. Toward this goal, first, the dataset is sorted by decreasing values of the points’ density. Then, for each point, , the points located at its left in the ordered sequence of data points are considered, and the distance, , is evaluated. Finally, the identity (index) and distance of point nearest to are stored in as its big brother. In the case of point that has the maximal density, its big brother is set, by convention, to the point that has the maximal distance from , as determined in the kNN() step.
In this step, dataset points are sorted by descending values of the density feature. In the next gamma() step, the dataset has to be sorted by decreasing values of the gamma feature of the dataset points. To cope with these conflicting issues and also to maintain the integrity of some fundamental information, like the index of the big brother of a point, ParDP behaves as follows. In no case are points moved within the dataset during a sorting operation. Rather, an data structure is used that indirectly references the points of the dataset. Therefore, the effects of sorting are reflected by the references kept in the . At the time of fixing the identity of the big brother of a point, , the absolute, never-changing physical index (that each point stores in itself) of the big brother (iDelta) and its distance (delta) are stored in . The technique makes it possible to sort, at different times and according to different ordering criteria, the dataset points.
The delta() step mainly costs for heap sorting the dataset by the descending density of the points, which includes the cost for searching the big brother of each point.
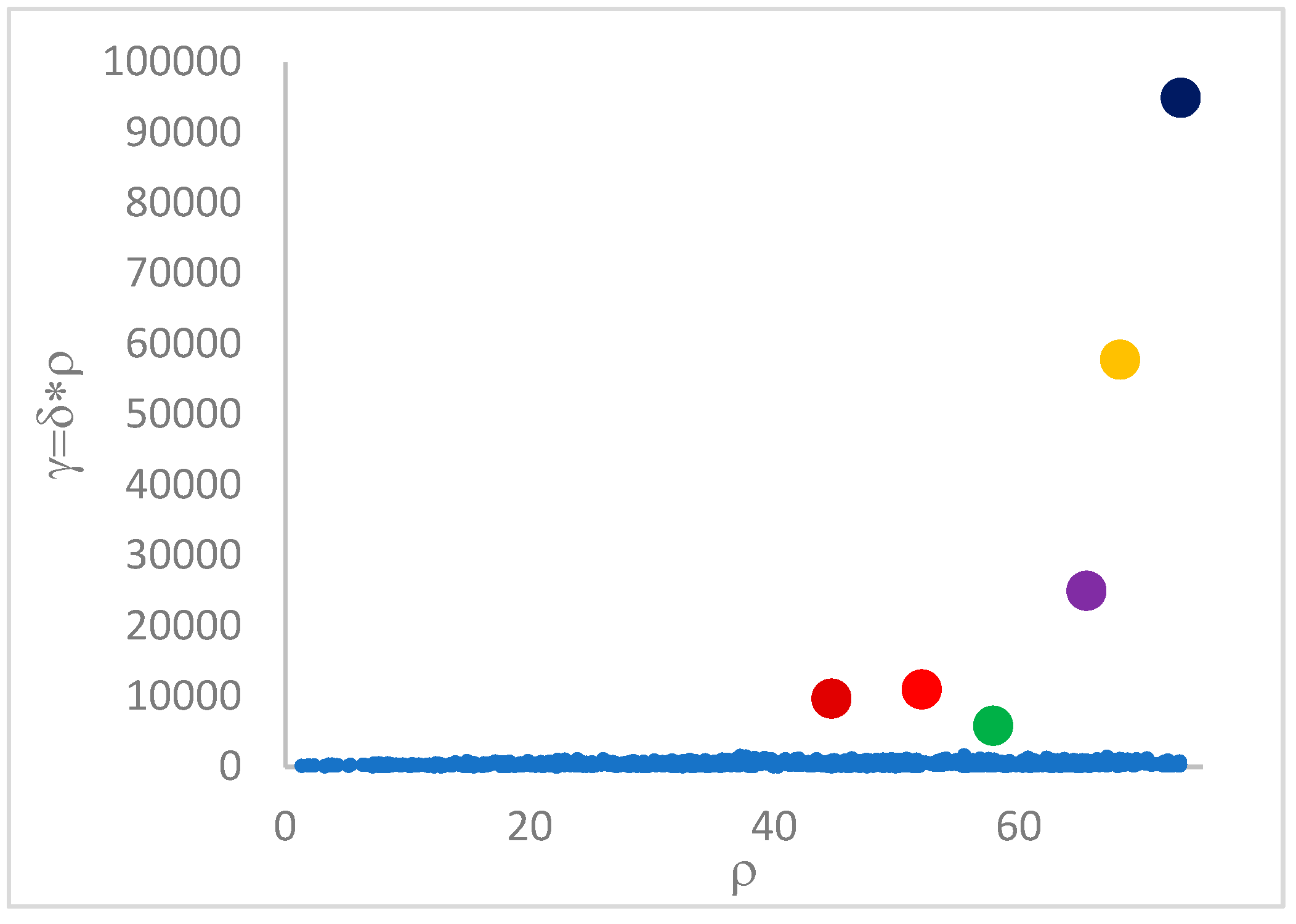
3.4. Step gamma()
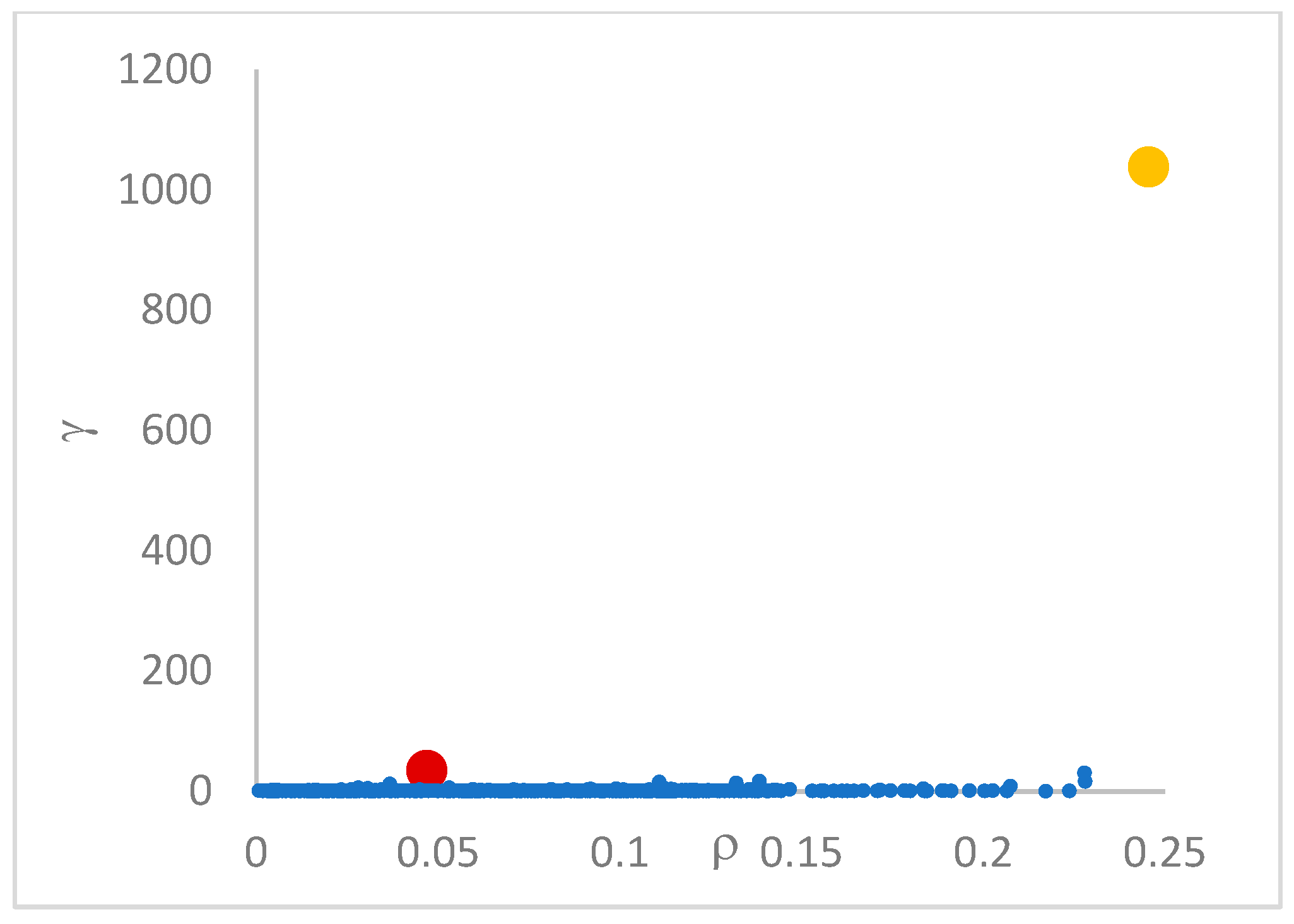
This step sorts the dataset by decreasing values, that is the product of ρ (density) per (delta or distance to big brother) of data points. Such ordering is a key to inferring the number of clusters suited for a dataset or to assessing the validity of the a priori known value of . All these activities can be carried out on the decision graph. Three equivalent types of decision graphs can be exploited: vs. ρ, vs. ρ, or vs. (sorted dataset point) index.
The gamma() step has the cost for heap sorting the dataset by the descending gamma values of points.
3.5. Step clustering()
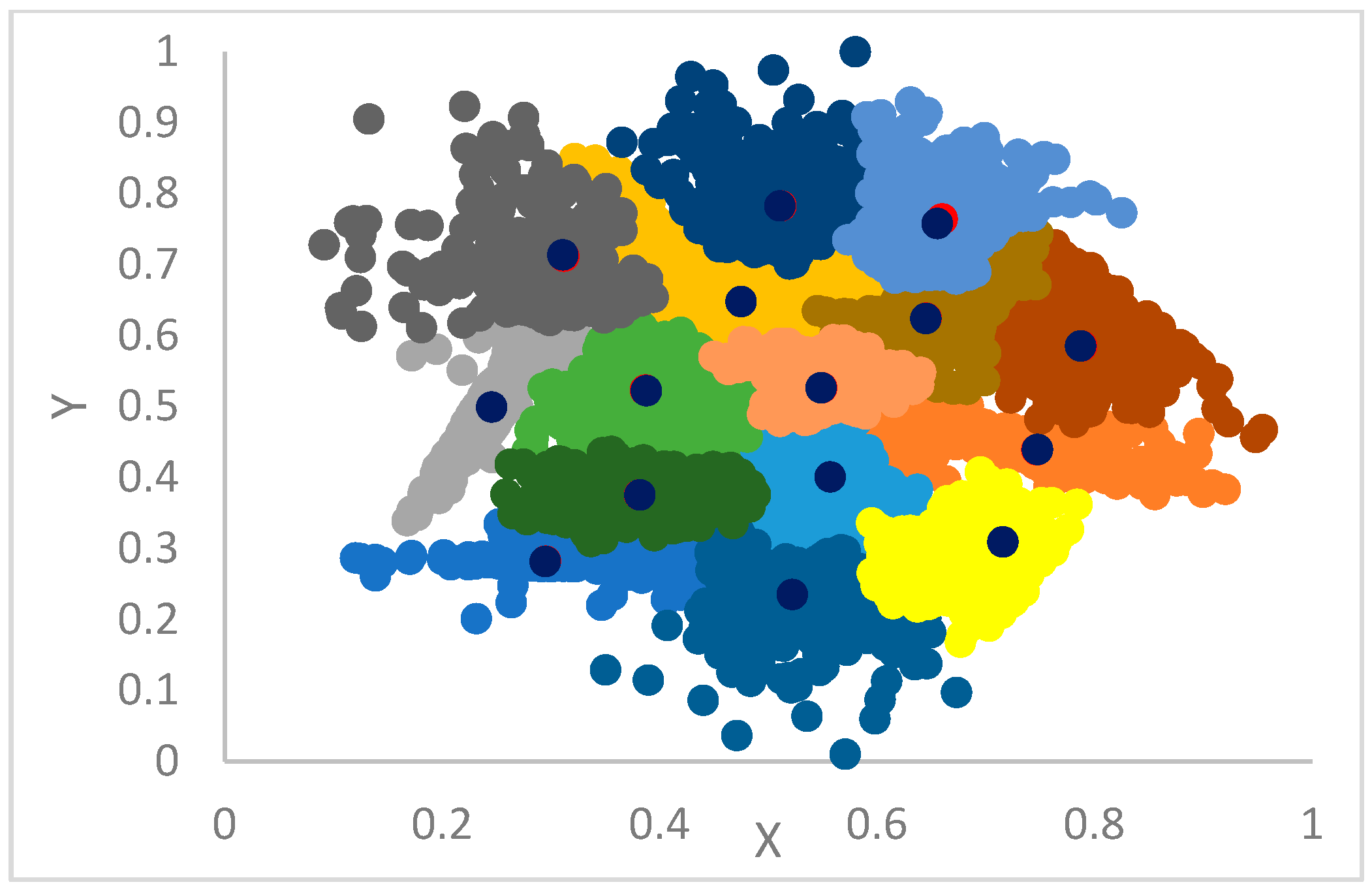
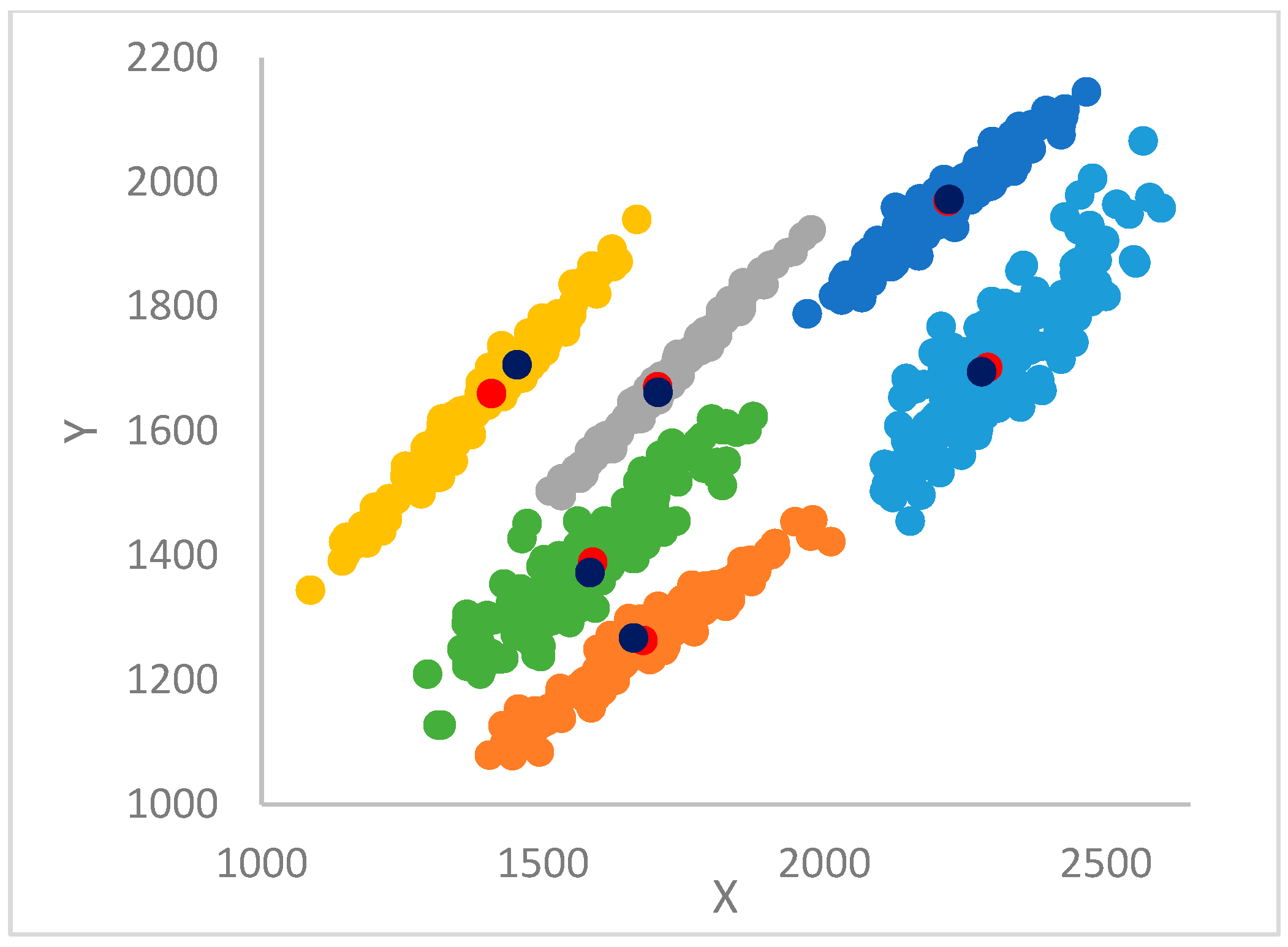
This step selects the first
points in the dataset, sorted by decreasing gamma values, as centroids (see
Section 3.4); assigns to each centroid a label identical to its index; and then proceeds with the final clustering as in the basic DP algorithm (see
Section 2) [
12]. In particular, each point is assigned the label (cluster) of its big brother. In the case the big brother is still unassigned, the procedure continues, recursively, by moving to its big brother and so forth until a centroid or an already assigned big brother is found from which its label is propagated back to all the points in the recursion return path.
The clustering step() costs about Overall, the ParDP execution costs .
3.6. Java Implementation Issues
The power and flexibility of ParDP derive from its implementation, which is based on Java parallel streams and lambda expressions [
19]. The same parallel and functional programming mechanisms were previously successfully applied, for example, to improve the execution of K-Means [
4] and Random Swap [
9,
11]. A stream is a
view of a data collection (e.g., an array like the dataset managed by ParDP) making it possible to express element operations functionally by lambda expressions. A parallel stream allows for the underlying fork/join mechanism to be exploited, which splits the data into multiple segments and processes segments by separate threads. Finally, it combines the partial results generated by the worker threads. A key benefit of the parallel stream concerns taking advantage of the high-performance computing environment provided by a multi/many-core machine, where lock-free parallelism is used, requiring lambda expressions of not modifying shared data.
The concise and elegant programming style ensured by parallel streams and lambda expressions is exemplified in Algorithm 2, where the non-cutoff-based approach of ParDP for predicting the points’ density is shown. First, a stream is opened on the dataset to process points in parallel. Computation is embedded in the map() operation, which works on a stream, transforms it, and returns a new stream. map() is an intermediate operation. The execution of the various map() operations is triggered by a terminal operation, like forEach(). Each map() receives a UnaryOperator<DataPoint> object whose apply() method is formulated by a lambda expression, which receives a point, p; transforms it (p is purposely mapped); and, finally, returns p. A key point of the realization in Algorithm 2 is that each point, p,
only modifies itself (the rho and dMax fields of p are modified). Absolutely no other changes are made to the shared data. The overall programming style only specifies
what to do. The parallel organization and execution of the code is delegated to the built-in fork/join control structure [
19].
| Algorithm 2. An excerpt of the kNN() method for the non-cutoff approach to density |
…
Stream<DataPoint> pStream = Stream.of(dataset);
if(PARALLEL) pStream = pStream.parallel();
pStream
.map(p->{
//detect distances to the first distinct k nearest neighbors of p
double[] dkNN = new double[k];
for(int j = 0; j < k; ++j) dkNN[j] = Double.MAX_VALUE;
double dMax = Double.MIN_VALUE; //maximal distance of p to any other point
for(int i = 0; i < N; ++i) {
if(i! = p.getID()) {
double d = p.distance(dataset[i]);
if(d > dMax) dMax = d;
int j = 0;
while(j < k) {
if(d < dkNN[j]) {
//right shift from j to k − 1
for(int h = k − 2; h >= j; --h) dkNN[h + 1] = dkNN[h];
dkNN[j] = d; break;
}
else if(d == dkNN[j]) break;
j++;
}
}
}
double ad = 0;
for(int j = 0; j < k; ++j) ad = ad + dkNN[j];
p.setRho(k/ad); //inverse of average kNN distances
p.setDMax(dMax);
return p;
})
.forEach(p->{}); //trigger of map computations |
Parallel streams are systematically adopted in ParDP, anywhere parallelism can be exploited. For example, in the delta() step of Algorithm 1, first the dataset is sorted by decreasing density values, then a parallel stream is opened and used to determine the big brother of each point, p. Parallel streams are also used for computing internal or external clustering accuracy measures.
4. Clustering Accuracy Measures
The validity or quality of a clustering solution generated by a given algorithm can be measured by some internal or external indexes [
9,
24]. Internal indexes are related to the internal composition of points of clusters. Two well-known indexes in this class are the
Sum-of-Squared Errors (
) and the
Silhouette coefficient (
) [
11,
25]. The
reveals how close the points assigned to the various clusters
are to their relevant centroids
:
The Silhouette coefficient,
, is a joint measure of the
internal compactness (
cohesion) of clusters and their
separation, which, in turn, is related to the overlapping degree of the clusters. The
of a point,
, is computed as:
where
is the average distance of
from the remaining points of the belonging cluster, and
is the minimal average distance of
to all the points of other clusters. The overall
is then defined as:
ranges in . A value close to 1 mirrors the well-separation of clusters in the achieved solution. A value close to 0 expresses the high overlapping of the clusters. A value that tends to −1 indicates wrong clustering.
It is worth noting that or the can directly be used as the optimization function in K-Means, Random Swap, and similar algorithms. DP, though, is not driven directly by the optimization of such internal indexes. DP, instead, is better evaluated by the use of external measures of the similarity (or dissimilarity) degree of two solutions: and . Each solution is composed of a vector of centroids and a vector of partitions, which are sets of points (indexes) assigned to the same cluster. Labels are the integers in [1..]. The number can be different in the two solutions, although it is usually the same value (assumed in the following). In some cases, one of the two solutions can be a ground truth () correct (“optimal”) solution. can be generated by the designer of a synthetic or benchmark dataset to allow the assessment of the accuracy of a particular solution () predicted by a clustering algorithm. In other cases, the ground truth can be a “golden solution” generated by an assumed correct algorithm. A correct solution can be available as a collection of ground truth centroids () or ground truth partition () labels assigned to all the points, e.g., of a real-world dataset. mirrors the best partition of the points into clusters.
External quality indexes include the
Cluster Index (
) [
26], the
Generalized Cluster Index (
) [
27], the
Adjusted Rand Index (
) [
24], the
accuracy index (
) [
28], and the
Normalized Mutual Information (
NMI) index [
24]. External indexes can be
pair-matching or
set-matching measures. In pair-matching measures, the centroids of the two solutions can be mapped onto each other and, possibly, in the two directions:
and
. Alternatively, pairs of points can be matched among the clusters (partitions) of the two solutions. In set-matching measures, partitions are mapped onto each other between the two solutions.
The
Cluster Index (
) maps each centroid of
to the nearest one in
, according to the Euclidean distance. The number of
orphans in
, that is the number of centroids upon which no centroid of
is mapped, is then counted.
continues by also mapping centroids of
onto
and by counting the resultant orphans in
. The maximal number of orphans in the two directions of mapping defines the value of the
.
ranges from 0 to
. The case where
indicates
is
structurally correct to
. A
mirrors the number of wrongly detected clusters/centroids.
can be practically applied to measure the similarity between the centroids of a predicted clustering solution and the ground truth centroids (
) available for the dataset. The set-matching-based
Generalized Cluster Index (
) extends
to the cases where the partitions of a particular solution are mapped to the ground truth partitions (
) available for a dataset. In this case, the Jaccard distance [
11,
27] between two partitions (sets of points) can be exploited for establishing the nearness:
The
pair-matching-
based index [
24,
29] maps pairs of points from
, practically, from detected to ground truth clusters. It is defined as:
where
counts the number of pairs of points that belong to the same cluster in
and the same cluster in
;
counts the number of pairs that are in the same cluster in
but in different clusters in
;
counts the number of pairs that are in distinct clusters in
but in the same cluster in
; finally,
counts the number of pairs that belong to different clusters in
and different clusters in
. The four quantities can be computed from the contingency matrix
[
24]. The
index ranges in [0..1]. Good clustering is mirrored by an
value close to 1.
The
accuracy index is defined as:
where
is the Kronechker operator which returns 1 if the two arguments are equals, 0 otherwise. The notation
indicates the cluster (partition) of the solution
to which the data point
is assigned. The function
maps the partition in
containing
to a partition of
. In this work, the set-matching-based bi-directional definition of the
proposed in [
28] is adopted. The measure exploits the contingency matrix separately built in the two directions of mapping.
ranges in [0..1], where values close to 1 mirror good clustering.
The
NMI index is an information theoretic measure that can be computed using the contingency matrix [
24] again. It is defined in terms of the
Mutual Information (
MI) and the
Entropy (H) measures of the partitions of the two solutions,
and
, to be compared.
where 0*log0 is assumed to be 0. The quantity
is the element <i,j> of the contingency matrix. The cardinality of a partition like
(
) is also the sum of the elements of the row
(column
) of the contingency matrix. The entropy of a solution,
, is defined as:
where the ratio between the cardinality of the partition,
, and the dataset size,
, expresses the probability of the cluster
. Finally, the
NMI can be defined, in a case (see [
24]), as:
Also, the
NMI ranges in [0..1] with values near to 1 denoting good clustering. As noted in [
28], although it is not normalized, the
ACC measure is very similar to the
NMI measure.
All the above accuracy indexes are implemented in Java and can be used to assess the quality of the clustering solutions computed by ParDP and similar tools. For simplicity, though, the ACC measure is preferred to the NMI index in the experiments reported in this paper.
6. Conclusions
Density peaks-based clustering (DP) [
12] proves effective in the handling of datasets with arbitrary cluster shapes. This paper proposes a new algorithm named ParDP, based on the k-Nearest Neighbors (kNN) technique [
13,
16], which can flexibly host different strategies for evaluating density, which is the core issue in DP operations. ParDP is currently implemented in Java using parallel streams and lambda expressions. The use of parallelism is a key to smoothing out the kNN computational cost of
required to compute all pairwise distances among points. This paper describes the design of ParDP, which includes the principal component analysis (PCA) for possibly reducing the number of point dimensions (coordinates) strictly needed for calculating the Euclidean distance. ParDP modularity is exploited, in a significant case, for building two alternative tools: ParDPC-kNN based on the proposal in [
13], and ParDPC-MNN, which rests on the concept of Mutual Nearest Neighbors (MNN) advocated in [
14,
15] for defining the point densities. These two tools, also provided by PCA, are used for comparison purposes with ParDP, in the clustering of several challenging synthetic and real-world datasets. ParDP appears capable of outperforming the competing tools in many practical datasets.
The work will continue according to the following points. First, to continue experimenting with ParDP to cluster irregular and complex datasets. Second, to develop additional strategies for density estimation, tailored to the needs of particular datasets. Third, to extend the application of ParDP to non-numerical datasets [
16], where the Euclidean distance has to be replaced by a different metric. Fourth, to compare the clustering capabilities of ParDP with those of powerful tools based on Fuzzy C-Means with Particle Swam Optimization [
32]. Fifth, to port ParDP to the Theatre actor system [
33] executing on top of the Spark distributed architecture to deal with larger datasets.